Abstract
Hypoxia-inducible factors (HIF)-1α and HIF2α are major transcription factors required for adaptive responses to hypoxia. HIFs form a complex with aryl hydrocarbon receptor nuclear translocator (ARNT) to bind to the regulatory regions of target genes. The acetylation of histones by histone acetyltransferases (HATs) is one of the epigenetic marks associated with active chromatin. Indeed, HIFs recruit p300 HAT to hypoxia response elements (HREs) within gene regulatory regions. Here, we report an unusual HIF-mediated transcriptional activation in ovarian clear cell carcinoma (CCC). While characterizing coagulation factor VII (FVII) gene induction during hypoxic conditions, we observed that the interaction of HIF2α with Sp1, but not with ARNT, could induce transcription of FVII in a HRE-independent manner. Unexpectedly, this gene activation is associated with histone deacetylation. We found that a class II HDAC, HDAC4, is recruited with HIF2α to the FVII promoter as a co-activator, while p300 HAT negatively regulated this process. Furthermore, this mechanism can be synergistically enhanced via a deacetylation-dependent pathway when cells are simultaneously exposed to hypoxic and serum-free conditions. These results suggest the presence of a stress-responsive transcription mediated by the HIF2α/Sp1/HDAC4 network and explain how CCC shed their procoagulant activity under hypoxia.
INTRODUCTION
The activation of various signaling pathways allows cells to adapt to be environmental stresses, such as O2 and/or nutrient deprivation. Hypoxia inducible factors, HIF1α and HIF2α, are two of the main nuclear transcription factors that permit the appropriate cellular responses to reduced oxygen tension surrounding cancerous tissues (1). Under normal oxygen levels, or normoxia, HIF expression levels are very low because these factors undergo proteasomal degradation. However, under O2-deprived conditions, HIFs escape from ubiquitination by E3 ubiquitin ligases and accumulate within the cell. HIFs form heterodimeric complexes with the constitutively expressed aryl hydrocarbon receptor nuclear translocator (ARNT, also called as HIF1β), enabling them to bind to hypoxia response element (HRE) sequences (1). This HIF–ARNT complex is associated with gene regulatory regions and functions to enhance transcription.
Histone acetylation is closely associated with transcriptional activation. Histone acetyltransferases (HATs) are recruited to regulatory regions of genes to catalyze the acetylation of histones and make chromatin structures more accessible for other transcriptional activators (2). Histone deacetylation, in contrast, is catalyzed by histone deacetylases (HDACs), which are associated with chromatin when transcription is down-regulated (2). One of the most important HATs, p300, is responsible for constitutive and inducible activation of numerous genes (2). Indeed, p300 is a transcriptional co-activator for HIFs, responsible for activating crucial events during cell survival, such as angiogenesis and metastasis (3).
Blood coagulation factor VII (fVII) is a key enzyme in the extrinsic coagulation cascade (4). Predominantly produced by hepatocytes, fVII interacts with its cellular receptor, tissue factor (TF) and, when bound to TF, is converted to its active form (fVIIa), thereby triggering a downstream coagulation cascade (5). This TF–fVIIa complex is known to initiate key pathogenic mechanisms in cancer, including cell motility, invasion, cell survival and angiogenesis (5), and is a major cause of thrombosis in cancer patients. In particular, in ovarian clear cell carcinoma (CCC), one of the most aggressive ovarian malignancies, the TF–fVIIa complex can cause venous thromboembolism (6), a common complication of cancer.
In CCC, fVII synthesis is induced in response to hypoxia (6,7), leading to the secretion of microvesicles that contain the TF–fVIIa complex; this secretion is likely to be one of the key causes of thrombosis in CCC-type patients with poor prognosis (6). We previously showed that various cancer cells could ectopically synthesize fVII (7). When this TF–fVIIa complex is derived from ectopically expressed fVII, cancer cells show enhanced motility and invasion (7). However, detailed mechanisms of the fVII induction are not clear. While we were characterizing the mechanism of hypoxic fVII-gene activation, we unexpectedly observed that HIF2α, but not HIF1α, binds to the FVII promoter region in the absence of HRE sequences (7), implicating that a novel cellular mechanism of adaptive response may exist. Thus, in this study, we sought to determine the detailed mechanisms of this unusual gene activation, including the involvement of histone acetylation, in an attempt to gain an understanding of the molecular mechanism of thrombosis in CCC patients. In addition, because cancer tissues with poor vascularization tend to be deficient in other serum factors in addition to molecular oxygen and nutrients (8), we further investigated how the deficiency of these factors affects this transcriptional regulation.
MATERIALS AND METHODS
Cell culture and reagents
Human cancer cell lines were maintained as previously described (7). Acriflavine (A8126) and tunicamycin (T7765) were purchased from Sigma (St Louis, MO, USA). Cell culture media used for experiments under glucose-free condition were from Invitrogen (11875 and 11879; Carlsbad, CA, USA).
Expression vectors
Expression vectors of HIF1α, HIF2α, and p300 were prepared by PCR amplification using PrimeScript 1st strand cDNA synthesis kit (Takara, Shiga, Japan) with the following primers: HIF1α, 5′-CTAGCTAGCACCGATTCACCATGGAGGGCG-3′ (forward; F) and 5′-GGGGTACCGCTCAGTTAACTTGATCCAA-3′ (reverse; R); HIF2α, 5′-TCGACGCGTCGACAATGACAGCTGACAAGGAG-3′ (F) and 5′-GCTCTAGAGCTCAGGTGGCCTGGTCCAG-3′ (R); p300, 5′-CCGCTCGAGCCTCGCTTGTATCTCCGAAAGAAT-3′ (F) and 5′-GGGGTACCGGTGTCTCTAGTGTATGTCTAGTGTACTCTGT-3′ (R). Fragments containing full-length cDNA derived from OVSAYO cells were inserted into the pCI vector (Promega, Madison, WI, USA) at the Nhe I/Kpn I (HIF1α), Mlu I/Xba I (HIF2α), and Xho I/Kpn I (p300) sites.
Small interference RNA experiments
Small interference RNAs (siRNAs) for HIFs were prepared as previously described (9). siRNAs for Sp1, p300, ARNT, and HDAC4were ON-TARGET plus SMART pool reagents (Dharmacon, Lafayette, CA). Silencer Negative Control 1 RNAi (Ambion) was used as non-specific siRNA. Transfection was performed using lipofectamine RNAi MAX (Invitrogen).
Quantitative RT-PCR analysis
We determined mRNA levels by quantitative RT-PCR, as previously described (7).
Construction of luciferase plasmids and reporter gene assay
A luciferase plasmid construct with the FVII 5′ full promoter region was prepared as previously described (10). Mutant constructs were prepared using the QuikChange mutagenesis Kit (Stratagene, La Jolla, CA). Luciferase assays were performed as previously described (10).
Western blot analysis
Western blotting was performed as previously described (10). Antibodies used for western blotting or immunoprecipitation were Flag M2 (F1804, Sigma), HA (H6908, Sigma), HDAC1 (sc-7872, Santa Cruz Biotechnology, Santa Cruz, CA), HDAC2 (Sc-7899, Santa Cruz), HDAC3 (Sc-11417, Santa Cruz), HDAC4 (ab1437, Abcam, Cambridge or 2072S, Cell Signaling, Danvers, MA), SIRT1 (1104-1, EPITOMICS, Burlingame, CA), ARNT (NB100-110, Novus Biologicals, Littleton, CO), Sp1 (NB600-232, Novus Biologicals), and CHOP (2895, Cell Signaling).
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analysis was performed as previously described (10). The primers, probes and antibodies used are described elsewhere (7). Other antibodies used in this study were as follows: ARNT (NB100-110, Novus Biologicals), acetyl-histone H3 (06-599, Millipore, Temecula, CA), acetyl-histone H4 (06-866, Millipore), histone H3 (ab1791, Abcam), HDAC4 (ab1437, Abcam), HDAC-1 (sc-7872, Santa Cruz) and HDAC-2 (Sc-7899, Santa Cruz). Primers used for ChIP analysis of VEGF-HRE region by real-time PCR were 5′-GCCTCTGTCTGCCCAGCTGC-3′ and 5′-GTGGAGCTGAGAACGGGAAGC-3′; hybridization probes were 5′-TTGCCAGACTCCACAGTGCATACGTGG-FITC-3′, and 5′-LCRed640-TCCAACAGGTCCTCTTCCCTCCCAG-3′.
Co-immunoprecipitation analysis
Expression vector of Sp1 tagged with a flag peptide at the N-terminus (pCI–Sp1–Flag) and an expression vector of HIF2α tagged with a hemagglutinin peptide (pCI–HIF2α–HA) were prepared using QuikChange kit by inserting corresponding DNA sequences into the pCI–Sp1 and pCI–HIF2α plasmids, respectively. Expression vectors of truncated HIF2α were also prepared using QuikChange kit as deletion mutants of pCI–HIF2α–HA. pCI–Sp1 was prepared by inserting the full-length cDNA fragment amplified from OVSAYO cells at the XhoI/KpnI cloning site. Primers used were 5′-CCGCTCGAGGACAGGACCCCCTTGAGCTTG-3′ (F) and 5′-GGGGTACCGGGGTATGGCCCATATGTCTCTG-3′ (R). After transfection (24 h), cells were lysed with RIPA buffer (Sigma, R0278) and incubated with normal IgG, anti-Flag, or anti-HA antibodies (see “Western blot analysis” section) at 4°C for 2 h. Antibody-protein complexes were captured using Dynabeads-protein G (Invitrogen). Immunoprecipitates were analyzed by western blotting. Immunoprecipitation of endogenous Sp1 was performed using an anti-Sp1 antibody (NB600-232, Novus Biologicals) as described above.
RESULTS
HIFs mediate FVII-gene activation under hypoxia
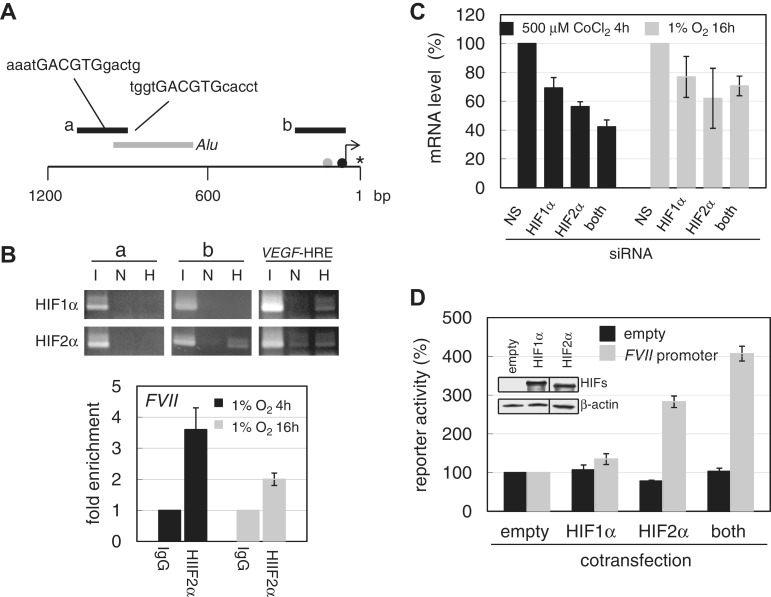
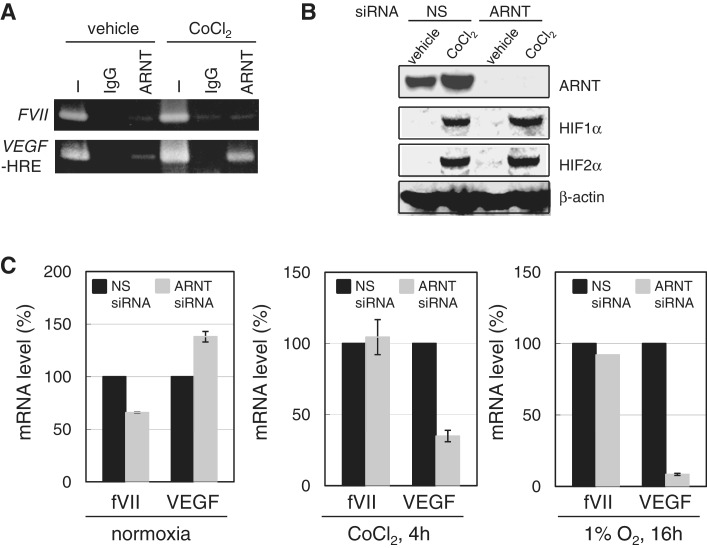
We previously reported that the FVII gene is inducible in cancer cells under hypoxia-mimetic conditions created with CoCl2 treatment (7), with highest FVII induction observed in the ovarian CCC cells, OVSAYO and OVISE. Thus, we used these cell lines to further examine the mechanism of the hypoxic gene activation. Using OVSAYO cells, we first performed a transcription factor-binding site analysis by inspecting an approximate 2500 bp region spanning an authentic FVII promoter and intron 1 (positions 3781–6180, GenBank accession number NG_009262; http://www.gene-regulation.com and http//:www.cbrc.jp/research/db/TFSEARCHJ.html). Our results identified the existence of candidate HREs upstream of the FVII 5′ region (positions 4104–4109 and 4281–4286, accession number NG_009262) close to predicted Alu elements (positions 4121–4417, accession number NG_009262 and Figure 1A). However, ChIP analysis showed that HIFs did not bind to this region (Figure 1A and B, bar a) in response to CoCl2 treatment (7) although both HIFs could occupy a HRE region of VEGF gene (Figure 1B). Unexpectedly, we found that HIF2α principally bound to the FVII promoter region (Figure 1A, bar b) in the absence of a HRE site in OVSAYO cells (7), and to the FVII promoter under real hypoxic (1% O2) culture conditions (Figure 1B); this suggests that HIF2α may be responsible for FVII gene induction. Finally, using western blotting and ChIP analysis, we confirmed the preferential binding of HIF2α to the FVII promoter region in an additional CCC cell line, OVISE (Supplementary Figure S1A and S1B).
Figure 1.
HIFs mediate FVII activation under hypoxia. (A) Schematic representation of FVII 5′-region. Black bars indicate the PCR amplicon for ChIP assay. Nucleotide sequence with capital letters represents the HRE sequence predicted by a computer search. Gray bar designates the position of predicted putative Alu repetitive elements. Gray and black circles indicate Sp1 and HNF-4 binding sites, respectively. A bent arrow and an asterisk indicate transcription and translation start sites, respectively, as previously determined (13). (B) ChIP analysis of HIFs. ChIP was performed for the FVII regions and a VEGF-HRE region in OVSAYO cells. PCR primers for FVII regions are as in Figure 1A. Primers for the VEGF-HRE region are as previously described (7). ‘I’, ‘N’, and ‘H’ indicate input sonicated DNA fragments without immunoprecipitation, normoxia, and 1% O2 for 16 h, respectively. Data were also estimated by real-time PCR. Fold enrichment of immunoprecipitated DNA for each transcription factor against background (templates due to non-specific binding with IgG) is shown. Data are mean of three independent experiments (hereafter n = 3) ± SD. (C) Effect of HIFs on the expression of FVII in OVSAYO cells. Cells were cultured in 500 µM CoCl2 for 4 h and 1% O2 for 16 h and analyzed with real-time RT-PCR. NS: non-specific. Data are the mean (n = 3) ± SD. (D) Luciferase reporter analysis of FVII activation by HIFs. A FVII-promoter construct (Supplementary Figure S2B) was transfected into OVSAYO cells with each HIF separately (400 ng) or together (200 ng of each). Inset: western blotting analysis of HIFs expressions 24 h post-transfection. β-actin shows equal protein loading. Relative promoter activities are shown as percentages of the activity from cells transfected with both empty vectors. Variations in transfection efficiency were corrected with a control Renilla luciferase vector. Data are the mean (n = 3) ± SD.
We next tested the effect of HIFs on FVII activation under CoCl2 or 1% O2 conditions. Using OVSAYO cells, we first suppressed HIF induction using HIF-specific RNA interference (RNAi) (9) and then examined its effect on fVII transcriptional induction using real-time RT-PCR analysis. We observed that fVII induction was suppressed by the knock-down of not only HIF2α but also HIF1α (Figure 1C), as compared to non-specific (NS) siRNA-transfected experiments, and this could be further amplified by the concurrent knock-down of both HIFs (Figure 1C). In addition, quantitative ChIP analysis by real-time PCR supported our earlier findings that HIF1α does not bind to the FVII promoter under hypoxia (Supplementary Figure S2A); this eliminated the possibility that trace amounts of HIF1α up-regulated FVII transcription through association with the promoter region.
To investigate whether the FVII promoter region is activated by HIFs, we next performed a reporter gene analysis. A FVII fragment (−400/+1), which covers the full FVII promoter activity (10), was fused to a luciferase vector (Supplementary Figure S2B). The reporter assay revealed that the promoter was activated by ectopic expression of HIF2α, whereas transfection of the same concentration (400 ng) of HIF1α induced a very weak activation (Figure 1D). Interestingly, the promoter was most activated by the simultaneous expression of equal concentrations of both HIFs (200 ng each; Figure 1D). Positive control experiments with a HRE sequence of the VEGF gene (Supplementary Figure S2B) showed enhanced luciferase activity with HIF induction (Supplementary Figure S2C). As previously reported, HIF2α caused the most activity, likely due to the differential regulation of HIF function and expression (11,12). Unlike with FVII activation, the activation of VEGF did not increase upon co-transfection of both HIFs. Together, these results suggest that HIFs may co-operatively activate the FVII promoter.
An Sp1-binding site is responsible for activation of FVII
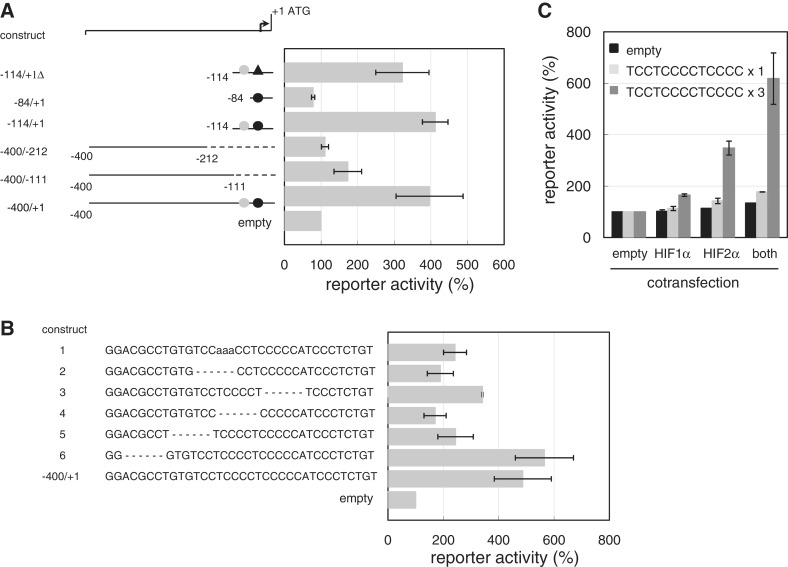
To further define the regions responsible for FVII activation, mutant luciferase constructs were co-transfected with both HIFs into OVSAYO cells. Constructs lacking a known Sp1-binding site demonstrated a weak response to the HIFs, similar to the response observed with the empty vector (Figure 2A). Using additional deletion constructs (Figure 2B), we found that promoter activation by HIFs was particularly impaired when the core Sp1-binding site of the FVII gene was mutated: the 5′-TCCTCCCCTCCCC-3′ sequence (13) (Figure 2B; construct 4). Constructs with single or triple copies of this sequence responded primarily to HIF2α, and when co-expressed with HIF2α, HIF1α co-operatively activated the luciferase expression (Figure 2C). Together, these findings suggest that the Sp1 site is responsible for FVII induction by HIFs.
Figure 2.
An Sp1 binding sequence is responsible for FVII activation by HIFs. (A) OVSAYO cells were co-transfected with FVII-promoter derived constructs and HIFs. Luciferase activity was estimated 24 h post-transfection. Gray and black circles indicate Sp1 and HNF-4 binding sites, respectively. Black triangles indicate a six base pair deletion of core HNF-4 binding sequence (10). (B) Construct with a region corresponding to −85 to −114 of the FVII promoter (Figure 2A), or its deletion mutants, were co-transfected with HIFs, and assessed for luciferase activity 24 h post-transfection. (C) Constructs inserted with single or triple copies of a Sp1-binding sequence (Figure 2B) were transfected with HIFs, and tested for luciferase activity after 24 h. For all experiments, relative promoter activities are shown as Figure 1D. Data are the mean (n = 3) ± SD.
HIF2α–Sp1 interaction induces FVII activation under hypoxia
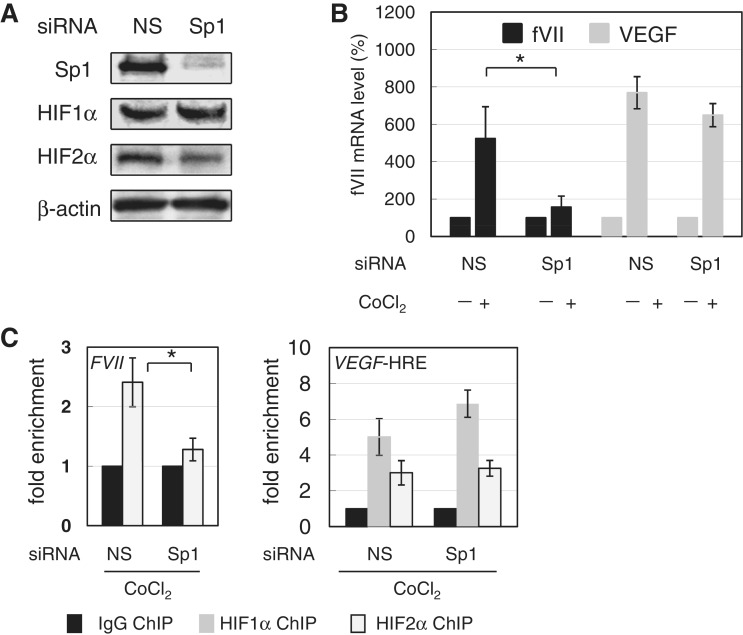
We previously showed that Sp1 binds to the FVII promoter in OVSAYO cells (10). Thus, we next tested whether Sp1 is required for FVII induction under hypoxia-mimetic and real-hypoxic conditions. In OVSAYO cells, RNAi was first used to suppress Sp1 without affecting HIF protein levels (Figure 3A), before the cells were cultured with CoCl2. Real-time PCR analysis revealed that Sp1 knockdown markedly reduced fVII mRNA induction in both OVSAYO (Figure 3B) and OVISE cells (Supplementary Figure S2D), and this same Sp1 dependency was observed under real-hypoxic conditions (Supplementary Figure S3A). Induction of the VEGF gene was not significantly influenced by a decrease in Sp1 (Figure 3B), even though Sp1 binds to the VEGF promoter, and is essential for basal promoter activity (14). Following this, we tested whether HIF2α binding to the FVII promoter is diminished in Sp1-siRNA-treated OVSAYO cells under CoCl2 treatment. Using ChIP analysis, we observed a decrease in the binding of HIF2α by Sp1-knockdown, although the binding of both HIFs to the VEGF-HRE region was not significantly altered (Figure 3C).
Figure 3.
HIF2α binds FVII promoter via interaction with Sp1. (A) Effect of Sp1 knockdown on the expression of HIFs induced in OVSAYO cells cultured in 500 µM CoCl2 for 4 h. (B) Effect of Sp1 on FVII expression in OVSAYO cells cultured with or without 500 µM CoCl2 for 4 h after Sp1 knockdown. FVII expression was examined by real-time RT-PCR. VEGF expression was performed as a control of HRE-dependent expression. NS: non-specific. Data are the mean (n = 3) ± SD. *P < 0.05 (two-sided Student’s t-test). (C) Effect of Sp1 on binding of HIFs to FVII promoter in OVSAYO cells cultured in 500 µM CoCl2 for 4 h after Sp1 knockdown. ChIP analysis for HIFs binding was performed for VEGF-HRE and FVII regions. Data were estimated by qPCR. Data are the mean (n = 3) ± SD.
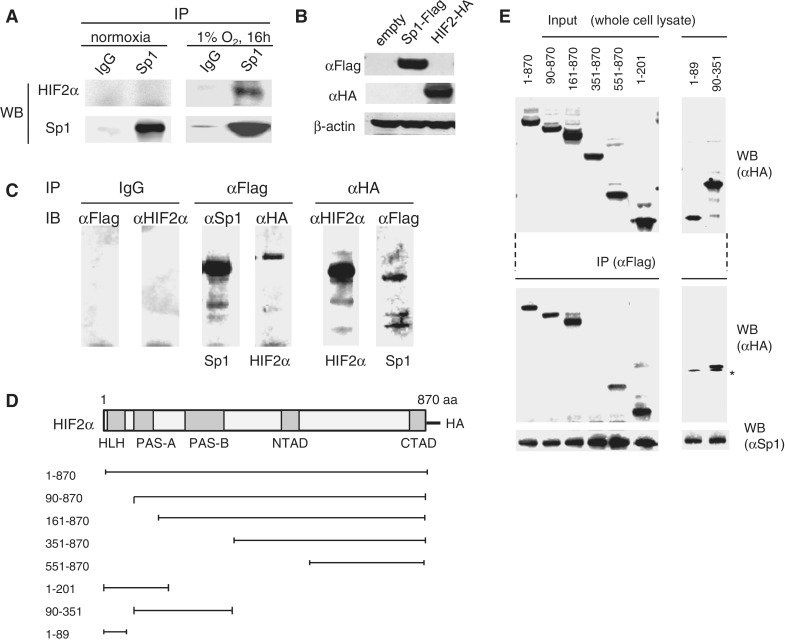
We next wanted to examine whether HIF2α interacts with Sp1. Immunoprecipitation analysis revealed that HIF2α induced in OVSAYO cells cultured under 1% O2 for 16 h can be precipitated with endogenous Sp1 (Figure 4A); this suggests that Sp1 and HIF2α interact to form a protein complex. Furthermore, HA-tagged HIF2 and Flag-tagged Sp1 were co-transfected into OVSAYO cells (Figure 4B), and immunoprecipitated with anti-HA or anti-Flag antibodies. Western blotting results showed that HIF2α–HA formed a protein complex with Sp1–Flag in OVSAYO cells (Figure 4C). To determine the binding sites, we then co-transfected various deleted mutants of HIF2α (Figure 4D) with Sp1–Flag. We observed that Sp1 binds to HIF2α, even in the absence of the HLH (90–870) and PAS-A (161–870) domains (Figure 4D and E), and interacts with the C-terminal region that includes the CTAD domain (551–870) of HIF2α, but not when this portion was fused with the middle region containing the NTAD domain. Sp1 also associated with the HLH-PAS-A portion of the protein (1–201) as well as both PAS domains (90–351), but not with the HLH domain alone.
Figure 4.
(A) Co-immunoprecipitation analysis of endogenous Sp1 and HIF2α expressed in OVSAYO cells. (B) Western blot analysis of Flag-Sp1 and HA-HIF2α expressed in OVSAYO cells. (C) HIF2α interacts with Sp1 in OVSAYO cells. Cells transfected with HIF2-HA and Sp1-Flag were subjected to immunoprecipitation 24 h post-transfection and analyzed by immunoblotting. (D) Truncated HIF2α proteins used for co-immunoprecipitation experiments. (E) Cells were transfected with truncated HIF2–HAs and Sp1–Flag and immunoprecipitated with anti-Flag antibody 24 h post-transfection. An asterisk indicates a non-specific band.
To verify the Sp1-dependent binding, we next performed ChIP analysis with an anti-HIF2α antibody on OVSAYO cells transfected with luciferase constructs and treated with CoCl2 (Supplementary Figure S3B). PCR analyses revealed that HIF2α binds to the full-length promoter, and that binding can be abrogated upon deletion of the Sp1 site. In addition, CoCl2 treatment did not affect the expression (6) or binding levels (Supplementary Figure S3C) of Sp1 to the FVII promoter, suggesting that HIF2α interacts with Sp1 that is constitutively associated with promoter region, and that HIF2α does not activate the FVII promoter by increasing Sp1 binding.
FVII activation does not require ARNT
HIFs induced under hypoxia form heterodimeric complexes with constitutively expressed ARNT to bind HRE sequences. Thus, we next sought to determine whether this complex formation is necessary for FVII activation. ChIP analysis revealed that ARNT did not bind to the FVII promoter in response to CoCl2 stimulation (Figure 5A); however, ARNT was recruited to the VEGF-HRE site under these conditions (Figure 5A). Following this, we assessed the effect of ARNT expression on FVII and VEGF gene inductions. Using RNAi, we first suppressed ARNT in OVSAYO cells (Figure 5B), in which induction of HIFs was not affected (Figure 5B). Using real-time RT-PCR analysis, we detected a decrease and an increase in fVII and VEGF mRNA levels, respectively, when ARNT-suppressed cells were cultured under normoxia (Figure 5C). However, there was a marked reduction in VEGF mRNA when OVSAYO cells were treated with CoCl2 or 1% O2 with no change in the percentage of fVII mRNA as compared with the negative control (Figure 5C). Finally, we tested whether inhibiting the formation of the HIF-ARNT complex influences FVII activation. OVSAYO cells were cultured with CoCl2 for 4 h in the presence of a heterodimerization inhibitor, acriflavine (ACF) (15). Real-time RT-PCR analysis showed that ACF treatment increased the fVII mRNA level via unknown mechanism (Supplementary Figure S4A), and reduced the mRNA levels of the HRE-dependent VEGF and ENO1, as expected (Supplementary Figure S4A).
Figure 5.
(A) ChIP for ARNT binding was performed for FVII and VEGF-HRE regions in OVSAYO cells cultured in the presence or absence of CoCl2. (B) Western blotting of ARNT expression in cells transfected with NS- or ARNT-siRNA. (C) Effect of ARNT on the expression of FVII and VEGF in OVSAYO cells cultured under normoxia, 500 µM CoCl2 for 4 h, or 1% O2 for 16 h. After ARNT knockdown, cells were cultured under above conditions. Real-time RT-PCR analysis was performed. Data are the mean (n = 2) ± SD.
Hypoxic FVII activation associates with deacetylation
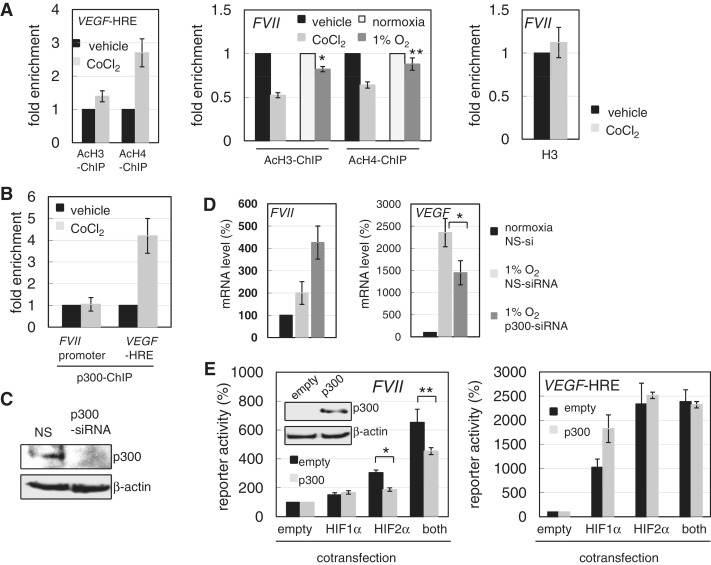
HIFs recruit p300 HAT to HREs and activate target genes. Therefore, we wanted to assess how histone acetylation is linked to FVII activation. First, we tested the effect of CoCl2 treatment on the acetylation of histones H3 and H4 within the FVII promoter region in OVSAYO cells. As expected, ChIP analysis of VEGF-HRE region showed an elevation in the histone acetylation levels (Figure 6A, left and Supplementary Figure S4B). However, we unexpectedly found that histones within the FVII promoter were deacetylated in response to CoCl2 treatment (Figure 6A, middle and Supplementary Figure S4B). ChIP analysis also showed that the association level of histone H3 within the FVII promoter was unchanged following CoCl2 stimuli (Figure 6A, right), further verifying that decreased acetylation levels did not result from histone eviction. We also confirmed that histones were deacetylated within the FVII promoter under CoCl2 treatment in OVISE cells (Supplementary Figure S4C), and under real hypoxic conditions in OVSAYO cells (Figure 6A, middle).
Figure 6.
Hypoxic FVII activation associates with deacetylation. (A) Effect of CoCl2 treatment or hypoxia on histone acetylation at VEGF-HRE and FVII loci in OVSAYO cells. ChIP analysis for acetylated histones (AcH3 and AcH4) and histone H3 was performed by real-time PCR. Data are the mean (n = 3) ± SD. *P < 0.005, **P < 0.08, compared with normoxia control. (B) p300 binding was analyzed by quantitative ChIP analysis for FVII and VEGF-HRE loci in OVSAYO cells treated with CoCl2 for 4 h. (C) Western blotting of p300 expression in OVSAYO cells transfected with NS- or p300-siRNA. (D) Effect of p300 on FVII and VEGF expressions in OVSAYO cells cultured under hypoxia. After p300 knockdown, cells were cultured under normoxia or 1% O2 for 16 h. Real-time RT-PCR analysis was then performed. Data are the mean (n = 3) ± SD. *P < 0.05. (E) Effect of p300 over-expression on activation of the FVII promoter and VEGF-HRE by HIFs. OVSAYO cells were transfected with a FVII-promoter construct, HIFs and p300, and examined by luciferase activity. Inset: western blotting analysis of p300 expression 24 h post-transfection. Relative activities are shown as a percentage of empty vector expression. Data were normalized to Renilla luciferase activity. Data are the mean (n = 3) ± SD. *P < 0.005, **P < 0.05.
Steady-state histone acetylation across chromatins is maintained by the dynamic equilibrium between HATs and HDACs (2,16). Although we observed p300 binding to the FVII promoter in OVSAYO cells, the association level was not altered after CoCl2 treatment (Figure 6B). Conversely, as expected, p300 binding to VEGF-HRE was elevated by CoCl2 treatment (Figure 6B). Since hypoxic FVII activation is associated with histone deacetylation, we tested the role of p300 on this transcriptional activation. Using RNAi, p300 was first suppressed (Figure 6C), before the cells were cultured under 1% O2 for 16 h. Using real-time RT-PCR, we observed an increase in the fVII transcript level with p300-knockdown, whereas VEGF transcript levels were diminished (Figure 6D). Furthermore, ectopic expression of p300 suppressed FVII promoter activation when HIF2α alone or both HIFs were expressed (Figure 6E). In contrast, there was a relatively low increase in VEGF-HRE activation by HIF1 with ectopic p300 expression, similar to the level observed by HIF2 expression (Figure 6E). These results demonstrate that deacetylation promotes FVII activation by HIFs.
HDAC4 is a co-activator of FVII induction
As HRE-dependent mechanisms recruit HAT to activate transcription, we surmised that HDACs may co-activate FVII under hypoxic conditions. Thus, we first examined recruitment of the ubiquitously expressed HDAC1 and HDAC2 to the FVII promoter region in response to CoCl2 treatment. ChIP analysis revealed no change in the association levels of HDAC1 and HDAC2 after CoCl2 exposure (Supplementary Figure S5A). A recent report found that SIRT1 deacetylated HIF2α (12); therefore, we tested whether class III HDAC(s) contributed to FVII induction. Whilst ChIP analysis revealed a weak increase in SIRT1 levels (Supplementary Figure S5B) and some binding to the FVII promoter in OVSAYO cells, this was not enhanced by CoCl2 stimulation (Supplementary Figure S5C). In addition, although SIRT1 associated with HRE of the VEGF gene, the binding was abrogated with CoCl2 (Supplementary Figure S5C). Similarly, the class III HDAC activator, resveratrol (12), did not promote FVII activation under CoCl2 treatment (Supplementary Figure S5D).
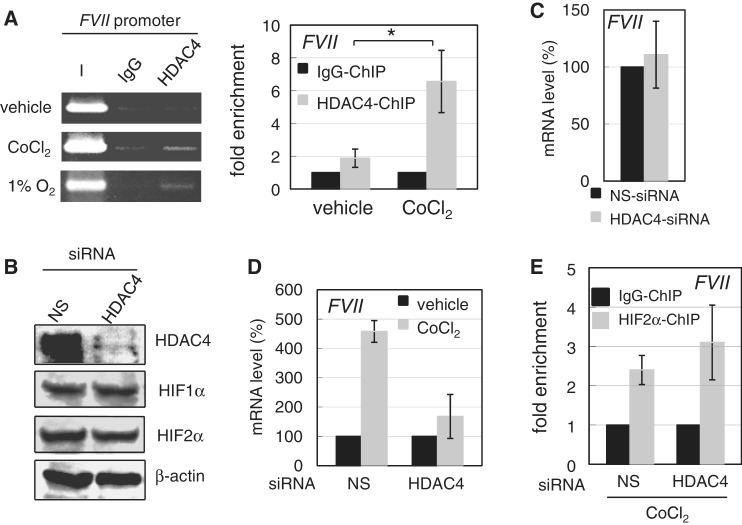
We next examined whether class II HDAC(s) (17) affect FVII activation. Western blotting showed that the class II HDAC, HDAC4 (Figure 7B) rather than HDAC5 (Supplementary Figure S5E), was predominantly expressed in OVSAYO cells. ChIP analysis showed the recruitment of HDAC4 to the FVII promoter in response to CoCl2 treatment or hypoxia (Figure 7A). Real-time RT-PCR analysis revealed that HDAC4-knockdown does not affect basal fVII transcript levels in OVSAYO cells (Figure 7C); however, we observed a significant reduction in its induction with CoCl2 treatment (Figure 7D) without influencing HIF expression levels (Figure 7B) or HIF2α binding to FVII promoter (Figure 7E). In addition, immunoprecipitation analysis revealed an association between HDAC4 and endogenous Sp1–HIF2α complex under hypoxia (Supplementary Figure S5F).
Figure 7.
HDAC4 is critical for FVII activation under hypoxia and its mimetic conditions. (A) ChIP analysis of HDAC4 binding was performed for cells cultured with vehicle, 500 µM CoCl2 for 4 h, or under 1% O2 for 16 h. ChIP was also performed by real-time PCR for cells cultured with vehicle or 500 µM CoCl2 for 4 h. Data are the mean (n = 3) ± SD. *P < 0.05. (B) Western blotting of HDAC4 expression in OVSAYO cells transfected with NS- or HDAC4-siRNA. Effect of HDAC4 knockdown the on expression of HIFs induced by CoCl2 treatment (500 µM for 4 h) is also shown. (C) Effect of HDAC4 on FVII expression in cells cultured under normoxia. After HDAC4 knockdown, real-time RT-PCR was performed. Data are the mean (n = 3) ± SD. (D) Effect of HDAC4 on FVII expression in OVSAYO cells cultured with CoCl2. After HDAC4 knockdown, cells were cultured with 500 µM CoCl2 for 4 h and analyzed by real-time RT-PCR. Data are the mean (n = 3) ± SD. (E) Effect of HDAC4 knockdown on binding of HIF2α to the FVII promoter in OVSAYO cells cultured in 500 µM CoCl2 for 4 h. ChIP analysis for HIF2α binding was performed for the FVII promoter. Data were estimated by qPCR. Data are the mean (n = 3) ± SD.
We also tested whether HDAC3 binds to the FVII promoter in response to CoCl2 treatment. Recently, it was shown that HDAC4 recruits and activates the transcription factor, FOXO, via deacetylation by HDAC3 (18). Although HDAC3 is expressed in OVSAYO cells (Supplementary Figure S5B), we only observed weak binding of HDAC3 to the FVII promoter region, which was not enhanced with CoCl2 treatment (Supplementary Figure S5G). In comparison, HDAC3 showed a strong interaction with HRE of the VEGF gene in OVSAYO cells (Supplementary Figure S5G).
Serum deprivation but not glucose deprivation synergistically enhances FVII activation under hypoxia
Cancer cells must adapt to environmental stress conditions, particularly in cases where inefficient vascularization within the tissue leads to O2 and nutrient deprivation (8). Thus, we first tested how glucose deprivation influences FVII-gene activation under hypoxia. Real-time RT-PCR analysis revealed that although glucose deprivation weakly increased fVII mRNA levels, the transcript levels were not further enhanced when cells were simultaneously exposed to 1% O2 (Supplementary Figure S6A). These results are consistent with data showing decreased HIF expression in cancer cells under glucose-free conditions (19) and the weak expression of HIF observed in OVSAYO cells under glucose-deprived conditions (Supplementary Figure S7B).
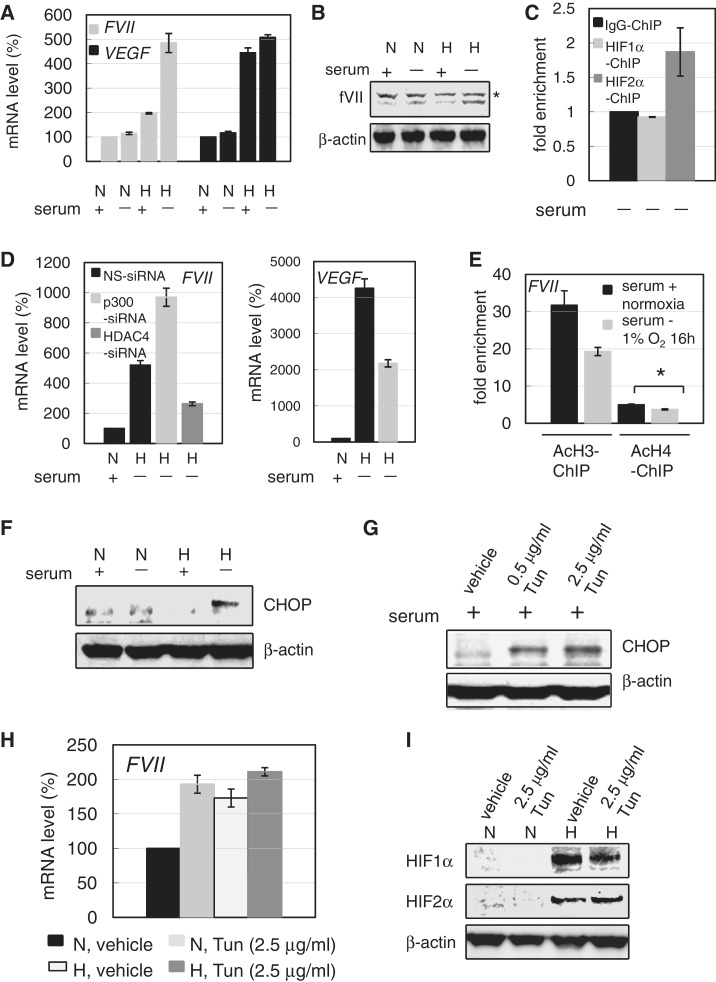
Serum starvation has been reported to induce a cellular response to hypoxia through the hypoxia-inducible factor-independent pathway (20). Hypoxia and serum deprivation are known to be components of ischemia (21,22), and we have previously demonstrated that CCC cells efficiently shed TF-fVIIa-containing microvesicles under both hypoxic and serum-free conditions (6). Since hypoxic cancer tissues should also be devoid of serum factors other than O2 and nutrients, we next examined whether serum starvation affects FVII activation. Cells were cultured with or without fetal calf serum (hereafter serum) under normoxia or 1% O2. Real-time RT-PCR analysis revealed a slight increase in fVII transcript levels in serum-deprived cells cultured under normoxia (Figure 8A); this is possibly owing to a weak induction of HIFs (Supplementary Figure S6B). However, fVII was synergistically increased at both the mRNA and protein levels when cells were exposed to both hypoxic and serum-free conditions (Figure 8A and B). In contrast, the VEGF mRNA levels were not significantly influenced (Figure 8A). ChIP analysis showed that HIF2α predominantly binds to the FVII promoter (Figure 8C), similar to that observed in serum-plus conditions (Figure 1B); this eliminated the possibility that an altered binding pattern of HIFs caused this synergistic activation. Furthermore, the association of Egr-1 with the FVII promoter under normoxia expression was unchanged by exposing the cells to hypoxic and serum-free conditions (Supplementary Figure S6C); Egr-1 is a transcription factor responsible for hypoxic activation of tissue factor gene by substituting Sp1 binding (23). Knockdown of p300 and HDAC4 increased and decreased hypoxic fVII mRNA induction, respectively, under serum-free conditions (Figure 8D). Furthermore, the acetylation level of histones within the FVII promoter was diminished under both hypoxic and serum-free conditions (Figure 8E), as in the case of CoCl2 treatment, demonstrating a synergistic activation associated with deacetylation.
Figure 8.
HDAC4 but not UPR is critical for synergistic FVII activation induced under both hypoxia and serum-free conditions. (A) Effect of serum deprivation on FVII and VEGF expressions in OVSAYO cells cultured under normoxia (N) or hypoxia (H: 1% O2 for 16 h). Results were measured by real-time RT-PCR analysis. Data are the mean (n = 3) ± SD. (B) Effect of serum deprivation on fVII expressions in OVSAYO cells cultured under normoxia or hypoxia as in (A). Asterisk indicates suspected non-specific background. (C) ChIP of HIFs binding to the FVII promoter in OVSAYO cells cultured under both hypoxia and serum-free conditions, performed using real-time PCR. Data are the mean (n = 3) ± SD. (D) Effect of p300 or HDAC4 on FVII and VEGF expression in OVSAYO cells cultured under both hypoxia and serum-free conditions. After 30 h of siRNA transfection, cells were further cultured for 16 h, as indicated, and analyzed by real-time RT-PCR. Data are the mean (n = 3) ± SD. (E) Decreased histone acetylation at the FVII promoter in OVSAYO cells cultured under both hypoxia and serum-free conditions. ChIP analysis was performed by real-time PCR. Data are the mean (n = 3) ± SD. *P < 0.05. (F) CHOP induction in OVSAYO cells. Cells were cultured under normoxia (N) or hypoxia (H; 1% O2 for 16 h) with or without serum for 16 h, and analyzed by western blotting. (G) Western blotting of CHOP induction in OVSAYO cells cultured with tunicamycin (Tun) in serum-plus medium for 16 h. (H) Effect of Tun on the expression of FVII in OVSAYO cells cultured under hypoxia in serum-plus medium via real-time RT-PCR. Data are the mean (n = 3) ± SD. (I) Effect of Tun on the expression of HIFs in OVSAYO cells cultured under hypoxia.
UPR signaling is not responsible for the synergistic transcriptional activation
Other than mechanisms mediated by HIFs, endoplasmic reticulum (ER) stress is a common cellular stress induced by stimuli, such as O2 and nutrient deprivation (24,25). Cells are able to adapt to such environments via unfolded protein response (UPR), which is mediated by the induction of various stress response proteins, such as CHOP, and can co-operate with HIF signaling. We hypothesized that UPR contributes to the synergistic FVII activation. We first examined whether cells cultured under our experimental conditions undergo ER stress. Western blotting showed that ER stress via CHOP induction only occurred when OVSAYO cells were exposed to both hypoxic and serum-free conditions (Figure 8F). We next tested whether UPR causes synergistic FVII induction. OVSAYO cells were cultured in the presence of an ER stress-inducing agent, tunicamycin (Tun) (26), in the presence of serum. The results confirmed that CHOP is induced by Tun treatment (Figure 8G). We next examined the effect of Tun on FVII expression under hypoxia. Real-time RT-PCR analysis revealed that, although Tun treatment elevated FVII transcript level under normoxia, it was not further up-regulated by hypoxia (Figure 8H). In addition, HIF levels induced under 1% O2 were largely unchanged by simultaneous Tun exposure (Figure 8I); these results suggest that UPR is not involved in the synergistic activation. This result is consistent with data showing that glucose deprivation under normoxia weakly enhanced FVII activation (Supplementary Figure S6A) with induction of CHOP (Supplementary Figure S7A), as UPR is known to be induced in cancer cells cultured under glucose-free condition (26).
DISCUSSION
Hypoxic activation of the HRE-dependent CAIX (27), RORA (28), and G3PDH (29) by HIF1α is modulated by Sp1 binding near the HRE. In addition, HIFs have been shown to interfere with the formation of the c-Myc/Sp1 and c-Myc/Max complexes that regulate gene expressions necessary for cell cycle (30) and mismatch repair (31) controls. Thus, Sp1 is generally considered to be an important regulator of gene expression under hypoxic conditions. Here, we showed a novel transcriptional activation of FVII mediated by HIFs that involves (i) the interaction of HIF2α with Sp1, but not with ARNT; (ii) an association with histone deacetylation; and (iii) a synergistic activation under serum-free conditions. Furthermore, it is plausible that genes potentially regulated by this mechanism may be important in the cellular response to severe hypoxic conditions with a poor supply of serum factor(s). Together, these findings have provided us with some explanation as to why CCC cells cultured under hypoxia/serum starvation can efficiently shed microvesicles enriched with fVIIa activity.
HIFs bind with ARNT through PAS domains at their N-terminus (32), whereas the C-terminus interacts with other transcriptional regulators, such as p300 and SIRT1 (33). In this study, we found that both ends of HIF2α can associate with Sp1; however, further studies are required to clarify the mechanisms of the interaction between Sp1 and the C-terminus of HIF2α. It was reported that cysteine (C)/histidine (H)-rich zinc finger domain of a Sp1/KLF family protein can bind to p300/CBP (34). Given that CTAD domain of HIF1α can associate with a C/H-rich zinc-binding motif (CH1) of p300/CBP (35), it seems likely that zinc-finger domain of Sp1 binds to the C-terminus of HIF2α through interaction with CTAD. However, the binding at the C-terminal region is abrogated by the presence of the middle region that contains the NTAD domain; this suggests that HIF2α has an inhibitory domain that affects the interaction between the C-terminal region of HIF2α and Sp1. It would be interesting to ascertain whether this inhibitory effect participates in the physiological regulation of HIF2α function. In addition, we also observed that the HLH region alone does not associate with Sp1. From these findings, we suggest that full-length HIF2α binds to Sp1 through its PAS domains. This is similar to a previous report showing that PAS domains of ARNT can associate with zinc-finger domain of Sp1 (36). Assuming that HIF2α binds with ARNT through its PAS domains, our data may explain why ARNT is not recruited with HIF2α to the FVII promoter under hypoxia.
Notably, the mechanism described in this study is similar to that of constitutive FVII expression under normoxia in breast cancer cells (10), where the same Sp1-binding site is critical for activation, and HIFs co-operatively activate transcription. We suggest that HIF1α may indirectly activate the FVII promoter region, as HIF1α does not directly bind to the FVII promoter, yet demonstrates co-operative effects when simultaneously expressed with HIF2α. The present mechanism is distinct from that of ABCC8-gene activation under hypoxia, in which there is a HIF1α-mediated up-regulation of Sp1 (37). This is interesting, as we showed that the expression and binding levels of Sp1 to the FVII promoter under normoxia in OVSAYO cells were not influenced by CoCl2 and hypoxia treatments.
Our finding that FVII activation associates with deacetylation was unexpected, because under normal circumstances, transcriptional activation closely associates with hyperacetylation of histones, not deacetylation. Indeed, previous work shows that basal FVII transcription is dependent on histone acetylation, with enhanced fVII mRNA levels in response to treatment of cancer cells with the HDAC inhibitor, trichostatin (10). Our data, showing that the expressions of p300 and HDAC4 correlate with a decrease and an increase in fVII transcript levels, respectively, suggests that the association between histone deacetylation and HIF2α recruitment may be a critical step for the formation of active chromatin.
Although this study has focused on the acetylation status of histones, it is also possible that deacetylation of HIF2α by HDAC4 may affect its function in the regulation of FVII transcription in CCC cells, since acetylation of lysine residues is critical to the functional regulation of HIF (33). However, it was recently reported in cancer cells that HDAC4 causes deacetylation of HIF1α, but not HIF2α, to enhance its stability (38). Since HIF1α participates in FVII activation in CCC cells, HDAC4 may affect FVII induction via modulation of HIF1α. Furthermore, HDAC4 was recently shown to deacetylate the transcription factor, FOXO, in liver cells to enhance its binding activity to target genes required for gluconeogenesis (18,39). However, it is unlikely that HDAC4 recruitment contributes to the interaction between HIF2α and Sp1 in the FVII promoter region, as we showed that HIF2α binding is not influenced by HDAC4 knockdown. It is also improbable that the synergistic activation of FVII is due to enhanced HIF2α binding, as the ChIP assay revealed that HIF2α binding under hypoxia is not enhanced by serum deprivation.
The tumor microenvironment is generally associated with hypoxia and poor nutrient supply due to inefficient vascularization (26). To inhibit apoptosis, cancer cells respond and adapt to these pressures by activating cellular mechanisms, such as angiogenesis, cell invasion, and signaling. Although the adaptive responses by HIFs are one of the major pathways induced under hypoxia, the co-operation of additional mechanisms and signaling by UPR assist cancer cell adaptation to hypoxia and nutrient deficiency, thereby promoting cell survival. In this study, we tested the hypothesis that cancer cells may sense serum deprivation and up-regulate transcription of those factors required for adaptation to hypoxia. The FVII-gene activation under real hypoxia in the presence of serum is rather moderate compared with that induced by CoCl2 exposure; however, this may be due to the relatively weak expression levels of endogenous HIFs (9). It is also possible that side effects of CoCl2 exposure including generation of reactive oxygen species and DNA damage (40) transmit signals to enhance Sp1–HIF2α-dependent transcription. We found that activation of the FVII gene is synergistically enhanced in a deacetylation-dependent manner under the serum-free conditions, in which ER stress is induced. Given that HIF expression in OVSAYO cells is dramatically reduced under glucose-free conditions and weak under glucose-deprived conditions, it is plausible that the synergistic activation does not occur by glucose deprivation as opposed to serum starvation. Therefore, serum starvation-induced stress may provide an additional environmental cue to which cells adapt. We suggest that HDAC4 may be a stress-responsive deacetylase, such as sirtuins (12), when cells are exposed to hypoxia. We show that UPR is not responsible for this synergistic activation and further identification of the mechanisms responsible for this synergism is currently ongoing in our laboratory.
This study presents a novel transcriptional activation that is mediated by the interplay between HIF2α/Sp1/HDAC4, enabling us to understand one of the mechanisms of thrombosis in CCC patients. Further exploration of the genes and cancer phenotypes regulated by this HRE-independent mechanism will provide a greater understanding of cellular adaptation that manifests in response to environmental stresses, and may potentially lead to the development of new therapeutics for cancers targeting class IIa HDACs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7 and Supplementary Methods.
FUNDING
Grant-in-Aid for the Encouragement of Basic Science and Technology from the Science and Technology Office of the Kanagawa Prefectural Government (to S.K.); Japanese Ministry of Education, Culture, Sports, Science and Technology (to Y.M. and S.K.); National Institutes of Health (HL60742 to W.R.). Funding for open access charge: Japanese Ministry of Education, Culture, Sports, Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furie B, Furie BC. The molecular basis of blood coagulation. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 5.Schaffner F, Ruf W. Tissue factor and PAR2 signaling in the tumor microenvironment. Arterioscler. Thromb. Vasc. Biol. 2009;29:1999–2004. doi: 10.1161/ATVBAHA.108.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokota N, Koizume S, Miyagi E, Hirahara F, Nakamura Y, Kikuchi K, Ruf W, Sakuma Y, Tsuchiya E, Miyagi Y. Self-production of tissue factor-coagulation factor VII complex by ovarian cancer cells. Br. J. Cancer. 2009;101:2023–2029. doi: 10.1038/sj.bjc.6605406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koizume S, Jin MS, Miyagi E, Hirahara F, Nakamura Y, Piao JH, Asai A, Yoshida A, Tsuchiya E, Ruf W, et al. Activation of cancer cell migration and invasion by ectopic synthesis of coagulation factor VII. Cancer Res. 2006;66:9453–9460. doi: 10.1158/0008-5472.CAN-06-1803. [DOI] [PubMed] [Google Scholar]
- 8.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 9.Koizume S, Yokota N, Miyagi E, Hirahara F, Tsuchiya E, Miyagi Y. Heterogeneity in binding and gene-expression regulation by HIF-2α. Biochem. Biophys. Res. Commun. 2008;371:251–255. doi: 10.1016/j.bbrc.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 10.Koizume S, Yokota N, Miyagi E, Hirahara F, Nakamura Y, Sakuma Y, Yoshida A, Kameda Y, Tsuchiya E, Ruf W, et al. Hepatocyte nuclear factor-4-independent synthesis of coagulation factor VII in breast cancer cells and its inhibition by targeting selective histone acetyltransferases. Mol. Cancer Res. 2009;7:1928–1936. doi: 10.1158/1541-7786.MCR-09-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracken CP, Fedele AO, Linke S, Balrak W, Lisy K, Whitelaw ML, Peet DJ. Cell-specific regulation of hypoxia-inducible factor (HIF)-1α and HIF-2α stabilization and transactivation in a graded oxygen environment. J. Biol. Chem. 2006;281:22575–22585. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- 12.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2α signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 13.Pollak ES, Hung H-L, Godin W, Overton GC, High KA. Functional characterization of the human factor VII 5′-flanking region. J. Biol. Chem. 1996;271:1738–1747. doi: 10.1074/jbc.271.3.1738. [DOI] [PubMed] [Google Scholar]
- 14.Pagés G, Pouyssegur J. Transcriptional regulation of the Vascular Endothelial Growth Factor gene-a concert of activating factors. Cardiovasc. Res. 2005;65:564–573. doi: 10.1016/j.cardiores.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl Acad. Sci. USA. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Katan-Khaykovich Y, Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–752. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol. Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 18.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuyama H, Endo H, Akashika T, Kato K, Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res. 2010;70:10213–10223. doi: 10.1158/0008-5472.CAN-10-2720. [DOI] [PubMed] [Google Scholar]
- 20.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 21.Chao W, Shen Y, Li L, Rosenzweig A. Importance of FADD signaling in serum deprivation- and hypoxia-induced cardiomyocyte apoptosis. J. Biol. Chem. 2002;277:31639–31645. doi: 10.1074/jbc.M204104200. [DOI] [PubMed] [Google Scholar]
- 22.Zhu W, Chen J, Cong X, Hu S, Chen X. Hypoxia and serum deprivation-induced apoptosis in mesenchymal stem cells. Stem Cells. 2006;24:416–425. doi: 10.1634/stemcells.2005-0121. [DOI] [PubMed] [Google Scholar]
- 23.Mackman N. Regulation of the tissue factor gene. FASEB J. 1995;9:883–889. doi: 10.1096/fasebj.9.10.7615158. [DOI] [PubMed] [Google Scholar]
- 24.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1α interactome. Mol. Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 26.Spiotto MT, Banh A, Papandreou I, Cao H, Galvez MG, Gurtner GC, Denko NC, Le QT, Koong AC. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 2010;70:78–88. doi: 10.1158/0008-5472.CAN-09-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaluz S, Kaluzová M, Stanbridge EJ. Expression of the hypoxia marker carbonic anhydrase IX is critically dependent on Sp1 activity. Identification of a novel type of hypoxia-responsive enhancer. Cancer Res. 2003;63:917–922. [PubMed] [Google Scholar]
- 28.Miki N, Ikuta M, Matsui T. Hypoxia-induced activation of the retinoic acid receptor-related orphan receptor α4 gene by an interaction between hypoxia-inducible factor-1 and Sp1. J. Biol. Chem. 2004;279:15025–15031. doi: 10.1074/jbc.M313186200. [DOI] [PubMed] [Google Scholar]
- 29.Higashimura Y, Nakajima Y, Yamaji R, Harada N, Shibasaki F, Nakano Y, Inui H. Up-regulation of glyceraldehyde-3-phosphate dehydrogenase gene expression by HIF-1 activity depending on Sp1 in hypoxic breast cancer cells. Arch. Biochem. Biophys. 2011;509:1–8. doi: 10.1016/j.abb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1α induces genetic instability by transcriptionally downregulating MutSα expression. Mol. Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Patel SA, Simon MC. Biology of hypoxia-inducible factor-2α in development and disease. Cell Death Differ. 2008;15:628–634. doi: 10.1038/cdd.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol. Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 34.Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G. Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J. Biol. Chem. 2002;277:7029–7036. doi: 10.1074/jbc.M108826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruas JL, Berchner-Pfannschmidt U, Malik S, Gradin K, Fandrey J, Roeder RG, Pereira T, Poellinger L. Complex regulation of the transactivation function of hypoxia-inducible factor-1α by direct interaction with two distinct domains of the CREB-binding protein/p300. J. Biol. Chem. 2010;285:2601–2609. doi: 10.1074/jbc.M109.021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi A, Sogawa K, Fujii-Kuriyama Y. Cooperative interaction between AhR/ARNT and Sp1 for the drug-inducible expression of CYP1A1 gene. J. Biol. Chem. 1996;271:12310–12316. doi: 10.1074/jbc.271.21.12310. [DOI] [PubMed] [Google Scholar]
- 37.Woo SK, Kwon MS, Geng Z, Chen Z, Ivanov A, Bhatta S, Gerzanich V, Simad JM. Sequential activation of hypoxia-inducible factor 1 and specificity protein 1 is required for hypoxia-induced transcriptional stimulation of Abcc8. J. Cereb. Blood Flow Metab. 2012;32:525–536. doi: 10.1038/jcbfm.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, Qian DZ. HDAC4 protein regulates HIF1α protein lysine acetylation and cancer cell response to hypoxia. J. Biol. Chem. 2011;286:38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR, Fischer WH, Thomas JB, Montminy M. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Hazra TK, Mitra S, Lee HM, Englander EW. Mitochondrial DNA damage and a hypoxic response are induced by CoCl2 in rat neuronal PC12 cells. Nucleic Acids Res. 2000;28:2135–2140. doi: 10.1093/nar/28.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.