Abstract
The RNA component of budding yeast telomerase (Tlc1) occurs in two forms, a non-polyadenylated form found in functional telomerase and a rare polyadenylated version with unknown function. Previous work suggested that the functional Tlc1 polyA− RNA is processed from the polyA+ form, but the mechanisms regulating its transcription termination and 3′-end formation remained unclear. Here we examined transcription termination of Tlc1 RNA in the sequences 3′ of the TLC1 gene and relate it to telomere maintenance. Strikingly, disruption of all probable or cryptic polyadenylation signals near the 3′-end blocked the accumulation of the previously reported polyA+ RNA without affecting the level, function or specific 3′ nucleotide of the mature polyA− form. A genetic approach analysing TLC1 3′-end sequences revealed that transcription terminates upstream of the polyadenylation sites. Furthermore, the results also demonstrate that the function of this Tlc1 terminator depends on the Nrd1/Nab3 transcription termination pathway. The data thus show that transcription termination of the budding yeast telomerase RNA occurs as that of snRNAs and Tlc1 functions in telomere maintenance are not strictly dependent on a polyadenylated precursor, even if the polyA+ form can serve as intermediate in a redundant termination/maturation pathway.
INTRODUCTION
Telomeres are nucleoprotein structures made of repetitive DNA sequences and bound by specialized proteins. They ensure the protection and complete replication of the ends of linear chromosomes and thereby maintain genome integrity (1). In most eukaryotes, maintenance of telomeric DNA is accomplished by the telomerase ribonucleoprotein that uses an RNA moiety as a template for the addition of species specific DNA repeats to the chromosome ends (2,3). In the yeast Saccharomyces cerevisiae, the core telomerase required for in vitro activity is composed of the reverse transcriptase catalytic subunit Est2p and the template RNA Tlc1 (4,5). Other factors, including the Est1p, Est3p, yKu70/80 and Sm proteins, interact with the telomerase RNA and influence the enzyme activity in vivo (2,6). The eventual average of telomeric repeats maintained, commonly referred to as average telomere length, is influenced by various factors that regulate telomerase activity as well as access of the enzyme to its substrate, the telomeres (6,7). However, little is known about how telomerase subunits are regulated. The expression of certain components of budding yeast telomerase, such as Est1p, was shown to be regulated both at the transcriptional and posttranscriptional level by defined signals embedded in the gene sequence (8–11). In contrast, virtually no information is available about the mechanism controlling the expression of the telomerase RNA moiety. Previous work established that in living yeast cells, fewer molecules of Tlc1 RNA than telomeres are present (12). This could mean that the amount of Tlc1 RNA in the cell is rate limiting for telomerase activity. Indeed, TLC1/tlc1Δ heterozygote diploid cells exhibit shorter telomeres than wild-type diploid cells do, consistent with a TLC1 haploinsufficiency (12). These data further imply that maintaining optimal telomere length requires a tight regulation of Tlc1 expression. In fact, it was shown that a significant number of genes affecting the regulation of steady-state telomere length do so by affecting TLC1 transcription (13). For example, the abundant RNA polymerase II associated PAF1C complex plays an important role in this, but specific cis- and trans-acting determinants for TLC1 expression remain elusive.
The Tlc1 RNA displays many features shared by small nuclear RNAs (snRNA) such as a 5′ 2,2,7-trimethylguanosine (TMG) cap and a Sm-protein binding site near the 3′-end (14). These features suggest that the generation of the Tlc1 RNA could follow a pathway related to that described for snRNAs (14). Indeed, it was observed that like snRNAs, Tlc1 shuttles between the nucleus and the cytoplasm and requires factors that also affect snRNA synthesis and stability (8,14,15). However, unlike all known snRNAs, Tlc1 is found in two forms inside yeast cells, a major non polyadenylated species (polyA− form; about 90% of total) and a minor polyadenylated (polyA+) version that accounts for about 10% of the RNA (16). These two Tlc1 forms not only differ by the presence of a polyadenylated tail, but also by the position of the last encoded nucleotide. The 3′ end of the major polyA− form was found to be considerably more upstream from the polyA addition sites (17). The purification of the telomerase RNP from S. cerevisiae revealed only the polyA− RNAs associated with an active complex (17), suggesting that the polyA+ form does not directly contribute to telomerase function. Therefore, answers to the open question as to how in fact the mature Tlc1 polyA− is generated become critically important if we are to understand telomerase regulation.
Expression of Tlc1 from a heterologous promoter such as pGal4 increases the overall amount of Tlc1 RNA, with the polyA+ version accounting for up to 60% of the total, but the actual level of the polyA− form appears unaffected (16). Furthermore, a conditional repression of Tlc1 expression and defects in the polyadenylation machinery block the expression of the polyA+ form and reduce the amount of the polyA− version (16). This led to the suggestion that Tlc1 is first transcribed as a polyA+ RNA precursor that is subsequently processed by de-adenylation and nucleolytic cleavage into the mature polyA− form. However, the functional relevance of this peculiar maturation pathway and its contribution to telomerase function remained unclear.
Here we analyzed the sequences near the 3′-end of the telomerase RNA gene with the goal of establishing the transcription termination pathways that are involved in the generation of the Tlc1 RNA in vivo and that may influence its expression levels. The results show that mutations in all probable or cryptic polyadenylation signals near the Tlc1 3′-end altered the synthesis of the polyA+ Tlc1 form without affecting the quality or quantity of the mature polyA− RNA. Moreover, a genetic approach identified sequences required for transcription termination upstream of the natural Tlc1 polyadenylation sites. These results indicate that the synthesis and function of mature Tlc1 are not dependent on prior polyadenylation at the previously documented sites. Our results also show that the newly discovered upstream termination is under the control of the Nrd1/Nab3 non-coding RNA transcription termination pathway.
MATERIALS AND METHODS
Strains and media
All yeast strains used in this study are listed in Table S1 and were manipulated using standard procedures (18,19). Strain CSHY76 was kindly provided by C. Greider (20). The phenotypes produced by the TLC1 3′-end mutation construct (Tlc1–3′MUT) were assayed using a tlc1Δ mutant spore derived from the diploid strain CSHY76. The cup1Δ strain (46a) and the temperature-sensitive (ts) nrd1 (nrd1-5), nab3 (nab3-11) and sen1 (sen1-E1597K) strains were described previously (21–23). Copper plates were made by adding a dilution of filtered CuSO4 solution after autoclaving and before pouring the plates.
Plasmid construction
Plasmids used in this study are listed in Table S2. The pADCEN36 plasmid containing wild-type TLC1 was described earlier (24). Point mutations in the polyadenylation signals found in the TLC1 3′-end were obtained by PCR (plasmid SLP149). The mutated polyA signal cassette was amplified by PCR and cloned into the SmaI site of pRS316 to generate plasmid SLP161. This plasmid was later used to produce the probe required for the ribonuclease protection assays described in Figure 1. The ACT1–CUP1 reporter plasmid pGAC24 used for transcription termination sequence analysis was described previously (23). Sequences from the TLC1 3′-end were amplified by PCR with primers containing XhoI sites and cloned into the XhoI site of the ACT1 intron in pGAC24. The TLC1 3′-end cloned sequences ranged from the NsiI site at position +1130 to the positions +1180, +1209, +1218, +1228, +1233, +1266, +1290 or +1311. The given positions are relative to the TLC1 transcription start site (+1) as determined in (24). The TLC1–1311–i construct represents the +1130 to +1311 region cloned in the inverted orientation. The pGAC24–SNR13 terminator construct was described previously (25). The Nrd1 binding site mutation consists of a G to C nucleotide substitution at the +1186 position and the Nab3 binding site mutation consists of a T to C nucleotide substitution at the +1165 position. These mutations where introduced into the TLC1–1233 construct by site-directed mutagenesis to generate the TLC1–1233Nrd1mBs and TLC1–1233Nab3mBs plasmids. The Nab3 binding site mutation was introduced in the TLC1–1233Nrd1mBs construct to yield the TLC1–1233Nrd1/Nab3mBs construct. Primers used for PCR amplifications and mutagenesis are listed in Table S3. All plasmid constructs used in the various termination assays were verified by sequencing.
Figure 1.
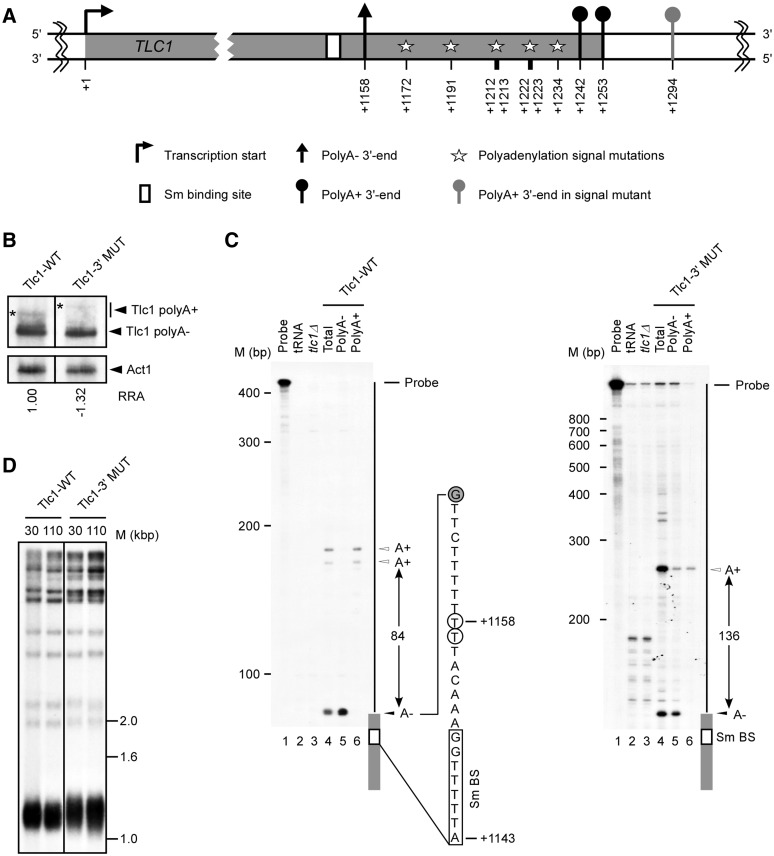
Disruption of the polyadenylation signals does not affect the synthesis of mature telomerase RNA. (A) Schematic representation of the TLC1 transcription unit with a focus on the 3′-end. Stars represent mutations in all probable/cryptic polyadenylation signals near the TLC1 3′-end (A1172:C, A1191:G, A1212:G, T1213:C, A1222:C, T1223:C, A1234:G). The numbers are relative to TLC1 transcription start site (+1). (B) Northern blot analysis of Tlc1 RNA isolated from cells harboring wild-type TLC1 (Tlc1-WT) and TLC1 carrying mutations near the 3′-end (Tlc1-3′MUT). Stars (*) represent polyadenylated Tlc1 species. The relative RNA amount is an average of three experiments with a standard deviation of ±0.30 or less and was determined using an Instant Imager. (C) Mapping of the Tlc1 3′-ends was achieved by T1 RNase protection assays. RNAs were hybridized to antisense RNA probes specific for Tlc1-WT or Tlc1-3′MUT, and digested by ribonuclease T1. The labeled undigested RNA probe (probe) and the probe digested in the presence of tRNA (tRNA) were included as controls. The results of digesting the different probes after hybridization to RNA extracted from cells carrying a complete deletion of the TLC1 gene (tlc1Δ) was also included to ascertain the specificity of the different protected bands. PolyA− and polyA+ indicate the digestion of probes hybridized to RNA found in the flow-through and eluted fractions of oligo (dT) column, respectively. The position of the probe, the different protected 3′-ends and the distance separating these ends are indicated on the right of each panel. Nucleotide positions (+1143 and +1158) are according to transcription start site. Size markers are indicated on the left of each panel. The precise position of the 3′-end was mapped using a sequence ladder run in parallel lanes (data not shown). The sequence shown represents the sequence adjacent the Tlc1 3′-end (gray boxes). The empty box indicates the Sm binding site (Sm BS). The open circles indicate the previously mapped 3′-ends and the closed circles indicate the position of the observed G specific T1 cleavage. (D) Southern blot analysis of telomere lengths in strains harboring constructs with wild-type (Tlc1-WT) or polyadenylation signals mutant TLC1 (Tlc1 3′MUT). Genomic DNA was extracted from individual yeast clones grown for 30 or 110 generations, digested with XhoI and separated on an agarose gel. The different fragments were visualized using a randomly labeled probe complementary to the telomere sequence.
Complementation analysis and Telomere length analysis
The effect of the TLC1 3′-end mutations (Tlc1-3′MUT) on cells was tested using senescence and telomere length assays as described previously (24).
Isolation of total and polyA+ RNA
Total RNA was prepared as previously described (8). Oligo (dT) based fractionation of polyA+ RNA was performed as described (26) using oligo (dT) type 7 matrix (GE Healthcare, Baie d’Urfé, Québec).
Northern blot analysis
Northern blots in Figures 1 and 4 were performed as previously described (8,27). Briefly, total RNA (15–20 µg) was fractionated on a 4% polyacrylamide gel and transferred to a nylon membrane (Hybond N+, GE Healthcare, Baie d’Urfé, Québec). These blots were hybridized to randomly labeled probes corresponding to the specific sequences as indicated in the figures and signals visualized by autoradiography. For the northern blot in Figure 1B, the RNA was quantified using an Instant Imager (Packard, Meriden, CT). For the northern blot in Figure 4, each strain (WT, nrd1, nab3 and sen1) was grown at permissive temperature (23°C) to reach a concentration of 1 × 107 cells/ml. RNA was extracted from half the culture and the other half was then diluted to 0.5 × 107 cells/ml in pre-warmed media and allowed to grow back to 1 × 107 cells/ml at semi-permissive temperature (32°C for nrd1 and sen1; 27°C for nab3). The cultures were diluted in the same way two more times before RNA was extracted. Cells have thus grown for three population doublings at semi-permissive temperature before RNA extraction. Relative quantification of polyadenylated and non-polyadenylated Tlc1 RNA species were obtained with a PhosphorImager and the IMAGE-QUANT (Molecular Dynamics) and PeakFit (Systat Software Inc) software. For the northern blot in Figure 3D, total RNA (10 µg) extracted from cells harbouring different ACT1–CUP1 constructs was fractionated on a 1.5% formaldehyde-agarose gel and transferred to a nylon membrane. RNA transcribed from the construct was detected by hybridization to a randomly labeled probe specific to the CUP1 coding sequence that is located downstream of the ACT1 intron and the signals visualized by autoradiography.
Figure 4.
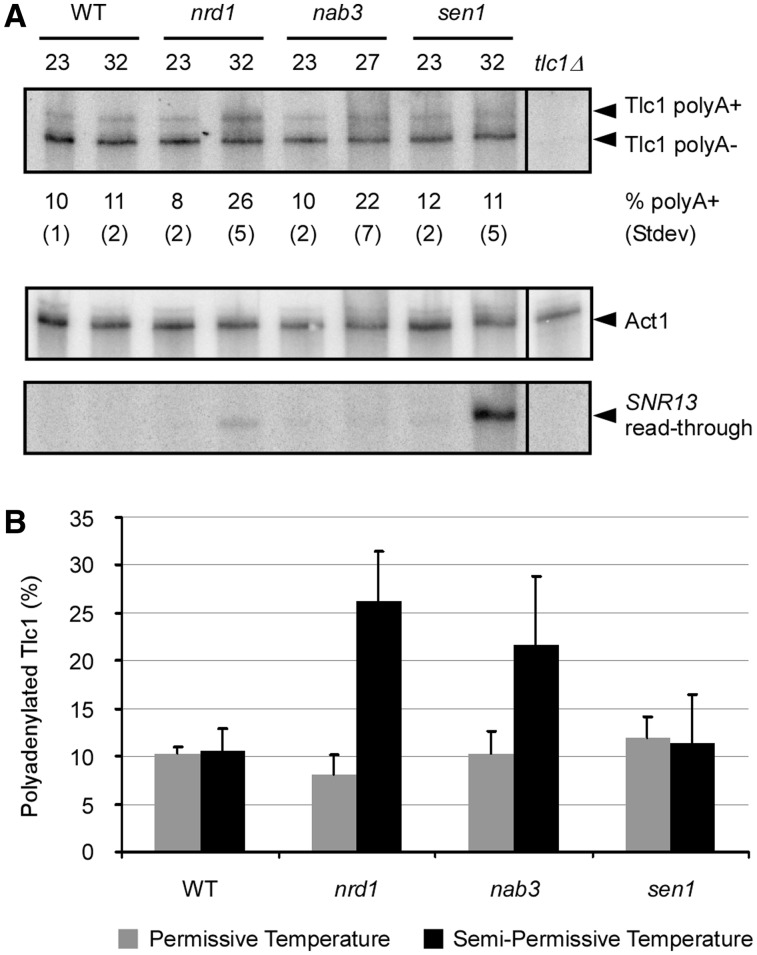
Ts alleles of members of the Nrd1/Nab3 transcription termination pathway disturb the Tlc1 polyA+/polyA− balance. (A) Northern blot analysis of the levels of Tlc1 polyA− and polyA+ RNA in wild-type (WT) or temperature-sensitive nrd1-5 (nrd1), nab3-11 (nab3) and sen1-E1597K (sen1) strains grown at permissive (23°C) or semi-permissive (27°C for nab3, 32°C for nrd1 and sen1) temperatures. Cells at semi-permissive temperature were grown for three population doublings before RNA extraction. The indicated percentage of polyadenylated Tlc1 (% polyA+) is an average of a minimum of four experiments with the indicated standard deviations below. The northern blot shown is a representative of results obtained. RNA quantifications were obtained with a PhosphorImager and the IMAGE-QUANT (Molecular Dynamics) and PeakFit (Systat Software Inc) softwares. In order to control for variations in total RNA amounts, the membranes were stripped of radioactivity and rehybridized to a probe specific for the Act1 mRNA. Loss of function of Nrd1p, Nab3p and Sen1p in the growth conditions used was determined by rehybridization to a probe specific for the snR13 snoRNA and detection of the level of read-through products. (B) Graphical representation of variations in the percentage of polyadenylated Tlc1 in Nrd1/Nab3 transcription termination pathway mutants observed by northern blot in (A).
Figure 3.
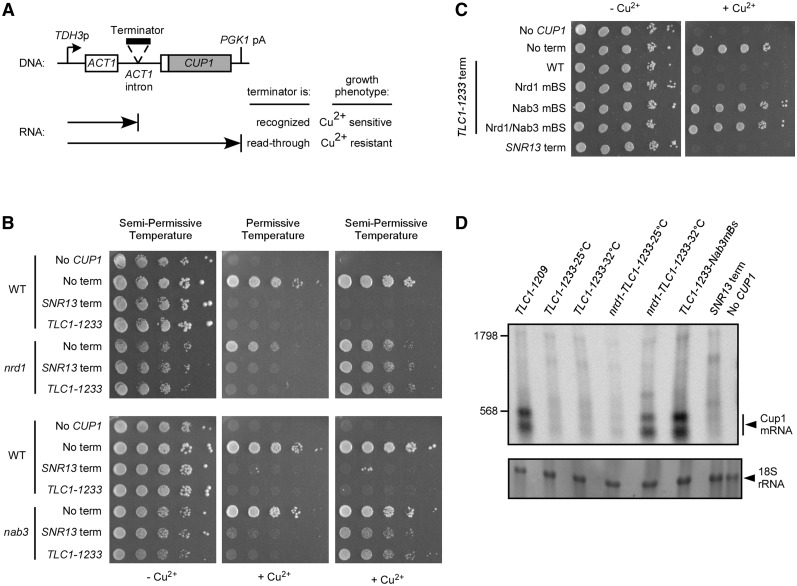
The Tlc1 upstream terminator depends on the Nrd1/Nab3 transcription termination pathway. (A) Diagram of the ACT1–CUP1 fusion reporter gene with a cloned intronic terminator and the expected phenotypes on copper plates for recognition/read-through of the terminator. (B) Tenfold serial dilutions of cultures with cup1Δ strain 46a (WT), nrd1-5 (nrd1) or nab3-11 (nab3) and containing ACT1–CUP1 constructs with no terminator (No term), snoRNA terminator (SNR13 term) or Tlc1 terminator (TLC1–1233). A strain harboring no ACT1–CUP1 construct is used as a control for copper sensitivity (No CUP1). Cultures were spotted on medium lacking leucine and containing 0 or 0.5 mM added copper and incubated for 3 days at permissive (23°C) or semi-permissive (27°C for nab3, 32°C for nrd1) temperatures. (C) Same serial dilutions spot test as in (B) with strain 46a containing ACT1–CUP1 constructs with no terminator (No term), snoRNA terminator (SNR13 term) or Tlc1 terminator (TLC1–1233 term). The Tlc1 terminator is either unchanged (WT) or contains point mutations in the consensus Nrd1p binding site (Nrd1 mBS), in the consensus Nab3p binding site (Nab3 mBS) or in both sites (Nrd1/Nab3 mBS). A strain harboring no ACT1–CUP1 construct is used as a control for copper sensitivity (No CUP1). (D) Northern blot analysis of RNAs produced from ACT1–CUP1 reporter gene constructs using a CUP1-specific probe. TLC1–1233-25°C and TLC1–1233-32°C: RNAs derived from wild-type strain (46a) carrying the TLC1–1233 construct grown at 25°C or 32°C. nrd1-TLC1–1233-25°C and nrd1-TLC1–1233-32°C: RNAs derived from cells carrying the nrd1-5 ts allele and the TLC1–1233 construct grown at permissive (25°C) or semi-permissive (32°C) temperature. TLC1–1233-Nab3mBs: RNA derived from wild-type 46a cells carrying the TLC1–1233 construct with a point mutation in the Nab3p binding site. The TLC1–1209 construct is used as a negative control for transcription termination and the SNR13 snoRNA terminator is used as a positive control for termination (SNR13 term). A strain harbouring no ACT1–CUP1 construct is used as a negative control for hybridization (No CUP1). The RNA was hybridized to a probe specific to the CUP1 coding sequence downstream of the ACT1 intron. Size estimations in nucleotides indicated on the left were obtained after visualization of the 18S rRNA by methylene blue staining and after rehybridization with a terminally labelled oligonucleotide specific for the U1 snRNA (data not shown). The 18S rRNA visualized by methylene blue staining was used as a loading control.
RNase protection assay
Total RNA (20 µg) was incubated with 0.5 fmol of riboprobe and incubated at 42°C in 80% formamide buffer for more than 8 h (28). The hybridization reaction was then digested with RNase T1 for 1 h at 37°C. The protected fragments were analyzed on polyacrylamide gels. The riboprobes complementary to Tlc1 3′-end were generated by T7 or T3 transcription from plasmids containing either the whole TLC1 sequence or bearing only the polyadenylation signal mutation cassette. In the case of the wild-type sequence, the plasmid was digested with the HinfI restriction enzyme inside TLC1 sequence, whereas the plasmid containing the polyadenylation signal mutation cassette was digested the with PvuII enzyme. In order to be able to identify specific bands at a nucleotide resolution, each gel also contained four lanes with the products of the sequencing reactions using DNA of the same area as assayed by the RNA protection assay as template. In addition, a 100 b end-labeled marker DNA ladder was also run in another separate lane on each gel.
Spot test analysis of copper sensitivity in ACT1–CUP1 constructs
The cup1Δ strains harbouring the ts alleles and containing ACT1–CUP1 constructs were grown in liquid media at 23°C to exponential phase. Cells were then diluted serially by 10-fold and spotted onto plates with or without added copper. The plates were incubated at 23, 27 or 32°C for 2–4 days before photographs were taken.
RESULTS
Disruption of Tlc1 polyadenylation signals does not affect synthesis of mature telomerase RNA
It was previously suggested that the primary 3′-end of the Tlc1 RNA is determined by polyadenylation signals about 1190–1240 bp downstream from the mature 5′-end (16 and Figure 1A). Indeed, it was shown that a particular mutation in this region affected polyadenylation but did not inhibit production of the mature polyA− Tlc1 (16). However, as polyadenylation signals in yeast are not well defined (29), we pushed further the identification of the sequences required for Tlc1 3′-end formation by disrupting all possible polyadenylation signals and monitoring the impact on the expression and function of the RNA. The impact of the mutations on RNA expression was examined by northern blot (Figure 1B) and the positions of the 3′-ends were mapped using RNase protection assays (Figure 1C). Thus, as expected from previous results, RNA extracted from wild-type cells revealed the presence of both the polyA+ and polyA− forms of Tlc1 (Figure 1B; Tlc1-WT). In contrast, the introduction of seven point mutations in between positions +1172 and +1234 (Figure 1A and Supplementary Figure S1), that were designed to eliminate all probable polyadenylation signals, abolished the expression of the normal length polyA+ Tlc1 RNA transcript (construct called Tlc1-3′MUT in Figure 1B–D). Instead, the presence of these mutations is associated with the appearance of a new and somewhat longer transcript without significantly affecting the expression of mature polyA− Tlc1 (Figure 1A, B). Fractionation of these RNAs on an oligo (dT) cellulose column indicated that the longer RNA species observed being expressed from the Tlc1-3′MUT template is polyadenylated (Figure 1C and data not shown). Therefore, in the situation of the Tlc1-3′MUT template, the steady-state level of the Tlc1 RNA remains the same and the fraction of polyA+ versus polyA− form also is in the same range as in Tlc1-WT, but the polyA+ form is longer and no polyA+ form of the length observed in wild-type cells is detectable (Figure 1B and data not shown).
In order to directly assess the impact of disrupting the polyadenylation signals on Tlc1 3′-end formation, we examined the RNA termini produced from the Tlc1-3′MUT template using RNase protection assays with RNaseT1. As expected and completely consistent with previous results, the 3′-ends of the wild-type Tlc1 RNA could be mapped to two different regions: the first is 15 nt and the second about 100 nt downstream from the Sm binding site (Figure 1C). Separation of the RNAs on oligo (dT) columns indicated that the first region corresponds to the 3′-end of polyA− Tlc1 while the second region identifies two very close positions of polyA+ 3′-ends (Figure 1C). These latter polyA+ sites correspond to two sites determined previously by RACE (16). RNA extracted from cells harboring the TLC1-3′MUT as template gene did not exhibit changes in the position of the first termination site (polyA− form), but yielded a different second termination site further downstream, consistent with the higher MW band detected on the northern blot (Figure 1B and D). The overall conclusions reached by these experiments were also confirmed by using RNaseA1 instead of RNaseT1 digestion (data not shown). These data therefore indicate that disruption of Tlc1 polyadenylation signals does not affect the quality of the polyA− 3′-end. Moreover, the RNA generated in cells harboring this mutated template did not affect telomere length (Figure 1D) or cell viability (data not shown). Therefore, while the mature polyA− Tlc1 RNA could still somehow be generated via processing of the now longer polyA+ Tlc1 RNA species, these results strongly suggest that the naturally occurring polyA+ Tlc1 species is neither required or important for telomerase function.
Sequence elements required for Tlc1 RNA transcription termination
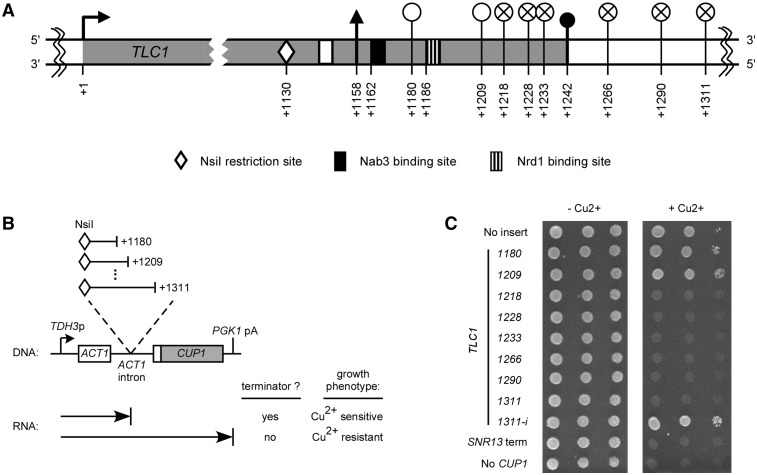
In order to precisely define the sequences required for Tlc1 RNA transcription termination, we used a previously developed ACT1–CUP1 fusion reporter gene assay (23). The Cup1p protein, required for growth in the presence of excess copper, is fused to the first exon and following intron of the gene encoding actin and the resulting composite gene is expressed via the promoter of the TDH3 gene (Figure 2B). This intron containing ACT1–CUP1 fusion gene transcribed on a plasmid (pGAC24) confers resistance to copper to a strain that lacks its endogenous CUP1 gene (Figure 2C and Supplementary Figure S2B, top lane). Insertion of sequences causing transcription termination in the intron and thus upstream of the CUP1 coding region will lead to copper sensitivity, allowing identification of terminator elements (Figures 2B, C and Supplementary Figure S2B). Using this setup and rationale, we inserted PCR amplified sequences from the TLC1 3′-end into the ACT1 intron of pGAC24 to search for elements required for Tlc1 RNA transcription termination. The cloned sequences all started at position +1130 (the NsiI restriction site marked with a white diamond in Figure 2A) and had endpoints at positions +1180, +1209, +1218, +1228, +1233, +1266, +1290 or +1311 relative to the TLC1 transcription start site (see graphical representation on Figure 2A and B). The different cloned TLC1 3′-end sequences are identified by their last nucleotide position (for example, TLC1–1233 represents the sequence spanning from +1130 to +1233). Tenfold serial dilutions of cultures with the cup1Δ strains harbouring the different ACT1–CUP1 constructs were spotted on copper plates. As mentioned above, an ACT1–CUP1 control plasmid without insert (No insert) conferred copper resistance and an ACT1–CUP1 construct with the SNR13 snoRNA terminator (SNR13 term) caused copper sensitivity (Figure 2C and Supplementary Figure S2B). The inserted TLC1–1180 and TLC1–1209 sequences still allowed transcription read-through, generating a functional Cup1p protein and hence resistance to copper. In stark contrast, the insertion of sequences TLC1–1218, TLC1–1228, TLC1–1233, TLC1–1266, TLC1–1290 or TLC1–1311 inhibited growth on copper plates, indicating the presence of a terminator element. Close inspection revealed that quite surprisingly, the minimum sequence causing transcription termination ends significantly upstream (+1218) of the first natural polyadenylation site documented at +1242 (Figures1C and 2A). The TLC1–1311–i construct, representing the +1130 to +1311 region cloned in the inverted orientation, did not cause copper sensitivity (Figure 2C and Supplementary Figure S2B). This result eliminates the possibility that the observed effect results from dysfunction of the ACT1–CUP1 fusion gene simply by insertion of a long sequence. A strain without the ACT1–CUP1 plasmid is used as a control for copper sensitivity (No CUP1). All strains showed similar growth on media without added copper (Figure 2C and Supplementary Figure S2B). The presence of a terminator element upstream of the natural polyadenylation sites strongly suggests that mature polyA− Tlc1 RNA can be produced independently from the polyadenylated forms at the +1242 sites.
Figure 2.
Sequences required for transcription termination upstream of the Tlc1 polyadenylation sites. (A) Schematic representation of the TLC1 transcription unit with a focus on the 3′-end. The numbers are relative to TLC1 transcription start site (+1). The white circles (empty or crossed) correspond to 3′-end positions of the various TLC1 sequences cloned in the ACT1–CUP1 reporter gene represented in (B). Empty white circles represent constructs containing a sequence that allows transcription read-through and crossed circles represent constructs carrying a sequence that cause transcription termination. See Figure 1 for symbols representing transcription start, polyA− 3′-end, polyA+ 3′-end and Sm protein binding sites. (B) Diagram of the ACT1–CUP1 fusion reporter gene with cloned intronic Tlc1 inserts and the expected phenotypes on copper plates for presence/absence of a terminator element. (C) Tenfold serial dilutions of a culture with the cup1Δ strain 46a containing ACT1–CUP1 constructs with no insert or with inserted sequences from Tlc1 3′-end. The cloned sequences start at the NsiI site at position +1130 and end at position +1180, +1209, +1218, +1228, +1233, +1266, +1290 or +1311, relative to transcription start site (+1). The different cloned TLC1 3′-end sequences are identified by their last nucleotide position. The TLC1–1311–i construct represents the +1130 to +1311 region cloned in the inverted orientation. The SNR13 snoRNA terminator is used as a positive control for transcription termination (SNR13 term). A strain harbouring no ACT1–CUP1 construct (but containing an empty LEU2 plasmid) is used as a control for copper sensitivity (No CUP1). Cultures were spotted on medium lacking leucine and containing 0 or 0.5 mM added copper and incubated for 3 days at 30°C.
The Tlc1 upstream terminator is under the control of the Nrd1/Nab3 transcription termination pathway
Using the same ACT1–CUP1 fusion reporter gene assay, we then searched for the trans-acting factors required for the newly discovered Tlc1 terminator function. If a given factor is involved in this particular termination event, one expects read-through of the involved terminator sequence (and copper resistance) when said factor is rendered defective by mutation (Figure 3A). Interestingly, the region between the polyA− Tlc1 3′-end (+1158) and the polyA+ 3′-end (+1242) contains sequences corresponding to Nrd1p and Nab3p consensus binding sites (30,31; Figure 2A and Supplementary Figure S1). The Nrd1 complex, composed of the Nrd1p, Nab3p and Sen1p proteins, is necessary for transcription termination of many small non-coding RNAs such as sn/snoRNAs (21,25,32). To verify if termination of Tlc1 transcription at the new site occurs via this Nrd1/Nab3 termination pathway, we used the same copper-resistance indicator strains as above but in addition introduced thermosensitive alleles of NRD1, NAB3 and SEN1. Tenfold serial dilutions of cultures with cup1Δ strains carrying the various ACT1–CUP1 constructs were spotted on copper plates and incubated at permissive or semi-permissive temperatures. Again as expected, strains harboring no ACT1–CUP1 plasmid (No CUP1) or an ACT1–CUP1 plasmid with no inserted terminator (No term) showed copper sensitivity and resistance, respectively, at all temperatures in all strains (Figures 3B and Supplementary Figure S2C). Furthermore, in the wild-type strain, the terminator for snoRNA (SNR13 term) as well as the newly discovered TLC1 terminator (TLC1–1233) caused copper sensitivity at all temperatures (see above and Figures 3B and Supplementary Figure S2C). Previous work had demonstrated that Nrd1p and Nab3p are required for the SNR13 snoRNA transcription termination (25,30). Accordingly, we observe SNR13 terminator read-through (copper resistance) when strains harboring nrd1-5 (nrd1) and nab3-11 (nab3) ts alleles are grown at semi-permissive temperature (27°C for nab3, 32°C for nrd1, Figure 3 and Supplementary Figure S2C). These cells are still sensitive to copper when grown at permissive temperature (23°C) where the Nrd1p/Nab3p factors and SNR13 terminator are fully functional (Figure 3B and Supplementary Figure S2C). The TLC1 terminator comprised in the TLC1–1233 fragment behaved exactly like the snoRNA terminator in this assay: termination is properly executed at 23°C, conferring copper sensitivity, but is no longer functional at higher temperatures, allowing read-through and growth in presence of copper (Figure 3B and Supplementary Figure S2C).
In order to further ascertain the implication of the Nrd1/Nab3 termination pathway in Tlc1 transcription termination, we introduced single point mutations that are predicted to abolish the function of the consensus Nrd1p and Nab3p binding sites into the TLC1–1233 ACT1–CUP1 construct and analyzed the effect on copper sensitivity. The wild-type TLC1 terminator (TLC1–1233 term WT) and the No CUP1, No term and SNR13 term controls again behaved as expected (see above and Figure 3C). Strikingly, the single point mutation in the Nab3p binding site (TCTT to TCCT) is sufficient to abolish termination and allow growth on copper plates comparable to the No term control (Figure 3C; TLC1–1233 term Nab3 mBS). However, the Nrd1p consensus binding site mutation (GTAG to CTAG) barely affected termination (Figure 3C; TLC1–1233 term Nrd1 mBS). A combination of the mutations in the Nrd1p and Nab3p sites yields the same effect than the single Nab3p-site mutation. These observations strongly support the conclusion that the newly identified Tlc1 terminator is under the control of the Nrd1/Nab3 transcription termination pathway.
Formally, it remained possible that the copper sensitivity caused by the TLC1–1233 terminator is the result of defective splicing, somehow caused by the exogenous sequences inserted in the actin intron, but not by transcription termination, as we concluded. In order verify this possibility, we analyzed the RNAs produced from the different ACT1–CUP1 reporter gene constructs by northern blot (Figure 3D). A probe specific to the CUP1 coding sequence located downstream of the ACT1 intron should reveal accumulation of unspliced precursor RNAs, if there was no transcription arrest but rather splicing interference. However, the results obtained with this assay do not reveal any discernable Cup1 mRNA produced in a wild-type strain carrying the TLC1–1233 construct (TLC1–1233-25°C and TLC1–1233-32°C) nor in a nrd1-5 strain carrying the TLC1–1233 construct grown at permissive temperature (nrd1-TLC1–1233-25°C, Figure 3D), which is consistent with a functional terminator. RNA derived from cells harboring the construct with the SNR13 snoRNA terminator (SNR13 term) is used as a positive control for termination and again, no clear specific signal for read-through RNA can be detected (Figure 3D). On the other hand, we used RNA derived from cells with the TLC1–1209 construct as control for read-through transcription followed by efficient splicing and there indeed were readily detectable specific mRNAs containing CUP1 sequences in this lane. Furthermore and also consistent with read-through, cells carrying the nrd1-5 ts allele and the TLC1–1233 construct grown at semi-permissive temperature (nrd1-TLC1–1233-32°C) show a significant increase in Cup1 mRNA. Likewise, cells carrying the TLC1–1233 construct with the point mutation in the Nab3p binding site (TLC1–1233-Nab3mBs) also produced Cup1 mRNA. The two Cup1 mRNA bands are also observed in the empty ACT1–CUP1 construct (data not shown) and are probably produced from alternative transcription start or termination sites. Regardless, these northern blots did not yield any evidence for detectable precursor RNAs containing CUP1 sequences in any of the cases where ACT1–CUP1 constructs caused copper sensitivity. Therefore, the presence of the Cup1 mRNA (Figure 3D) clearly correlates with copper resistance (Figure 3B and C) and absence of signal with copper sensitivity, which strongly supports the conclusion that these effects result from transcription termination and not splicing interference.
The results obtained with the ACT1–CUP1 fusion reporter gene assay predict that affecting the members of the Nrd1/Nab3 termination pathway should disturb the balance between polyA+ and polyA− Tlc1 RNA species. In order to verify this prediction, the expression of Tlc1 RNA was analyzed in RNA derived from ts mutants of the Nrd1/Nab3 complex by northern blot. Cells were grown at semi-permissive temperature (27°C for nab3, 32°C for nrd1 and sen1) for three population doublings before RNA extraction. RNA quantifications were obtained as described in materials and methods and are presented as an average of a minimum of four independent experiments. Consistent with previous data, the Tlc1 RNA in all strains grown at permissive temperature (23°C) contained 8–12% of polyA+ RNA (16 and Figure 4). At semi-permissive temperature, a strain expressing the nrd1-5ts allele (nrd1 in Figure 4) displayed a significant increase in the percentage of polyA+ Tlc1, up from 8% at permissive temperature to 26%. The results are similar for RNA derived from cells with the nab3-11ts allele (nab3 in Figure 4), but the changes are less significant (10–22%). The percentage of Tlc1 polyA+ RNA was practically unchanged for cells carrying the sen1-E1597Kts allele (sen1 in Figure 4) grown at semi-permissive temperature (12 and 11%). The WT strain also showed no significant change in the fraction of polyA+ Tlc1 when grown at various temperatures. Similar results were observed with cultures exposed to restrictive temperature (37°C) for 3 h (data not shown). Membranes stripped and rehybridized to a probe specific for actin mRNA revealed minimal variations in total Tlc1 RNA amounts between samples (Figure 4B). As a positive control for Nrd1p, Nab3p and Sen1p inactivation in the growth conditions used, the same membranes where rehybridized to a probe specific for the snR13 snoRNA. It was shown previously that affecting members of the Nrd1/Nab3 termination pathway causes read-through of the natural SNR13 terminator, eventually yielding an accumulation of an extended RNA (25,30). Our experiment described above indeed also revealed an increase of the read-through RNA from the snR13 RNA locus in ts cells grown at semi-permissive temperature (Figure 4A, bottom panel). The relative increases for this positive control RNA parallel quite well the increases for the Tlc1 RNA, i.e. robust increase in the nrd1 mutant and a rather marginal one in the nab3 mutant. Taken together, these results allow us to conclude that the newly discovered Tlc1 terminator located upstream of the natural first polyA+ sites is under the control of the Nrd1/Nab3 transcription termination pathway.
DISCUSSION
Despite the continuously increasing number of new types of RNAs, the telomerase RNAs remain in a class of their own. For example, the budding yeast Tlc1 RNA is similar to snRNAs with a TMG cap and Sm-protein binding at its 3′-end. However, due to its size and particularly to the occurrence of a polyadenylated (polyA+) form, it also has striking features of an mRNA. The picture does not get clearer when the RNA of other species are considered. The RNA from ciliates is transcribed by RNA polymerase III with a promoter resembling that of U6 snRNA genes (33,34) and human telomerase RNA shares features with H/ACA snoRNAs (35,36). Furthermore, the mechanisms controlling the expression of the telomerase RNA moiety needs to be clarified since the abundance of telomerase components is critical for telomere stability. We therefore began investigating the regulation of Tlc1 RNA generation by an analysis of the TLC1 3′-end sequences in order to identify regulatory elements controlling its transcription termination and 3′ maturation.
Previously it was shown that only the polyA− form of the Tlc1 RNA is associated with the active telomerase RNP (17). The physical 3′-end of that RNA is located only 15 nt downstream of the start of the Sm-protein binding site (17 and Figure 1C). While these data could not completely exclude the possibility that a small minority of the active telomerase RNPs may also contain the polyA+ form of the Tlc1 RNA, most likely the polyA− form by in large is predominant and the above question becomes how this polyA− RNA ending at nucleotides +1157 or +1158 is generated. The current tenet in the field was that Tlc1 is first produced as a longer polyA+ RNA that is then processed into the mature polyA− form (16). This idea was based on the fact that the level of both the polyA+ and polyA− forms of Tlc1 are reduced upon the depletion of the polyadenylation termination factors Pcf1p and RNA14p/15p (16). Nevertheless, the possibility of a direct formation of a polyA− Tlc1 without a need for the polyA+ precursor was hinted to by the fact that even in this experiment, a certain amount of the shorter Tlc1 polyA− RNA continues to accumulate hours after the disruption of RNA polyadenylation (16 and data not shown). In addition, when the Tlc1 polyadenylation signals are fused to another non-coding RNA (snoRNA), the generated polyA+ RNA was not processed into polyA− RNA (37). Our experiments described here show that mutating all probable or cryptic polyadenylation signals near the Tlc1 3′-end altered the synthesis of the polyA+ form without affecting the quantity or quality of the mature polyA− RNA. This result demonstrates that accumulation of the naturally occurring polyA+ Tlc1 is not required for the biogenesis or function of telomerase (Figure 1).
The data thus lead to the suggestion that other sequences near the 3′-end of TLC1 may be required for the generation of the mature RNA. To explore this possibility, we inserted sequences from the TLC1 3′-end into the ACT1–CUP1 fusion reporter gene. This approach allowed the identification of sequence elements that prevented the production of a Cup1 coding RNA, yielding copper sensitivity of the cells (Figures 2 and 3D). Most surprisingly, the terminator elements defined by these experiments are located upstream of the previously documented Tlc1 polyadenylation sites (Figure 2) and included a consensus binding site for the Nab3p terminator protein and several weak, but conserved consensus sites for the Nrd1p terminator protein (Supplementary Figure S1). This finding raised the possibility that mature polyA− Tlc1 RNA indeed could be generated via the Nrd1/Nab3-dependent RNA maturation pathway. Indeed, in yeast and in most metazoan cells, the mature 3′-ends of the majority of the non-coding RNAs are non-polyadenylated and are produced through this Nrd1/Nab3 termination pathway that is distinct from that generally acting on mRNAs (38–40). In yeast, the central Nrd1 complex required for this pathway is composed of Nrd1p, Nab3p and Sen1p (21,25,32). It is important to note that in the process of transcription termination, the Nrd1 complex acts in concert with the TRAMP complex to add a very short oligo(A) tail of about 2–4 adenosines that subsequently promotes RNA maturation or degradation by the exosome (41,42). However, the actual terminal DNA encoded nucleotide on the RNA can be at various distances from the Nrd1p or Nab3p binding sites.
Quite remarkably, a recent study in fact documented binding of Nrd1p and Nab3p to the very same Tlc1 3′-end region and quite convincingly determined Tlc1 RNA 3′-ends with short oligo(A) tails in the area (43). In fact, the 3′ position of the minimal sequence required for transcription termination as determined here (+1218) coincides almost to the nucleotide with the most downstream of the Nrd1-mediated 3′-ends determined by Clip-sequencing [+1221; see Supplementary Figure S1 and (43)]. Furthermore and most importantly, these positions clearly are upstream from the first polyA+ addition site determined previously [+1242; (16)]. These data therefore strongly suggest that the Nrd1/Nab3-dependent non-coding RNA termination pathway causes transcription termination around nucleotide +1220.
Further supporting this conclusion are our findings that in the absence of fully functional Nrd1p or Nab3p, read-through occurs on the new Tlc1 terminator in the ACT1–CUP1 reporter gene assay (Figure 3). In fact most revealing is the fact that a single point mutation in the Nab3p binding site is sufficient to abolish the function of the Tlc1 terminator (Figure 3C). As predicted by these results, analyses of RNAs extracted from cells carrying ts alleles of NRD1 or NAB3 grown at semi-permissive temperatures revealed an increase in the proportion of the polyA+ form of Tlc1 (Figure 4). Consistent with this latter observation, RNA derived from cells harboring a different NRD1 mutant allele (nrd1-102) and that were incubated at restrictive temperature also showed a near twofold increase in longer Tlc1 forms, most likely the polyA+ forms (43).
Our hypothesis stipulates that the Tlc1 RNA is produced by a RNA polymerase II transcription complex that terminates upon recognition of the Nrd1/Nab3 termination signals (Figure 5). Short oligoA addition and processing of that transcript back to the +1158 will create the polyA− Tlc1 RNA without the necessity of a polyA+ precursor. Formally, we cannot exclude the possibility that part of the mature polyA− RNA could be formed through processing of the longer polyA+ form. Thus, Tlc1 RNA 3′-end formation could occur via two redundant pathways: the first constitutive and major pathway directly produces the necessary polyA− RNA via the Nrd1p-dependent snRNA pathway, as shown by the results presented here. The second pathway would be based on some read-through of the Nrd1p/Nab3p sites, generating polyA+ RNA at the next available cryptic polyA+ sites and this polyA+ RNA could be processed, at least in part, into the same polyA− form as above (see Figure 5). There is indeed precedence for this secondary pathway as well: a transcription complex that bypasses a Nrd1p/Nab3p-dependent snRNA termination signal usually terminates in the nearest next stretch of A/U rich sequence (44,45). Indeed, many snoRNAs and snRNAs become polyadenylated upon the deletion of ribonucleases that normally degrade aberrant RNAs (44).
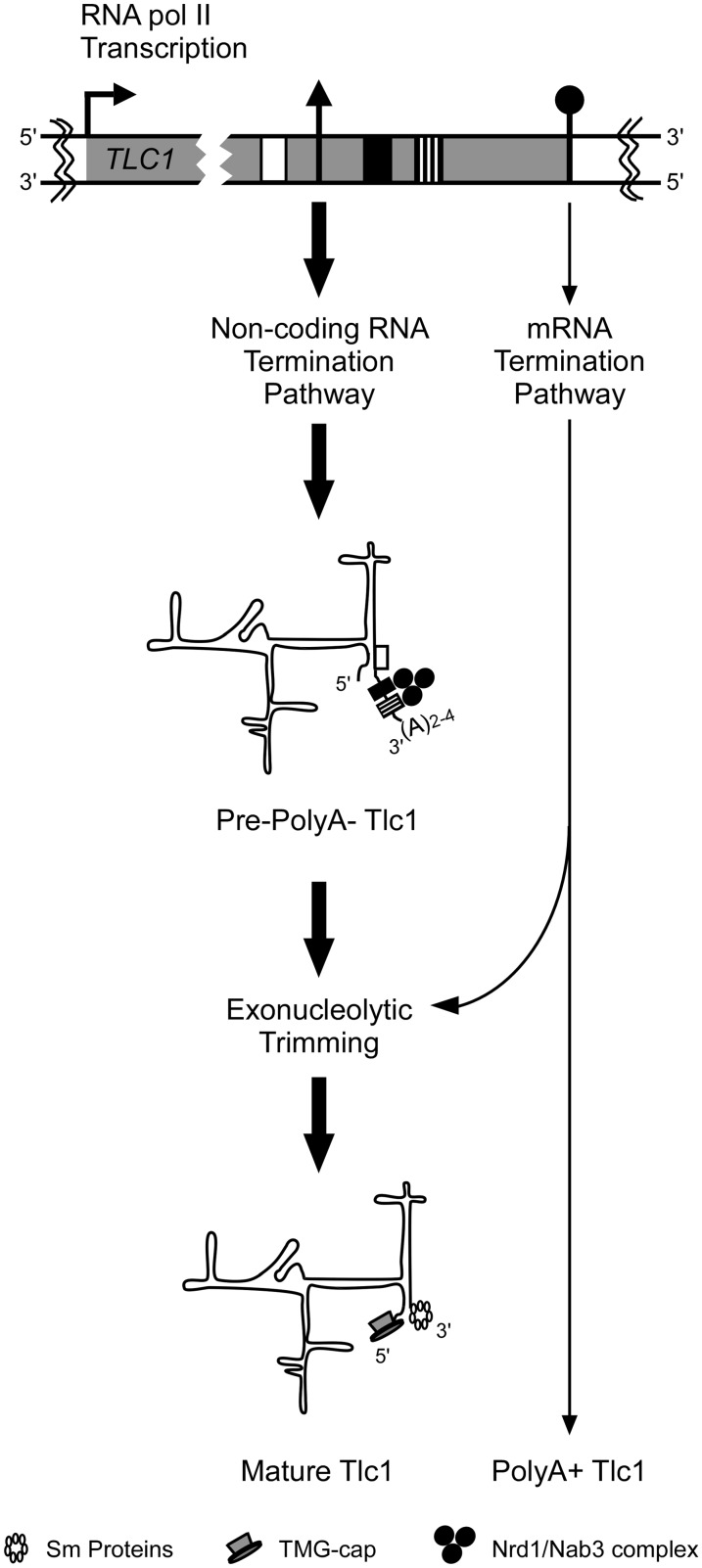
Figure 5.
Graphical illustration of maturation pathways for Tlc1 RNA. A schematic representation of the TLC1 transcription unit, with a focus on the 3′-end, is shown on top. The transcription elongation complex of TLC1 either terminates near the mature polyA− 3′-end via the non-coding Nrd1/Nab3 RNA termination pathway when encountering Nrd1p/Nab3p binding sites or continue to the nearby polyadenylation signals. Termination near the mature end will result in a TMG-capped and non-polyadenylated RNA that is trimmed by 3′ to 5′-end exonucleases to the mature 3′-end defined by the position of the Sm binding proteins. Termination near a polyadenylation signal via the cleavage/polyadenylation machinery will produce a polyadenylated RNA that will normally be deadenylated and degraded by exonucleases, unless stabilized by the binding of Sm proteins in the cytoplasm that also facilitates Tlc1 import to the nucleus. Therefore, this polyadenylated Tlc1 can also be processed to the mature form. According to this model, individual mutations in either pathway may not dramatically affect the steady-state level of mature Tlc1 RNA.
The Tlc1 RNA maturation pathway, including the fail-safe termination by a polyA+ form, may thus be the same as that used for snRNAs and consequently, together with the known other Tlc1 properties that are similar to snRNAs, the findings presented here strongly suggests that the budding yeast telomerase RNA belongs to the snRNA class of RNAs. It has been reported that termination at Nrd1p/Nab3p-dependent sites may be inefficient for a relatively long RNAs (32). Indeed, the only known polyA− RNA of the approximate size of Tlc1 in yeast (U2 snRNA) uses endoribonucleolytic cleavage by the dsRNA-specific ribonuclease Rnt1p to suppress polyadenylation (44). However, there still are numerous cases where the Nrd1/Nab3 complex seems to function more than 1-kb downstream of the transcription start site (43,46,47). Given that the Tlc1 RNA termination mechanism relies strongly on the Nab3p-binding site, less so on the Nrd1p sites and is independent of Sen1p, the Tlc1 terminator could be a long snRNA terminator variant similar to the one after the snR20 gene (32).
Although both the polyA+ and polyA− species of Tlc1 coexist in normal cells, the functional importance of the polyA+ form remains unclear. Both forms of Tlc1 are conserved in different ‘sensu stricto’ Saccharomyces (data not shown) and even in distant yeast relatives like Kluyveromyces lactis (16). However, different mechanisms have evolved for telomerase RNA 3′ end formation. For example, in Schizosaccharomyces pombe a particular spliceosomal cleavage generates a mature polyA− RNA (48–50) and in humans, the RNA could be considered a variant H/ACA snoRNA (35,36). We propose that the persistence of Tlc1 polyadenylation in closely related yeast species simply reflects the limitation of the termination mechanisms generating non-polyadenylated RNA. This idea also underscores the very high importance of transcription termination for generating adequate steady-state levels of RNA. In order to make sure proper amounts of RNA are present in the cells at the required time-points, cells have evolved redundant pathways ensuring termination at every single transcription unit and these include the fail-safe mechanism for mRNA termination using the Nrd1/Nab3 complex or Rnt1p cleavage (51), or the bipartite terminator at the 3′ end of some genes of snoRNAs (37,42,52–54).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. Supplementary Figures 1–2.
FUNDING
Canadian Institutes of Health Research (grant MOP97847 to R.J.W.). Support for the RNA group core was provided by CIHR. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We are especially grateful to David Brow (University of Wisconsin) for suggesting the copper sensitivity assay and providing strains, plasmids and support. We also thank Carol Greider for strain CSHY76 and François Ménard for technical help. S.A. is a Chercheur National of the Fonds de la Recherche en Santé du Québec (FRSQ) and R.J.W. holds the Canada Research Chair in Telomere Biology.
REFERENCES
- 1.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 3.Collins K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 5.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 6.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 7.Gilson E, Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell. Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 8.Larose S, Laterreur N, Ghazal G, Gagnon J, Wellinger RJ, Elela SA. RNase III-dependent regulation of yeast telomerase. J. Biol. Chem. 2007;282:4373–4381. doi: 10.1074/jbc.M607145200. [DOI] [PubMed] [Google Scholar]
- 9.Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 10.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 11.Goudsouzian LK, Tuzon CT, Zakian VA. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol. Cell. Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 15.Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. Embo J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapon C, Cech TR, Zaug AJ. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 17.Bosoy D, Peng Y, Mian IS, Lue NF. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- 19.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. New York: Cold Spring Harbor; 1990. [Google Scholar]
- 20.Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 25.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 26.Perbal B. A Practical Guide to Molecular Cloning. 2nd edn. New York: John Wiley and Sons, Inc; 1988. [Google Scholar]
- 27.Zappulla DC, Goodrich KJ, Arthur JR, Gurski LA, Denham EM, Stellwagen AE, Cech TR. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA. 2011;17:298–311. doi: 10.1261/rna.2483611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons, Inc; 1997. [Google Scholar]
- 29.Irniger S, Braus GH. Saturation mutagenesis of a polyadenylation signal reveals a hexanucleotide element essential for mRNA 3′ end formation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1994;91:257–261. doi: 10.1073/pnas.91.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 33.Yu GL, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 34.Orum H, Nielsen H, Engberg J. Structural organization of the genes encoding the small nuclear RNAs U1 to U6 of Tetrahymena thermophila is very similar to that of plant small nuclear RNA genes. J. Mol. Biol. 1992;227:114–121. doi: 10.1016/0022-2836(92)90686-e. [DOI] [PubMed] [Google Scholar]
- 35.Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol. Cell. Biol. 2010;30:2775–2786. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morlando M, Greco P, Dichtl B, Fatica A, Keller W, Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell. Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA. 2011;17:2011–2025. doi: 10.1261/rna.2840711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abou Elela S, Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. Embo J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol. Cell. Biol. 2007;27:4157–4165. doi: 10.1128/MCB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciais D, Bohnsack MT, Tollervey D. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 2008;36:3075–3084. doi: 10.1093/nar/gkn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. Embo J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 50.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 51.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinmetz EJ, Ng SB, Cloute JP, Brow DA. cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol. 2006;26:2688–2696. doi: 10.1128/MCB.26.7.2688-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fatica A, Morlando M, Bozzoni I. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. Embo J. 2000;19:6218–6229. doi: 10.1093/emboj/19.22.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.