Abstract
Plasmodium falciparum origin recognition complex 1 (ORC1) protein has been implicated in DNA replication and silencing var gene family. However, the mechanism and the domain structure of ORC1 related to the regulation of var gene family are unknown. Here we show that the unique N-terminus of PfORC1 (PfORC1N1–238) is targeted to the nuclear periphery in vivo and this region binds to the telomeric DNA in vitro due to the presence of a leucine heptad repeats. Like PfORC1N1–238, endogenous full length ORC1, was found to be associated with sub telomeric repeat regions and promoters of various var genes. Additionally, binding and propagation of ORC1 to telomeric and subtelomeric regions was severely compromised in PfSir2 deficient parasites suggesting the dependence of endogenous ORC1 on Sir2 for var gene regulation. This feature is not previously described for Plasmodium ORC1 and contrary to yeast Saccharomyces cerevisiae where ORC function as a landing pad for Sir proteins. Interestingly, the overexpression of ORC1N1–238 compromises the binding of Sir2 at the subtelomeric loci and var gene promoters consistent with de-repression of some var genes. These results establish role of the N-terminus of PfORC1 in heterochromatin formation and regulation of var gene expression in co-ordination with Sir2 in P. falciparum.
INTRODUCTION
Origin recognition complex (ORC), a six-protein complex is essential for DNA replication initiation from yeast to mammals. ORC, a member of the pre-replicative complex (pre-RC) serves as a landing pad for the subsequent loading of other members of pre-RC like Cdc6 and minichromosome maintenance protein family (1). Apart from DNA replication, ORC, as a complex or its individual subunits has been implicated in various ranges of cellular functions (2,3). Transcriptional silencing in the yeast Saccharomyces cerevisiae HM mating type loci is dependent on the binding of ORC protein to the HM silencer region containing autonomously replicating sequence (4,5). Both ORC1 and ORC2 have been shown to interact directly with heterochromatin protein HP1 that is important for heterochromatin organization and transcriptional regulation of heterochromatin genes in eukaryotes ranging from Drosophila to Xenopus and mammals (6–8). ORC6, another member coordinates chromosomal DNA replication with segregation suggesting a possible role during cytokinesis (9). These observations suggest that although ORC is implicated mainly in DNA replication, it takes part in various other cellular functions.
The largest subunit of ORC, ORC1 has a regulatory role in DNA replication and other functions since it comes on and off the chromatin during the cell cycle. It contains a conserved C-terminal domain and an N-terminal domain that exhibits poor homology among species. The C-terminal region contains an AAA+ motif that binds and hydrolyses ATP, a hallmark of origin function (10–13). The extreme C-terminus contains a helix-turn-helix motif that may be responsible for origin binding (14). The N-terminal region of ORC1 shows a diverse function. In yeast, this region interacts with Sir1 which might be essential for the recruitment of other SIR proteins (Sir3 and Sir4) leading to the establishment of the silenced HM mating type loci (4,5). The N-terminal region of human ORC1 contains a BAH (bromo adjacent homology domain, also present in other ORC1) domain that may facilitate association of ORC with chromatin (15,16). Although, domain mapping of ORC1 responsible for interaction with HP1 leading to the regulation of heterochromatin gene transcription is not reported, it is highly possible that this interaction is mediated through the N-terminus of ORC1 since the exclusive function of the C-terminus is related to DNA replication.
During its replication in erythrocytes, the human malaria parasite Plasmodium falciparum proceeds through several developmental stages that can be classified as ring stage (preparatory stage before DNA replication), trophozoite stage (DNA replication) and the schizont stage (chromosomal segregation and nuclear division). Unlike other eukaryotic cells, multiple rounds of DNA replication takes place in the parasites before cytokinesis at the end of schizogony. Plasmodium falciparum contains homologs for ORC1, ORC5 and a putative homolog for ORC2. Interestingly, no clear homologs for other ORC subunits can be identified by BLAST analysis or are annotated in the Plasmodium Database (PlasmoDB). Therefore, Plasmodium ORC may have limited subunits or there are functional homologs of these subunits that are yet to be identified.
Both PfORC1 and PfORC5 form replication foci co-localizing with proliferating cell nuclear antigen (PCNA) during replicating trophozoite stage. This might occur through a direct interaction between PfORC1 and PCNA mediated through the presence of a PCNA-interacting protein (PIP) motif in ORC1 at the C-terminus (17,18). This region also contains the conserved AAA+ motif and the helix-turn-helix domain and might therefore mediate DNA replication in the parasite (17).
PfORC1 contains a unique N-terminal extension that shows no homology with any other ORC1 proteins. Except a leucine heptad repeat region (present in PfORC1 and other related Plasmodium species) and two putative CDK phosphorylation sites (present at the extreme N-terminus), no other putative functional motif can be detected. Therefore, the question arises regarding the function of this extension.
Chromosome ends of Plasmodium contain distinct structural domains that include the telomere and the polymorphic subtelomeric region. The telomeres are composed of degenerate G-rich heptamer repeats 5′-GGGTT(T/C)A-3′. The telomere and subtelomeric regions form 20–40 kb non-coding regions with lower A + T content (∼70%) compared to the internal chromosome regions (∼82%) (19). The subtelomeric repeat sequences or telomere associated repetitive elements (TAREs) vary among different species of Plasmodium.
In P. falciparum, the subtelomeric region is composed of six different TAREs (TARE1-6), their length may vary among different strains. TARE-1 (∼0.9–1.9 kb) is present nearest to the telomere with complex tandem repeats (20). The length of TARE-2 is 1.6 kb. It contains a 135 bp degenerate sequence that is repeated 12 times. These repeat sequences are interspersed by two distinct 21 bp sequences. TARE-3 contains three to four consecutive 0.7 kb elements (21,22). The length of TARE-4 ranges from 0.7 to 2 kb. It contains highly degenerate and short repeats which are interspersed by non-repetitive 230 bp sequences. TARE-5 (1.4 to 2 kb) contains moderately degenerate tandem repeats with 12 bp (5′ACTAACA(T/A)(C/G)A(T/C)(T/C)). TARE-6 is also known as Rep20 elements. The length of TARE6 varies from 8.4 to 21 kb with 21 bp degenerate sequence (23).
PfEMP1 (P. falciparum erythrocyte membrane protein 1), a major parasite virulence factor is encoded by sixty-member var gene family. The majority of these var genes is located adjacent to the subtelomeric region. The var genes are classified by promoter sequence in to five different types (upsA, upsB, upsC, upsD and upsE) based on their chromosomal locations and orientation of transcription. upsC var genes are found in central chromosomal regions. However, all but one upsB var genes are located in sub-telomeric regions. The orientation of transcription of subtelomeric upsA, upsD and upsE var genes is opposite to upsB var genes (24,25). It is believed that telomeres and TAREs play a major role in switching the expression of var gene family leading to the immune evasion and severity of the disease (24–29).
Recently, PfORC1 has been shown to bind to telomeric and subtelomeric repeat regions along with PfSir2, a histone deacetylase involved in the silencing of some virulence genes present at the chromosome end loci (30). There is an increase in the protein level of ORC1 and Sir2 as the parasites differentiate from ring to trophozoite and schizont stage. Both ORC1 and Sir2 are localized to the nuclear periphery containing telomeric clusters during the ring or early trophozoite stage. However, both proteins reorganize themselves at the onset of DNA replication and coincide with the partial dissociation of the telomeric clusters from the nuclear periphery leading to the spreading of the proteins in the nucleus and cytoplasm (30). These results suggest the possible role of PfORC1 in var gene silencing in P. falciparum along with PfSir2. Nevertheless, the domain structure of PfORC1 with the possible involvement in the regulation of var gene expression is still elusive.
Other than PfSir2, PfHP1 is also involved in the regulation of var gene expression by binding to the H3K9me3 (trimethylated H3K9) marks at the silenced loci (31,32). HP1 may favor the formation of heterochromatin at the perinuclear chromosome end clusters. Another Sir2 paralog (Sir2B) shows silencing of var gene repertoire that are different and mutually exclusive from the genes controlled by the PfSir2 as described earlier (33). In fact, Sir2B is involved in the regulation of var genes controlled by the upsB promoter whereas PfSir2 controls var genes under upsA/E promoter (33,34).
There are numbers of proteins involved in mutually exclusive transcription of var gene family. However, it is not known how these proteins are recruited at the telomeres, subtelomeric repeat regions and the ups promoters. Is there any pattern of sequential loading and dependence of loading of one protein with the others? Investigation of these processes will provide additional insights into the underlying molecular mechanisms of var gene regulation and pathogenicity of the malaria parasite.
In order to find out whether the N-terminus of ORC1 has a unique role in targeting this protein to the nuclear periphery containing telomeric clusters and its possible involvement in var gene regulation, we generated several transgenic parasites expressing GFP-fusion proteins containing wild-type and deletion mutants of the N-terminus of PfORC1. We find that the N-terminus alone (PfORC1N1–238) can target this protein to the nuclear periphery and occupies the identical positions in the telomeric, subtelomeric regions and within var gene promoters as shown by the full length endogenous ORC1. PfORC1N1–238 binds directly to the telomeric DNA in vitro, an activity that is dependent on its oligomeric property due to the presence of unique leucine heptad repeats. Further, we have used the Sir2 knock out parasite line to investigate the sequential loading of the telomeric and subtelomeric region associated proteins involved in var gene silencing. We find that the binding of ORC1 but not HP1 to the subtelomeric repeat regions in vivo is dependent on loading of Sir2 on these regions. Moreover, the overexpression of ORC1N1–238 compromises the binding of Sir2 at the telomeric loci and var gene promoters consistent with de-repression of some var genes. These results establish the importance of the unique N-terminal region of PfORC1 in var gene regulation and show a unique pattern of loading the telomeric and subtelomeric repeat region-associated proteins that may be central to the var gene regulation important for pathogenicity.
MATERIALS AND METHODS
Parasite culture
Plasmodium falciparum 3D7 strain was cultured as described earlier (35). Synchronization of the culture was achieved by using 5% sorbitol treatment of the parasites (18).
Bacterial strain
Escherichia coli strain DH10β was used for cloning purposes. BL21 Codon plus (RIG) (17) cells were used for the expression of the recombinant proteins.
DNA manipulation
Cloning for the expression in bacteria
Different domains of N-terminal region of ORC1 (ORC1N1–238, ORC1N1–182, ORC1N25–238, ORC1N1–53; Figure 1A), C-terminus of ORC1 (ORC1C; residues 689–1022 of full length ORC1) and coding regions of PfHP1, PfSir2 and PfPCNA were amplified by PCR using P. falciparum 3D7 genomic DNA as template and cloned into pET28a or pMALc2X vector. The details of restriction sites, primer sequences and site directed mutagenesis primers (for making point mutation in the leucine heptad repeats of ORC1N1–238; ORC1NL11–238) are listed in Supplementary Table S1.
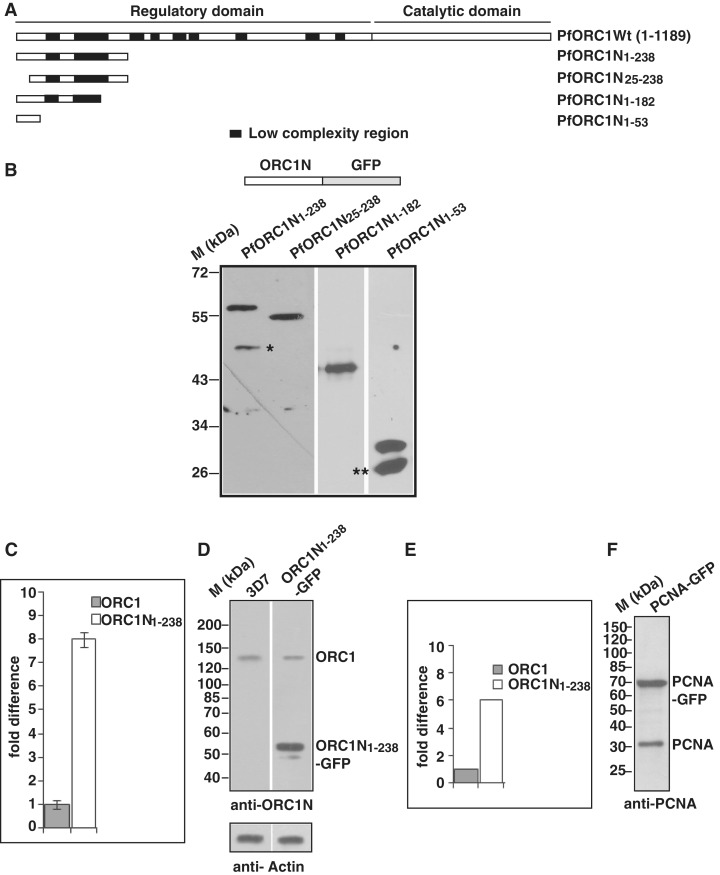
Figure 1.
Expression of the N-terminus of PfORC1 as GFP-fusion protein. (A) Schematic diagram of full length PfORC1. The putative regulatory domain (1–783 residues) and the catalytic domain (784–1189) containing the ATP binding and hydrolysis domain are shown. The black blocks indicate the low complexity regions with asparagine and lysine repeat rich regions. (B) The upper panel shows the schematic diagram of different forms of PfORC1N as GFP-fusion protein. The bottom panel shows the western-blot analysis of parasite lysate obtained from different parasite lines as indicated above using anti-GFP antibodies. Asterisk and double asterisk indicate likely degradation products. (C) Comparison of expression pattern of endogenous ORC1 and ORC1N1–238–GFP transgene in ORC1N1–238–GFP expressing parasites at the transcript level by real time PCR analysis. gapdh was used as control. ORC1N1–238–GFP shows several fold higher expression than endogenous ORC1 (D) Comparison of expression of endogenous PfORC1 and PfORC1N1–238–GFP at the protein level. Equal amount of lysate (∼100 µg) obtained from 3D7 wild-type or PfORC1N1–238–GFP expressing trophozoite stage parasites were resolved in SDS–PAGE followed by western-blot analysis by polyclonal antibodies against N-terminus of PfORC1. Top panel shows the expression of the respective proteins under the same experimental conditions. The control western blot using PfActin antibodies shows equal loading of proteins in each lane (bottom panel). (E) The intensity of the bands corresponding to endogenous ORC1 and PfORC1N1–238-GFP were quantified densitometrically and plotted accordingly. (F) Approximately 100 µg lysate obtained from PfPCNA-GFP expressing parasites were resolved in SDS–PAGE followed by western-blot analysis using polyclonal antibodies against PfPCNA. The results indicate that PCNA1-GFP is over-expressed compared to endogenous PCNA.
Generation of GFP constructs
The DNA fragments corresponding to PfORC1N1–238 (1–714 bp), PfORC1N25–238 (73–714 bp), PfORC1N1–182 (1–546 bp), PfORC1N1–53 (1–159 bp), PfORC1L11–238 (with a substituted leucine 137 residue to alanine) and PCNA were amplified by PCR and cloned into pARL–GFP vector using the KpnI and AvrII restriction sites (36). Fusion gene expression is driven by crt (P. falciparum chloroquine resistance transporter) gene promoter. The resulting recombinant clones were sequenced to exclude unwanted mutations. The primer sequences are listed in Supplementary Table S1.
Transfection
Transfection of the 3D7 parasites using the pARL fusion constructs were performed using the protocol described elsewhere (37). The transfected parasites were selected using the drug WR99210 (Jacobus Pharmaceuticals). Expression was verified by western-blot analysis using transfected parasite lysate in the presence of anti-GFP antibodies and fluorescence microscopy.
Purification of recombinant proteins
Protein purification was performed using the protocol as described elsewhere (38) and in Supplementary Materials and Methods.
Polyclonal antibody production
Raising polyclonal antibodies against PfORC1C (C-terminal 689–1189 residues), PfORC5, PfPCNA and PfSir2 have described elsewhere (18,39). Polyclonal antibodies against PfHP1 and PfORC1N1–238 were raised in mice using purified His6-PfHP1 and His6-PfORC1N1–238, respectively, as antigen essentially following the protocol as previously described (18).
Electrophoretic mobility shift assay
As previously described (17) DNA binding activity of PfORC1N1–238 and other proteins were investigated using 20 µl reaction mixture containing DNA binding buffer [10 mM Tris–Cl (pH 7.5), 100 mM KCl, 5 mM Mgcl2, 2 mM dithiothreitol, 6% glycerol, 50 µg/ml bovine serum albumin]. The respective proteins were incubated in DNA binding buffer containing the appropriate [γ-32P] ATP labeled DNA fragment. Telomere DNA (175 bp) was amplified from a plasmid DNA kindly provided by Dr Rosaura Harnandez-Rivas (19,30) using the primers (Telomere F and R) as listed in Supplementary Table S1. These primer sequences correspond to the vector DNA sequences flanking the cloned telomeric DNA. AT-rich DNA (150 bp) was obtained by PCR amplification of a DNA fragment containing PfGyrA sequence. GC-rich DNA (242 bp) was obtained by PCR amplification of a part of tetr gene from plasmid pBR322. The corresponding primers are listed in Supplementary Table S1. Both telomere DNA and AT-rich DNA was used as probe following radiolabeling in the presence of [γ-32P] ATP. Approximately 2 ng radiolabeled DNA probe was used per reaction. Cold DNA was used as competitors in electrophoretic mobility shift assay (EMSA) experiments.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed as recommended by Upstate Biotechnology ChIP Assay Kit (26225) with some modifications (30) using ring/early trophozoite stage parasites. Briefly, chromatin was cross-linked using 1% formaldehyde for 10 min at 37°C and the parasites were collected after two washes with 1× PBS followed by saponin lysis. Parasites were resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.1) containing protease inhibitor cocktail and incubated on ice for 10 min. Parasites were then sonicated 10 times for 10 s at maximum setting with 1-min interval on ice between each pulse in order to obtain DNA fragments in the range of ∼200 to ∼1000 bp. After 15 min of centrifugation at 10000g, 15 µl of the supernatant was used as input and the remainder was diluted 10-fold in ChIP Dilution Buffer. This diluted fraction was subjected first for pre-clearing with 60 µl of Protein A Agarose beads (SIGMA) followed by immunoprecipitation by incubating overnight using respective antibodies (2 µl rabbit anti-ORC1C, 2 µl rabbit anti-GFP, 5 µl mouse anti-Sir2, 4 µl mouse anti-HP1, 2 µl mouse anti-PCNA1, 2 µl mouse anti-SSB immune sera). The immunoprecipitated DNA was analyzed for semi-quantitative PCR amplification of TAREs and var gene promoters using primers listed in Supplementary Table S1. PCR primers were designed carefully and PCR conditions were optimized so that ∼500–600 bp products are obtained for all the TARE regions except TARE-6 (∼1.2 kb). PCR amplifications were performed for 20–22 cycles to avoid saturation of the products as shown in the results section. The band intensities were quantified using the Image J software.
Immunofluorescence
Localization of various endogenous proteins like ORC1, HP1, Sir2 was performed using immunofluorescence assay with the help of respective antibodies as described in Supplementary Materials and Methods and elsewhere (18).
Combined IFA/FISH
Combined IFA/FISH was performed as described previously (32). Parasites were fixed with 4% formaldehyde and permeabilized with 0.1% Triton X. Parasites were incubated with anti-GFP antibody (Abcam) (1:200) followed by fluorescein-tagged appropriate secondary antibodies (1:200). FISH was carried out using a TARE-6 (rep20) probe as described previously (32).
Glutaraldehyde cross-linking assay
To stabilize the dimer/oligomer forms of ORC1N1–238 that can be distinguished from the monomer form following SDS–polyacrylamide gel electrophoresis (PAGE) analysis, cross-linking experiments were performed as previously published (38). The reaction was carried out in a buffer containing 10 mM Tris–HCl pH 8.0, 100 mM KCl, 4 mM DTT, 4 mM MgCl2 using two different concentrations (0.0004 and 0.0006%) of glutaraldehyde at 25°C for 2 h. The reaction was terminated by the addition of 2× SDS sample buffer and boiling at 95°C for 3 min. The samples were loaded onto a 10% SDS–PAGE gel and subsequently western-blot analysis was performed by using anti-His6 antibodies.
Real-time polymerase chain reaction
Real Time PCR analysis was performed essentially using the protocol as described elsewhere (40). To achieve this, tightly synchronized 8 - to 12-h ring stage parasites (3D7 wild-type, PfSir2 KO and ORC1N1–238–GFP expressing parasites) were subjected to saponin lysis and pellets were recovered subsequently. Parasite pellets were resuspended in TRIzol (Invitrogen) and RNA was extracted using manufacture’s protocol (Invitrogen) including digestion with RNase free DNase I (Fermentas) and preparation of cDNA using SuperScript III Reverse transcriptase (Invitrogen). RT-PCR reactions were performed using cDNA and specific primers (Supplementary Table S1). For the comparative analysis of endogenous ORC1 and ORC1N1–238–GFP transcript levels, same forward primer (PfORC1F589) was used along with reverse primer PfORC1R765 or GFPR51 for the endogenous or transgene, respectively (Supplementary Table S1). The amount of cDNA obtained from different parasite lines was first normalized against the gapdh control. Real-time RT-PCR was performed using an Applied Biosystem equipment and absolute SYBR Green Mix (Applied Biosystem). All samples and controls were performed in triplicate using the following cycle conditions: 95°C, 15 min followed by 40 cycles of 94°C, 30 s; 60°C, 40 s and 68°C, 50 s.
RESULTS
The N-terminal region of PfORC1 is targeted to the nuclear periphery
The N terminal region of P. falciparum ORC1 is unusually long with several low complexity regions containing asparagine and lysine rich repeats as shown in Figure 1A. However, the role of the N-terminus is not known yet.
In order to investigate the putative function of the N-terminus of PfORC1 for its subcellular localization, we generated transgenic parasites expressing GFP-fusion proteins containing wild-type and deletion mutants of the N-terminus of PfORC1 (PfORC1N). The presence of low complexity regions within the N-terminus limited the maximum length to 714 bp (1–238 amino acid residues, ORC1N1–238–GFP, Figure 1A) for the expression of this region as GFP-fusion protein and resulted in three deletion mutants ORC1N25–238–GFP (deletion of 24 amino acid residues from the N-terminus), ORC1N1–182–GFP (1–182 residues) and ORC1N1–53–GFP (1–53 residues) (Figure 1A). Expression of the fusion proteins were verified with western-blot analysis using antibodies against GFP (Figure 1B).
We investigated the expression level of the transgene under the control of the crt promoter and compared it with the endogenous ORC1 both at the transcript level and protein level (Figure 1C and D). First we analyzed the relative abundance of the endogenous ORC1 and ORC1N1–238–GFP transcript level in ORC1N1–238–GFP expressing parasites by quantitative real time PCR analysis. qPCR shows approximately seven to eight times more transcript level for the transgene relative to the endogenous ORC1 (Figure 1C). Second, western-blot analysis of equal amount of lysate obtained from 3D7 wild-type or ORC1N1–238–GFP expressing parasites using polyclonal antibodies against N-terminus of ORC1 shows ∼5- to 6-fold increased expression of the GFP fusion protein compared to endogenous ORC1 (Figure 1D and E). This was further confirmed by an additional transgenic cell line using the same transfection vector but expressing the nuclear antigen PCNA as a GFP fusion protein. PCNA–GFP expressing parasites show distinct nuclear replication foci similar to endogenous PCNA as described earlier (Supplementary Figure S1A) (17,18). Once again, western-blot analysis indicates that the expression level of PCNA–GFP is higher than the endogenous PCNA (Figure 1F).
We investigated the subcellular localization of ORC1N1–238–GFP (1–238) and other deletion mutants by fluorescence microscopy (Figure 2A and B). We find that ORC1N1–238–GFP shows distinct punctate staining at the nuclear periphery during the ring stage of the parasites (panel 1, Figure 2B) and persists in later stages (panel 2, Figure 2B).
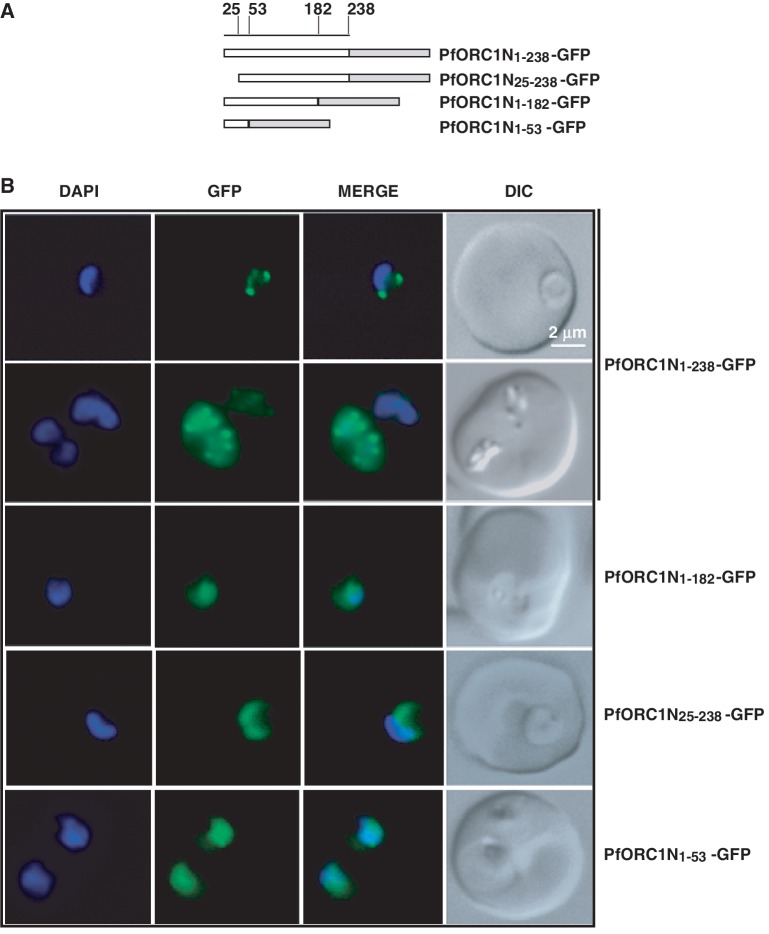
Figure 2.
Fluorescence microscopy of different N-terminal domains as GFP-fusion proteins. (A) Schematic diagrams of different N-terminal domains fused with GFP as indicated in the figure. The numbers on the top indicate the positions of the amino acid residues. (B) Live cell imaging of different parasite lines as indicated on the right. ORC1N1–238–GFP predominantly shows punctate staining at the nuclear periphery either at the ring stage (first row) or at the later stage (second row). None of the other constructs show nuclear punctate staining pattern. ORC1N25–238–GFP shows diffused staining pattern all over the parasite while both ORC1N1–182–GFP and ORC1N1–53–GFP shows enrichment of GFP signal in the nucleus.
In contrast, ORC1N1–182–GFP (1–182), the mutant comprising only the 182 N-terminal amino acids, shows diffused signal enriched within the nucleus but without the punctate pattern suggesting the role of downstream sequences (183–238) for tethering to the nuclear periphery (panel 3, Figure 2B).
Interestingly, the deletion of the first 24 amino acid residues (ORC1N25–238–GFP) also yields diffused staining where majority of the signal is seen in the cytoplasm suggesting a putative role of the extreme N-terminus residues for nuclear targeting (panel 4, Figure 2B). Subsequently, expression of only the first 53 N-terminal amino acids (ORC1N1–53–GFP) appears to be sufficient to direct some protein to the nucleus as shown by diffused staining pattern with enhancement of the fluorescence signal in the nucleus (panel 5, Figure 2B).
The 3D dissection using confocal microscopy further confirms the distinct punctuate staining of PfORC1N1–238 in the nuclear periphery and nuclear diffused staining pattern of ORC1N1–182 (Supplementary Figure S1B).
The N-terminal region of PfORC1 binds to DNA directly with a preference to the telomeric DNA
One possibility for the restricted distribution of PfORC1N1–238 in the periphery of the parasite nucleus could be its direct binding to telomeric repeat regions. This hypothesis is supported by earlier reports that have shown that: (i) chromosomal ends containing telomeric and subtelomeric regions are tethered to the nuclear periphery in P. falciparum (41,42) and (ii) ChIP reveals the loading of endogenous PfORC1 at the telomeric and subtelomeric repeat regions (TAREs) (30).
In order to test the hypothesis, we purified both ORC1N1–238 and ORC1N1–182 as His6 fusion proteins (Figure 3A and B). The recombinant proteins were used for EMSA using telomeric DNA as a radiolabeled probe. We find that ORC1N1–238 binds to DNA directly while ORC1N1–182 fails to do so. Moreover, we detected multiple bands following EMSA experiment that may represent different oligomeric forms of ORC1N1–238 (Figure 3C). These results indicate that the intrinsic DNA binding property of ORC1N1–238 may help it tethering to the nuclear periphery. ORC1N1–182 has lost the DNA-binding property as well as tethering to the nuclear periphery (Figure 2B).
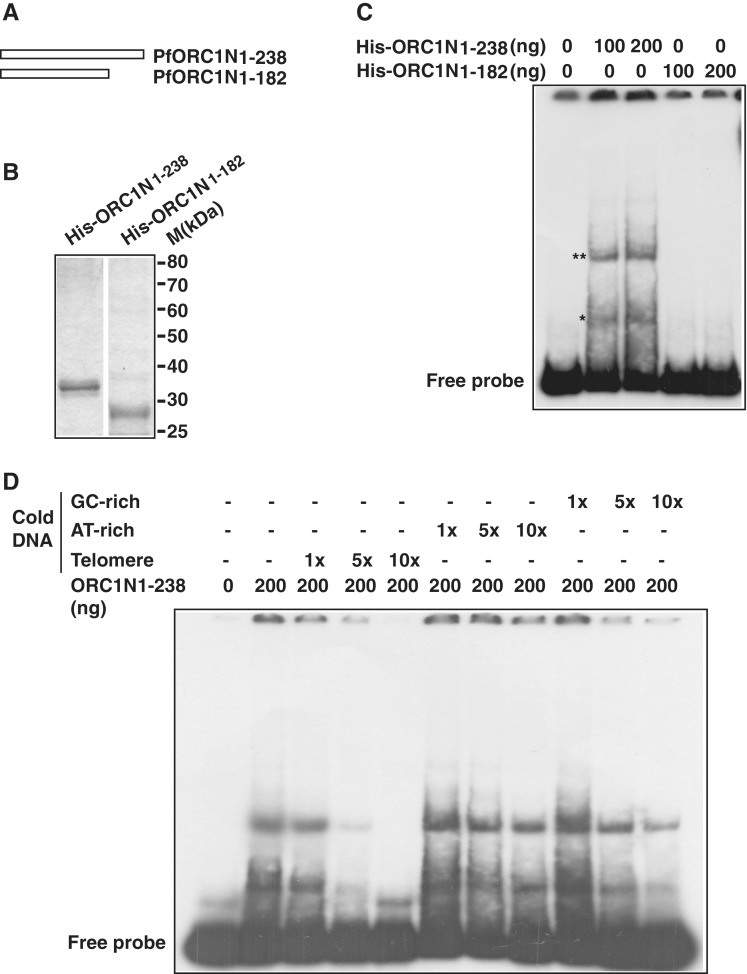
Figure 3.
PfORC1N1–238 binds specifically to telomeric DNA. (A) Schematic diagram of PfORC1N1–238 and PfORC1N1–182. (B) SDS–PAGE analysis of recombinant purified His6-tagged PfORC1N1–238 or PfORC1N1–182 proteins (1 µg each). (C) PfORC1N1–238 but not PfORC1N1–182 binds directly to telomeric DNA as shown by gel retardation assay (asterisk). The position of free probe is indicated. The higher order DNA–protein complexes are shown by ‘double asterisk’. (D) EMSA was performed using radioactive labeled telomeric DNA and purified PfORC1N1–238. Competition was performed using cold telomeric, AT-rich or GC-rich DNA of relatively similar length and varying concentration (1×, 5× and 10×, respectively). The reaction mixtures were separated in PAGE and the gel was dried and autoradiographed subsequently. Telomeric DNA competed most efficiently compared to AT-rich DNA and GC-rich DNA under the similar experimental conditions.
Does ORC1N1–238 show specificity to telomeric DNA? To address this issue, we have performed competitive EMSA with radiolabeled telomeric probe and different types of cold competitor DNA. We find that binding of ORC1N1–238 to radiolabeled telomeric DNA can be efficiently competed using cold telomeric DNA as competitor in a concentration dependent manner. Neither AT-rich nor GC-rich cold DNA can compete efficiently for the telomeric DNA binding activity of ORC1N1–238 suggesting the preferential binding of ORC1N1–238 to telomeric DNA over AT/GC rich DNA (Figure 3D).
ORC1N1–238 forms dimers in solution and a putative leucine heptad repeat at the N-terminus is essential for DNA binding
Leucine heptad repeats often form coil–coil domains that may be responsible for protein–protein and protein–DNA interaction and oligomerization. PfORC1N1–238 contains a perfect leucine heptad repeat (Figure 4A). We were interested to know whether the leucine heptad repeat would be essential for oligomerization and ORC1N1–238 DNA-binding. For this purpose, we changed the first leucine of the leucine heptad repeat into alanine (ORC1NL11–238) (Figure 4A). His-tagged wild-type and the mutant proteins were purified under the same experimental conditions (Figure 4B).
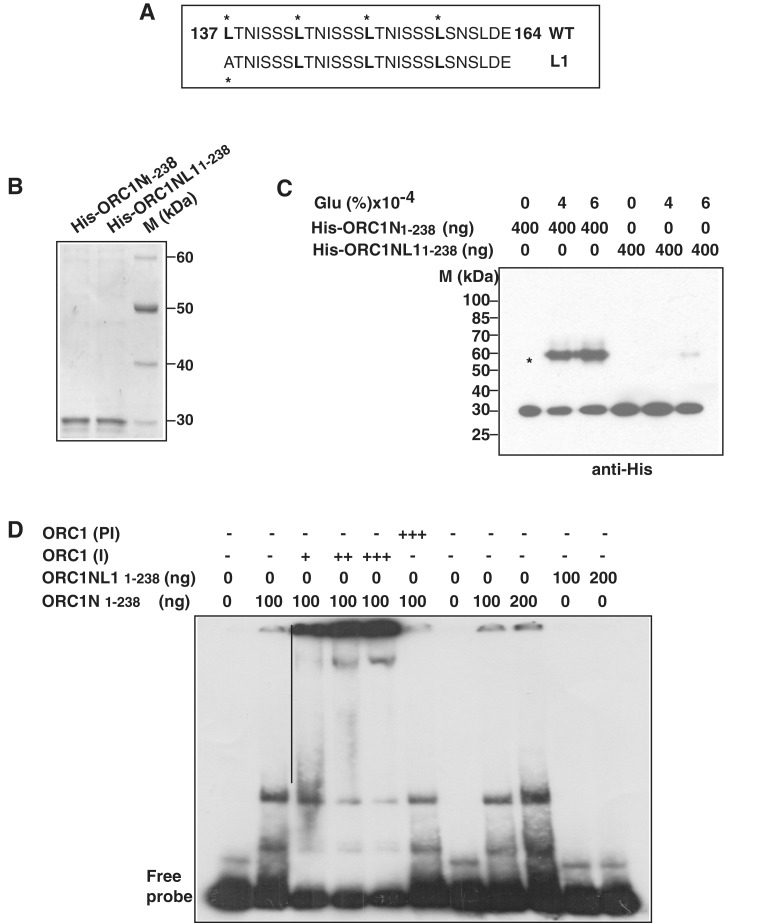
Figure 4.
The leucine heptad repeat in PfORC1N1–238 is essential for oligomerization and DNA binding. (A) The putative leucine heptad repeats of PfORC1N1–238. The leucine at every seventh position is marked. L1 shows the mutation at the first leucine residue. (B) Purification of His6-tagged PfORC1N1–238 wild-type and PfORC1NL11–238 mutant. (C) Cross-linking of His6-PfORC1N1–238 and His6-PfORC1NL11–238 using glutaraldehyde followed by western-blot analysis using anti-His antibodies shows strong dimerization of the wild-type protein (asterisk) but not for the mutant protein. (D) Specificity of ORC1N for DNA binding. EMSA was performed using purified PfORC1N1–238 and radiolabeled telomere DNA probe followed by supershift using different amount of antibodies against PfORC1N (I) (lanes 3–5; 0.5, 1.0 and 1.5 µl, respectively) or pre-immune sera (PI) (lane 6, 1.5 µl). The straight line shows the positions of different supershifted bands. EMSA using PfORC1N1–238 wild-type and mutant form shows that the wild-type but not the mutant form binds to telomeric DNA (lanes 7–11). Two different concentrations of each protein (100 and 200 ng) were used.
In order to investigate the oligomeric properties of ORC1N1–238, we performed cross-linking experiments using recombinant His6-ORC1N1–238 and His6-ORC1NL11–238 in the presence of glutaraldehyde. Cross-linking of proteins followed by western-blot analysis using anti-His antibodies indicates that ORC1N1–238 forms a dimer in the presence of the cross-linking agent (Figure 4C). The uncross-linked protein shows a band around ∼30 kDa whereas the cross-linked product shows a band ∼60 kDA suggesting the oligomeric property of ORC1N1–238. However, ORC1NL11–238 failed to show robust oligomerization property under the similar experimental conditions suggesting that leucine heptad repeats indeed are involved in oligomerization of the protein. The oligomerization property of ORC1N1–238 may also explain the higher molecular mass containing shifted bands in the EMSA experiment as shown before (Figure 3C).
Further, we performed EMSA experiments using the above proteins in the presence of radiolabeled telomere probe (Figure 4D). The authenticity of the EMSA results was confirmed using immune and pre-immune sera against ORC1. We find that wild-type ORC1N1–238 binds to DNA and increasing the concentration of immune sera against ORC1 results in a supershift of the protein–DNA complex whereas pre-immune sera do not yield any supershift band confirming the specificity of EMSA results (Figure 4D, lanes 3–6). Subsequently, EMSA was performed using ORC1N1–238 and ORC1NL11–238 proteins. ORC1N1–238 shows band shifts whereas ORC1NL11–238 fails to do so under the same experimental conditions. These results clearly indicate that the leucine heptad repeat present in ORC1N1–238 is essential for DNA binding activity.
Endogenous ORC1 binds to TARES and var gene promoters like ORC1N1–238
The binding of recombinant ORC1 to telomeric DNA and its peripheral nuclear localization leads to two important questions. Firstly, whether ORC1N1–238 is loaded at the telomeric region and TAREs in vivo and secondly, whether ORC1N1–238 shares the similar sites at the telomeric loci/TAREs as the endogenous ORC1.
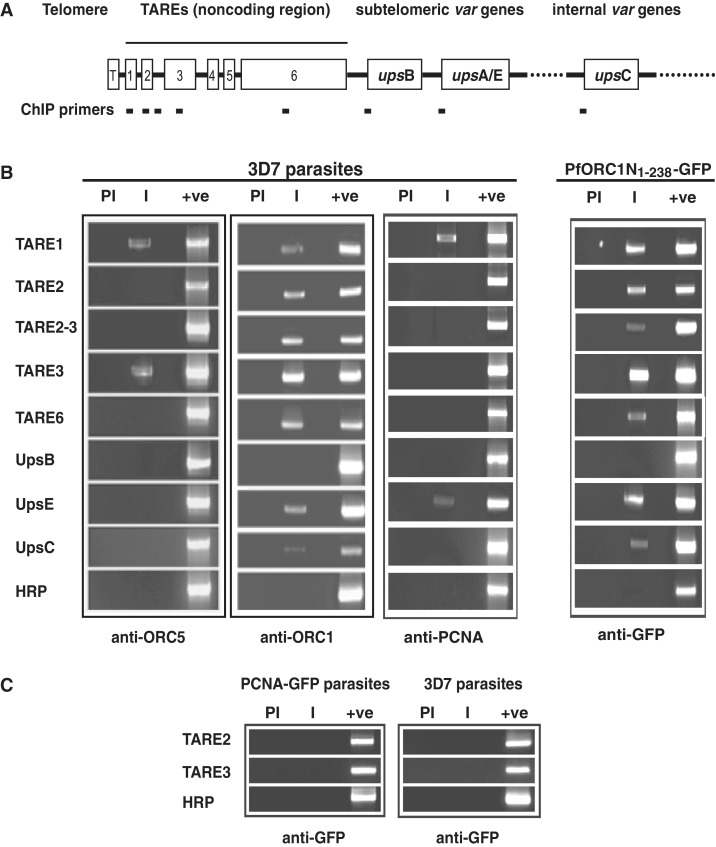
For this purpose, we performed ChIP experiments using immunoprecipitated chromatin from the ring/early trophozoite stage parasites with the help of polyclonal antisera against ORC1. This also enabled us to map the binding sites of endogenous ORC1. We included PCR primers for TARE regions and also from the promoters of different var gene family (Figure 5A). Since TARE regions are repetitive, there is a possibility that PCR will yield multiple bands. However, our primers for the PCR reactions (see Supplementary Tables S1 and S2; Figure 2A) resulted only in major PCR products ranging from ∼500–600 bp for majority of the TARE regions (1, 2, 2–3, 3). TARE-6 was the exception, where we have been able to design PCR primers resulting in a product of ∼1.2 kb. The location of the primers for TAREs and var gene promoters and their expected sizes are indicated in Supplementary Table S2. Semi-quantitative PCR reactions were performed for ∼20–22 cycles to avoid saturation (Supplementary Figure S2B).
Figure 5.
Endogenous ORC1 and GFP–ORC1N1–238 binds to subtelomeric repeat regions and var gene promoters. (A) Schematic diagram of P. falciparum chromosomal ends containing telomere (T), TARE regions (1–6) and subtelomeric as well as internal var genes (upsB, upsA/E and upsC, respectively). (B) Chromatin immunoprecipitation (ChIP) reactions were performed by immunoprecipitation of chromatin fraction using different antibodies as indicated in the figure followed by PCR amplification using different primer sets as shown on the left. The results indicate that both endogenous ORC1 and PfORC1N1–238 bind to most of these regions whereas ORC5 and PCNA bind to only few of them. HRP was used as control. (C) Control ChIP experiments using anti-GFP antibodies from GFP-PCNA1 parasites and 3D7 parasites followed by PCR does not show any product specific to TARE-2 and -3 regions. ‘PI’ and ‘I’ indicate pre-immune and immune sera, respectively, and ‘+ve’ indicates the PCR amplification using genomic DNA as control.
Following standardization of the PCR conditions (Supplementary Figure S2A and S2B), we performed PCR reactions using immunoprecipitated chromatin DNA as template. ChIP-PCR reactions indicate that ORC1 binds to all TARE regions as well as at least two var gene promoters (upsE and upsC) with a variable efficiency (Figure 5B). Primer targeting the genomic HRP region was used as a control for the specificity of the ChIP reactions.
It has been shown earlier that endogenous ORC1 binds to TAREs along with Sir2 (30). However, the promoters of the var genes have not been tested for the recruitment of the ORC1 and Sir2 proteins in the earlier study. Interestingly, majority of the TAREs and var gene promoters were unoccupied by either ORC5 (another member of Plasmodium ORC) or PCNA as shown by ChIP experiments using corresponding antibodies suggesting the exclusive presence of ORC1 at the telomeric region and var promoters (Figure 5B). It is important to note that both ORC5 and PCNA polyclonal antibodies are capable of immunoprecipitating the endogenous proteins (18). Pre-immune sera immunoprecipitated DNA failed to show any signal following PCR reactions in all the cases suggesting the specificity of the antibodies used. All the PCR primers can amplify the specific regions from the genomic DNA as shown in the control lanes for each case (Figure 5B).
After establishing that endogenous ORC1 binds to TAREs and var gene promoters, we investigated the status of ORC1N1–238 for their occupancy in those regions. Chromatin was immunoprecipitated using anti-GFP antibodies from the ORC1N1–238–GFP expressing parasite line followed by PCR reaction as used above. We find that like endogenous ORC1, ORC1N1–238 occupies majority of the TARE regions as well as the var gene promoters (Figure 5B). These striking similarities between these proteins for their occupancy at the TAREs and var gene promoters are in good agreement with the previous localization studies (Figure 2B).
As a control, we performed ChIP experiments from PCNA–GFP expressing parasites and 3D7 parasites using anti-GFP antibodies for immunoprecipitation. PCNA–GFP expressing parasites showed punctate nuclear replication foci (Supplementary Figure S1A) similar to the endogenous PCNA as reported earlier (18). ChIP reactions using anti-GFP antibodies from PCNA–GFP and 3D7 parasites followed by PCR experiments using primer sets positive for endogenous ORC1 and ORC1N1–238–GFP immunoprecipitated DNA failed to amplify any of these regions suggesting the specificity of the ChIP reactions. Representative ChIP-PCR reactions for TARE-2 and -3 regions are shown in Figure 5C.
After establishing the ChIP experiment successfully, the effect of L1 mutation was investigated in vivo. We find that wild-type ORC1N1–238 shows punctate foci pattern at the nuclear periphery as shown earlier whereas ORC1NL11–238 shows diffused staining pattern (supplementary Figure S3A). The loading of these proteins at the TAREs was tested by ChIP PCR experiments. We observe that TARE regions are occupied by ORC1N1–238 but not by ORC1NL11–238 (Supplementary Figure S3B). These experiments together indicate that the mutation in the leucine heptad region indeed affect the oligomerization of the protein leading to the loss of DNA binding activity both in vitro and in vivo.
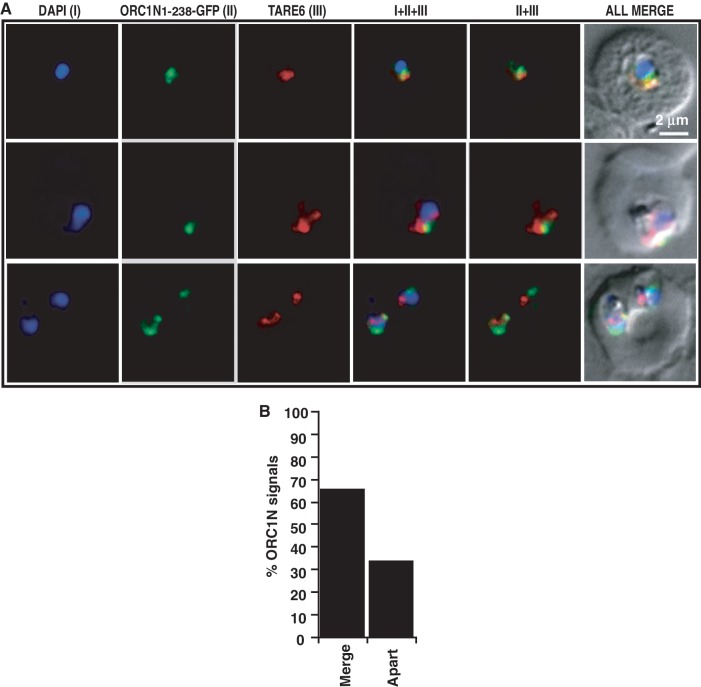
To further prove that ORC1N1–238 is indeed recruited to the TARE regions, we performed fluorescence in situ hybridization coupled immunofluorescence assay (FISH–IFA) using ORC1N1–238–GFP expressing parasites. TARE-6 region was used as FISH probe (19,30,32) and antibodies against GFP were used to detect ORC1N1–238–GFP expression. Previously FISH–IFA was used for the localization of PfHP1 protein at the TARE regions (32). Both FISH and IFA showed distinct signals for the presence of TARE-6 and ORC1N1–238–GFP (Figure 6A). Analysis of the FISH–IFA results indicate that TARE-6 and ORC1N1–238–GFP signals are either co-localized or they are positioned very close to each other in a large number of parasites (∼65%) (Figure 6A and B). These results demonstrate that ORC1N1–238–GFP is indeed localized to the nuclear periphery occupied by chromosomal ends containing telomeric sequences and TARE regions.
Figure 6.
Fluorescence in situ hybridization coupled immunofluorescence assay (FISH–IFA). (A) FISH–IFA was performed using fluorescence analog labeled TARE-6 DNA probe and antibodies against GFP in ORC1N1–238–GFP expressing parasites as described in the ‘Materials and Methods’ section. The results indicate that majority of the TARE-6 and ORC1N1–238–GFP signals are overlapping or closely associated with each other. Top panel corresponds to ring stage parasite and bottom two panels correspond to trophozoite stage parasites. (B) More than 60 images of FISH–IFA were taken with clear signal for ORC1N1–238–GFP and TARE-6. The percentage (%) of overlapping/closely associated (merge) ORC1N1–238–GFP and TARE-6 signals and non-overlapping (apart) were represented graphically. The scale bar is shown inset.
ORC1 is localized to the telomeric region in a Sir2 dependent manner
Plasmodium falciparum Sir2 has been implicated in the regulation of var gene expression by its inherent property of propagation through telomeric and TARE regions into the var gene promoters in vivo. We find ORC1 also binds to these regions as well as var gene promoters in vivo. ORC1 and Sir2 co-localize with each other during ring/early trophozoite stage parasites suggesting ORC1’s possible role in var gene silencing (30). We were interested to find out whether Sir2 has any role in directing ORC1 to the telomeric and subtelomeric region. For this purpose, we used the Sir2 knock out parasite line where the genomic copy of Sir2 has been deleted (34). We have previously shown stage specific expression of PfSir2 in different erythrocytic stages at the transcript and protein level (39).
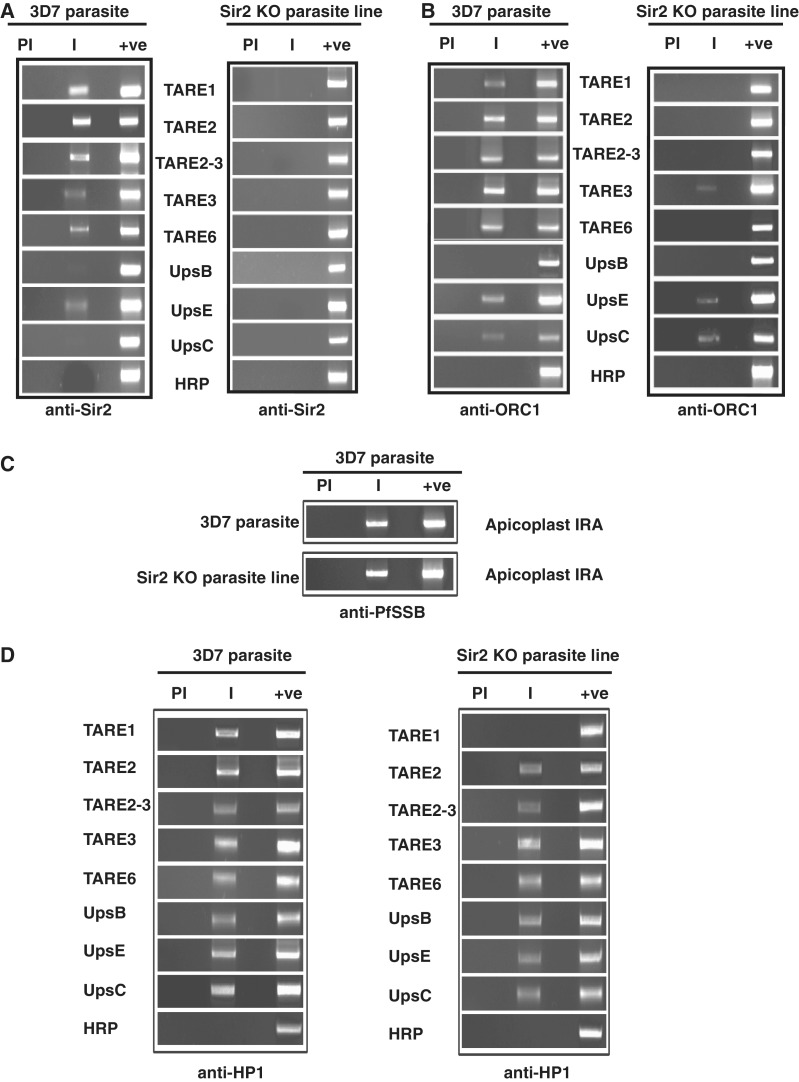
Immunofluorescence analysis using specific antibodies against PfSir2 shows distinct perinuclear spots at the nuclear periphery in 3D7 parasites whereas no signal can be found in Sir2 KO parasites (Supplementary Figure S4A). Comparison of ChIP PCR results for the occupancy of Sir2 and ORC1 in the TAREs and var gene promoters between 3D7 wild-type and Sir2 knock out lines show striking results (Figure 7A and B). As expected, Sir2 binding could not be detected at TAREs and var gene promoters in the Sir2 KO parasite line as Sir2 is not expressed in this parasite line. However, the binding of ORC1 to TAREs and var gene promoters was severely compromised in the Sir2 knock out parasite line. The compromised binding affinity of ORC1 to TAREs and var promoters was not due to any defect of the chromatin isolation from the mutant line. ChIP PCR experiments showed the presence of Plasmodium single strand DNA binding protein at the apicoplast (a plastid-like DNA containing essential organelle) ori region with similar efficiency for both the wild-type as well as mutant parasite line (Figure 7C). These results suggest the importance of Sir2 for the localization of ORC1 at the telomeric and subtelomeric region as well at the var gene promoters in vivo.
Figure 7.
Binding of ORC1 but not HP1 is dependent on Sir2 at the TAREs. (A) Chromatin immunoprecipitation using antisera against PfSir2 from wild-type and Sir2 knock out parasite line shows that Sir2 binds to TAREs and var gene promoters in wild-type but not in the Sir2 knock out parasite line. (B) ChIP experiments using antisera against PfORC1 as described above shows that PfORC1 binds to TAREs and var gene promoters in the wild-type 3D7 parasites but the binding is severely affected in Sir2 knock out parasites. (C) ChIP experiments using antisera against apicoplast targeted PfSSB followed by PCR amplification using primer sets from apicoplast IRA (internal repeat A) shows the binding of SSB to the IRA region in both 3D7 and Sir2 knock out line suggesting that the ChIP experiments are equally efficient in both the cases. (D) ChIP experiments using antisera against PfHP1 as described in (A) and (B) above shows that PfHP1 binds to TAREs and var gene promoters in the wild-type 3D7 parasites as well as in Sir2 knock out parasites.
To expand our study on other proteins involved in var gene regulation we investigated the occupancy of HP1 at TAREs and var gene promoters in the wild-type as well as Sir2 KO lines (Figure 7D). We find that the occupancy of HP1 is not affected at the above regions in the Sir2 KO line indicating that the binding of HP1 in telomeric/subtelomeric region is independent of Sir2.
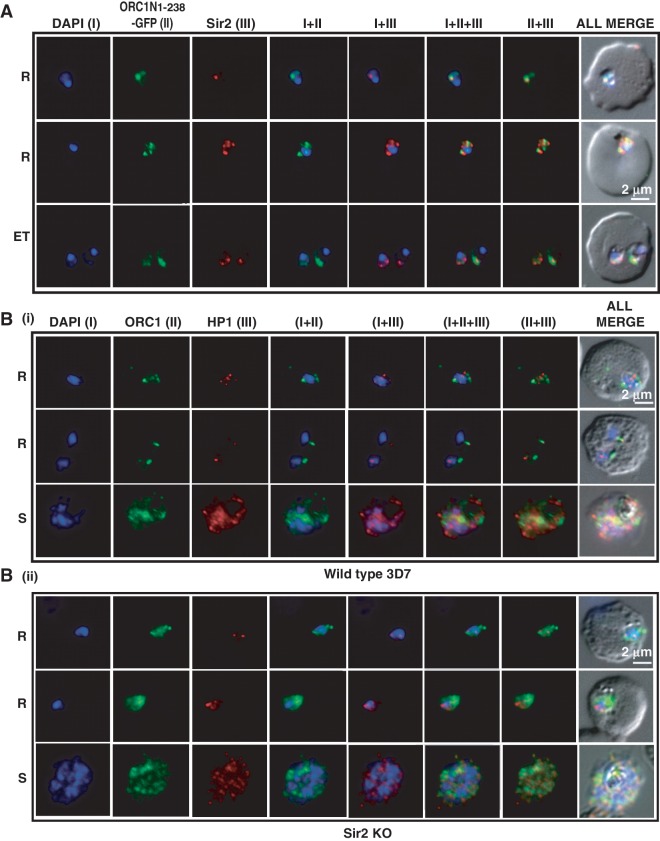
The recruitment of both ORC1N1–238–GFP and Sir2 at the TAREs in ORC1N1–238–GFP expressing parasites raises the question whether these two proteins interact with each other. Immunofluorscence assays using antibodies against GFP and Sir2 indicate co-localization of ORC1N1–238–GFP and Sir2 signals in the parasites expressing ORC1N1–238–GFP (Figure 8A) reflecting the wild-type situation (Supplementary Figure S4B). These results are consistent with previously published results showing co-localization of endogenous ORC1 and Sir2 at the nuclear periphery during the early stages of development (30). These results clearly indicate that ORC1 and Sir2 may be closely associated with each other. However, our efforts to co-immunoprecipitate these two proteins from Plasmodium lysate have failed. It is possible that these two proteins may not interact directly with each other or the interaction is not stable enough to allow their co-purification.
Figure 8.
Co-localization of ORC1N1–238–GFP and Sir2 and immunolocalization of ORC1 and HP1 in 3D7 and Sir2 KO parasite lines. Glass slides containing the parasite smears from ORC1N1–238–GFP expressing parasites or 3D7 wild-type or Sir2 KO parasite lines were treated for immunolocalization using respective antibodies as indicated in the panels (A) and (B). (A) Co-localization of GFP and Sir2 in ORC1N1–238–GFP parasites during early stages of development as indicated on the left. ‘R’ indicates ring stage and ‘ET’ indicates early trophozoite stage parasites. DAPI shows the nuclei. [B(i)] Both ORC1 and HP1 show nuclear punctate staining in 3D7 wild-type parasites during the early stages of development. [B(ii)] ORC1 shows more diffused and distributed pattern while HP1 shows punctate staining in the Sir2 KO parasites. ‘R’ indicates ring stage and ‘S’ indicates schizont stage parasites.
To further support the close association of ORC1 and Sir2 at the nuclear periphery, we performed immunofluorescence assays using parasites obtained from 3D7 wild-type or Sir2 KO parasite line [Figure 8B(i–ii)]. We find that ORC1 localization differs in Sir2 KO parasite line [first two panels, Figure 8B(i–ii)] suggesting that the presence of Sir2 is required for punctate staining pattern of PfORC1. Punctate staining pattern at nuclear periphery of ORC1 was not visible during schizont stages due to the redistribution of this protein at later stages (last panel, Figure 8Bi). This is consistent with earlier observations where both ORC1 and Sir2 have been shown to be redistributed during the late trophozoite and schizont stages (30). As a control, we performed immunofluorescence experiments in 3D7 versus Sir2 KO parasite line using antisera against HP1. We find that HP1 shows punctate staining pattern at nuclear periphery in wild-type parasites as well as in the Sir2 KO parasite line as previously described {(31,32) [Figure 8B(i–ii)]}.
The presence of Sir2 helps the formation of higher order DNA–protein complex of ORC1N1–238
The diminished presence of ORC1 at the TAREs and var gene promoters in the Sir2 KO parasite line reflects the dependence of ORC1 on Sir2 for in vivo occupancy at the above regions. Either the presence of Sir2 creates a favorable conformation for binding of ORC1 at the TAREs or ORC1 may need close association with Sir2 leading to the binding at the TAREs.
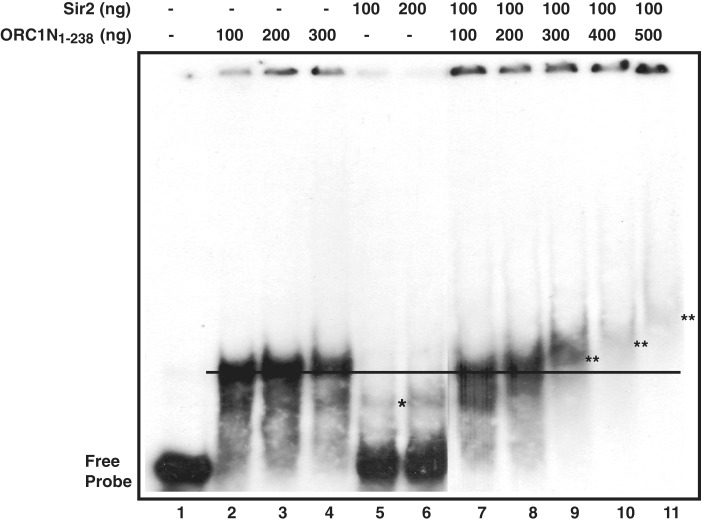
To investigate if Sir2 mediates the DNA binding activity of ORC1N1–238, we analyzed the DNA binding property of ORC1N1–238 in the absence and presence of purified Sir2 in a concentration dependent manner (Figure 9). The purification profile of His6-Sir2 is shown in Supplementary Figure S5A. We find that increasing concentration of ORC1N1–238 in the absence of Sir2 does not apparently supershift the DNA–protein complex band (Figure 9, lanes 2–4 and Supplementary Figure S4B, lanes 2–5). Sir2 by itself shows very poor DNA binding activity at the concentrations where ORC1N1–238 shows strong DNA binding activity. Next, purified Sir2 was incubated with radiolabeled telomeric DNA. The protein–DNA mixture was further incubated with purified ORC1N1–238 protein with increasing concentration. We find that protein–DNA complex is shifted more efficiently with increasing concentration of ORC1N1–238 in the presence of Sir2 (lanes 7–11, Figure 9). As a control, we performed the supershift experiment of ORC1N1–238 in the presence of recombinant MBP-ORC1C (C-terminal region of ORC1; residues 689–1022 of full length ORC1, Supplementary Figure S5A). Unlike Sir2, MBP-ORC1C failed to supershift of DNA–protein complex of ORC1N1–238 under the same experimental condition (Supplementary Figure S5B, lanes 8–11). These results indicate that the presence of Sir2 may facilitate the formation of a higher molecular mass containing DNA–protein complex mediated by ORC1N1–238. This phenomenon may be pre-requisite for the formation of higher order nucleoprotein complex essential for heterochromatin establishment and silencing var gene promoters. It is possible that the presence of Sir2 may help propagating ORC1 in vivo. Interestingly, ORC1N1–238–DNA complex cannot be supershifted in the presence of increasing concentration of Sir2 (data not shown). These results may indicate that the presence of Sir2 makes it conducive for ORC1 binding and propagation at the telomeric/TARE regions. This may also explain why the absence of Sir2 also affects the loading/propagation of ORC1 at the TAREs and var gene promoters.
Figure 9.
The presence of Sir2 helps forming higher order DNA–protein complex of PfORC1N1–238. EMSA was performed by incubating radio-labeled telomeric probe with PfSir2 followed by addition of increasing amount of PfORC1N1–238 with subsequent separation of DNA–protein complex in PAGE. Sir2 shows weak DNA binding (asterisk) (lanes 5 and 6) compared to ORC1N1–238 (lanes 2–4). Interestingly, supershift of ORC1-DNA band was observed in the presence of Sir2 (lanes 9–11). ‘Double asterisk’ indicates the supershifted bands relative to the black straight line.
Expression of ORC1N1–238 compromises Sir2 loading and derepresses some selected var genes
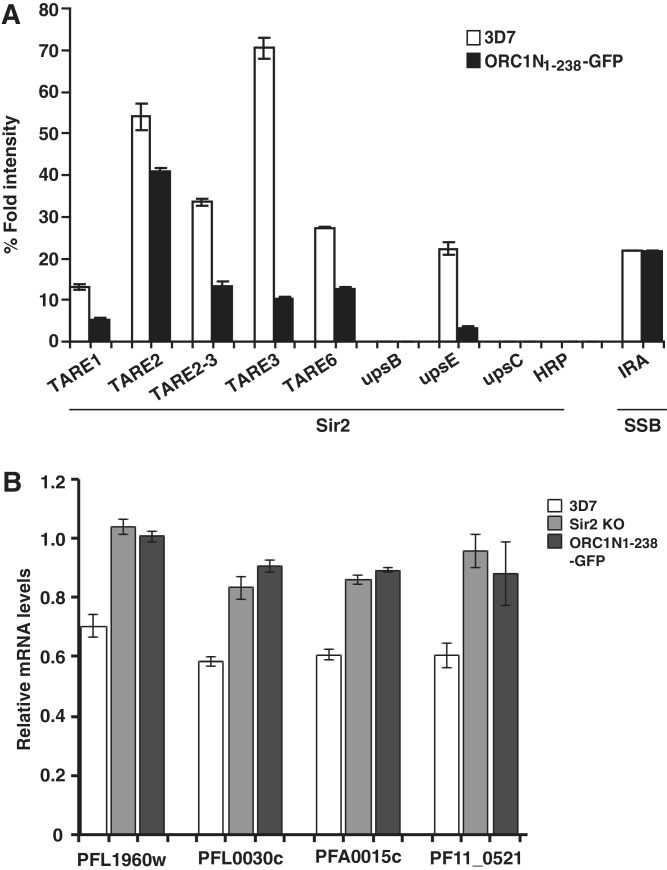
We have shown that the expression level of ORC1N1–238–GFP is higher than the endogenous ORC1 (Figure 1C). Moreover, ORC1N1–238 binds to DNA with higher affinity than Sir2 in vitro under the same experimental conditions (Figure 9). We were interested to see whether the expression of ORC1N1–238 would lead to modulation of Sir2 loading at the TAREs and var gene promoters leading to the change in var gene expression. We first tested the effect of ORC1N1–238 expression on Sir2 loading and function. ChIP results from different independent experiments indicate that the loading of Sir2 was indeed compromised in majority of the TAREs and var gene promoters in ORC1N1–238–GFP expressing parasite lines compared to the 3D7 control parasite lines. As a control, the loading of SSB at the apicoplast DNA was not affected in ORC1N1–238–GFP expressing parasites compared to the wild-type 3D7 parasites (Figure 10A). As Sir2 is a HDAC that represses some of the var gene families, compromised loading of Sir2 may lead to the de-repression of var gene expression. We find a substantial decrease in Sir2 protein loading in upsE promoter in ORC1N1–238–GFP parasite line.
Figure 10.
Overexpression of ORC1N1–238 causes diminished loading of Sir2 at the TAREs and var gene promoter leading to the de-repression of some var genes. (A) ChIP using anti-Sir2 antibodies followed by semi-quantitative PCR using primer sets from TAREs and var gene promoters suggest compromised loading of Sir2 at the TAREs and var gene promoters of ORC1N1–238 over-expressed parasite line compared to the 3D7 control parasites. ChIP experiments were repeated at least three times and results are represented graphically. The loading of SSB at the apicoplast IRA region is also shown. (B) Comparison of expression pattern of some selected var genes as indicated at the bottom between 3D7 wild-type, Sir2 KO and ORC1N1–238–GFP expressing parasites by real time PCR analysis. gapdh was used as control. All the var genes tested showed increased expression at the mRNA level for both Sir2 KO parasites and ORC1N1–238–GFP parasites compared to the wild-type 3D7 parasites.
In order to find out whether the higher level of expression of ORC1N1–238–GFP over endogenous ORC1 would indeed modulate the var gene expression, we compared the expression level of various var genes in 3D7 wild-type with ORC1N1–238 expressing parasites and Sir2 KO parasites by quantitative real time PCR analysis. These var genes include PFL0030c and PFA0015c under upsA/E promoter, PFL1960w under upsC promoter and PF11_0521 under upsA promoter. The first two genes are regulated in a Sir2 dependent manner as reported previously (34). We have selected PFL1960w since we have seen that both the endogenous ORC1 as well as ORC1N1–238–GFP occupy the promoter of this gene (Figure 5B). The real time PCR results indicate similar levels of stimulation of expression of different var genes both in the Sir2 KO parasite line as well as in ORC1N1–238–GFP expressing parasites compared to 3D7 wild-type parasites normalized against gapdh gene expression (Figure 10B). We observe an overall 30–40% increase of various var genes in the above two parasite lines compared to the 3D7 wild-type parasites. No such stimulation of var gene expression was observed in ORC1N1–182–GFP parasites (data not shown). These results suggest that the defect in loading Sir2 in the TAREs and var gene promoters in the ORC1N1–238 expressing parasites indeed modulate the expression of some of the var genes and indicates an intricate balance between ORC1 and Sir2 to regulate the var gene expression in P. falciparum.
DISCUSSION
PfORC1 contains a leucine heptad repeat region at the N-terminus with the possibility of formation a coiled coil domain. However, the functional significance of this domain is not yet established. Classical leucine heptad repeats containing proteins are bZIP type of transcription factors like jun and fos which form oligomeric structures, essential for binding to DNA (43,44). Geminin, an inhibitor of DNA replication also contains leucine heptad repeats that form coiled-coil domain, essential for binding to CDT1, a replication-licensing factor (45). The oligomerization property and the DNA binding activity of PfORC1N1–238 and the lack of these activities in the mutant PfORC1NL11–238 reflect the functional significance of the repeat region in ORC1. This is the first demonstration of a functional leucine heptad repeat containing protein in Plasmodium. The leucine heptad repeat is also found in the ORC1 proteins from related species P. yoelii (PY01316; ORC1) and P. vivax (PVX084195; ORC1) suggesting a conserved function.
We find that ORC1N1–238 binds to DNA and forms nuclear periphery punctate foci whereas ORC1N1–182 fails to do so suggesting that residues between 182 and 238 are involved in DNA binding. A detailed mutational analysis will be required to identify the key residues for DNA binding. The lack of DNA binding activity in PfORC1NL11–238 suggests that the loss of oligomerization may also affect DNA binding. Therefore, both the leucine heptad repeats and residues between 182 and 238 may contribute to DNA binding. This may be similar to the bZIP type of transcription factors where the presence of basic residues and leucine zipper regions are involved in DNA binding (46). The oligomerization property of ORC1 may also favor the aggregation of nucleosome leading to the formation of heterochomatin, a prerequisite for var gene silencing.
In yeast, ORC binds to mating type locus as a complex. The absence of PfORC5 at the majority of TAREs suggests that PfORC1 may have its exclusive function at the telomeric/TARE regions for the control of var gene expression. It is possible that there are different sub-complexes of ORC present in Plasmodium responsible for DNA replication and the control of var gene expression, respectively. The poor presence of PCNA at the TARE regions indeed indicates that the presence of ORC1 in these regions may not be due to the replication function. It is possible that telomeric regions in Plasmodium are replicated through the progression of forks into this region originated outside this region.
We have shown that PfORC1N1–238 and PfSir2 co-localizes with each other in vivo during the ring/early trophozoite stage of the parasite development. These results are similar to the earlier published results (30). We also find that the loading of ORC1 at the TAREs and var gene promoters is dependent on the presence of Sir2 in vivo and the presence of Sir2 favors the higher order DNA–protein complex of ORC1 in vitro. Together, these results indicate that ORC1 and Sir2 may associate with each other. Although we have not been able to show direct interaction between these proteins, it is possible that the interaction may be mediated through some unknown protein or DNA. The interaction between yeast ORC1 and Sir1 protein is mediated through the BAH domain present at the N-terminus of the ORC1 (15,16). BAH domain is conserved in majority of the ORC1 reported so far including human, Drosophila, and Xenopus (15), although no BAH domain is predicted at the N-terminus of the Plasmodium ORC1. However, only detailed mutational analysis will reveal the essential regions in the N-terminus of PfORC1 that are responsible for Sir2 interaction and therefore might represent a functional homologue to the BAH domain.
The functional analysis of PfORC1 reported in this work has a striking similarity with a recently published work performed in the yeast species Kluyveromyces lactis (47). In S. cerevisiae, the formation of heterochromatin silencing region at the yeast mating type locus is initiated by ORC followed by recruitment of Sir proteins (Sir 1–4) and subsequent spreading of Sir2–4 proteins facilitating the maintenance of the silenced zone. However, in K. lactis, the spreading of ORC1 at the silenced locus is dependent on Sir2 and the N-terminal BAH domain of K. lactis ORC1. Moreover, KlORC1 appears to act independently in the absence of KlORC4 and 5 subunits for the silencing function (4,5). Similarly in P. falciparum, the N-terminal region of ORC1 is involved in telomeric/TARE localization of ORC1 in a Sir2 dependent manner that may lead to the silencing of var genes. Interestingly, we find that the presence of PfORC5 is very limited compared to the PfORC1 loading at the TAREs suggesting PfORC1’s exclusive role in var gene regulation independent of other ORC subunits.
Co-incidentally, both K. lactis and P. falciparum does not contain a Sir3 homolog. Sir3 is a gene duplication product of ORC1 in S. cerevisiae (47,48). It spreads along with Sir4 protein over the silenced chromatin by virtue of its affinity towards deacetylated nucleosome (as created by Sir2 deacetylase). In K. lactis that diverged before gene duplication, ORC1 retains the spreading function of Sir3 along the heterochromatin to maintain the silenced zone. Similarly, we find that PfORC1 is spread through the TAREs that may be crucial for Plasmodium var gene silencing.
It will be interesting to investigate whether there is a link between the status of nucleosome (acetylated or non-acetylated) and the recruitment of PfORC1 and PfORC1N1–238 at the telomeres/TAREs and var gene promoters. In vitro binding of ORC1 and Sir2 to chromatin templates rather than naked DNA will be useful in this purpose.
The data points towards a more complex mechanism of var gene silencing as it has been described previously. It is still not clear how a particular var gene in a family will bypass this process of silencing and it is expressed in an allele specific manner. A local chromatin remodeling around the promoter of the silenced gene may induce gene expression while keeping the other cascade of genes in the silent form. Whether this needs translocation of a particular set of var gene from a transcriptionally silenced zone to an active zone remains to be elucidated.
Our data establish the regulatory role of the N-terminus of ORC1 in dimer formation, association with the telomeric DNA/TARE regions and directing ORC1 to the nuclear periphery region in association with Sir2 leading to the silencing of the var genes. These findings are summarized in a preliminary model illustrated in Supplementary Figure S6. Further studies have to establish whether this function is independent of the replication function of ORC1 that possibly lies within the C-terminus of the protein as it contains ATP binding and hydrolysis domains and PIP motif.
ChIP analysis suggests the presence of Sir2 at the upsE promoter (transcribed towards telomere) but not at the upsB (transcribed away from telomeres) and upsC (internal var genes) promoters. This is consistent with earlier observations showing that PfSir2 controls the expression of var genes transcribed towards the telomeric region (upsA/E) (34). In the contrary, we find that ORC1 is present both at the upsE and upsC regions. Interestingly, in the Sir2 KO line, although the presence of ORC1 was severely affected in the TAREs, it could still be detected at the upsE and upsC regions. It is possible that Sir2 may not influence the binding of ORC1 away from the telomeric region that efficiently. Alternatively, the presence of ORC1 in these regions may be linked with the replication function of ORC1.
It has been demonstrated that ORC1 is reliant on Sir2 for the recruitment at the telomere/TARE regions. However, overexpression of ORC1N1–238 diminishes the loading of Sir2 at the telomeres/TAREs followed by de-repression of var genes. ORC1N1–238 may do this job either by virtue of its stronger binding affinity towards DNA compared to Sir2 (Figure 9, lanes 2 and 3 versus 5 and 6) or by competing with endogenous ORC1 that may affect the co-ordination between Sir2 and ORC1 essential for spreading of heterochromatin region required for var gene silencing. Alternatively, the over-expression of ORC1N1–238 may show dominant negative effect by disrupting the complexes of heterochromatin proteins that are necessary for heterochromatin assembly.
Overall, the findings of this article strongly suggest that both ORC1 and Sir2 co-ordinate with each other and keeps an intricate balance for spreading heterochromatin like silencing zone leading to the repression of var gene family. This balance can be disturbed either by removing Sir2 or by over-expression of ORC1N1–238 leading to the de-repression of some var gene family members.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials and Methods, Supplementary Figures 1–6, Supplementary Tables 1 and 2 and Supplementary References [30,49].
FUNDING
Wellcome Trust, London, European Commission (MALSIG) and Swarnajayanti Fellowship awarded by the Department of Science and Technology, Government of India. Funding for open access charge: European Commission (MALSIG) and Swarnajayanti Fellowship awarded by the Department of Science and Technology, Government of India.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
S.K.D acknowledges the Humboldt Foundation, Germany for the award of Alexander von Humboldt fellowship that has been extremely useful to complete this work. Tobias Spielmann (BNI, Hamburg) is acknowledged for his help to perform the fluorescence and confocal microscopy. We thank Alan Cowman for providing us the Sir2 KO parasite line and Jennifer Volz for her help with the FISH–IFA experiments. A.D. and S.S. acknowledge CSIR, India for fellowships. The authors thank Vijay Verma for his help with the EMSA experiments and Nidhi Gupta for the purification of recombinant proteins. Ashis Nandi and Mrunmay Giri (SLS, JNU) are acknowledged for their help with Real Time PCR analysis. A.D. performed the ChIP, EMSA as well as Real Time PCR analysis, S.S. performed immunofluorescence assays and FISH–IFA, S.K.D. and S.H. generated all the parasite lines and characterized them with the help of A.G. and P.M. S.K.D., A.D., S.S. and T.G. analyzed the data. S.K.D. and T.G. wrote the manuscript.
REFERENCES
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki T, Gilbert DM. The many faces of the origin recognition complex Curr. Opin. Cell. Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou Z, Bernstein DA, Fox CA, Keck JL. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc. Natl Acad. Sci. USA. 2005;102:8489–8494. doi: 10.1073/pnas.0503525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 6.Shareef MM, King C, Damai M, Badagu R, Huang DW, Kellum R. Drosophila Heterochromatin Protein 1 (HP1)/Origin Recognition Complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin induced silencing. Mol. Biol. Cell. 2001;12:1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Romanowski P, Botchan MR. Association of the Origin Recognition Complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 8.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 10.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 11.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of, ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 13.Klemm RD, Bell SP. ATP bound to the origin recognition complex is important for preRC formation. Proc. Natl Acad. Sci. USA. 2001;98:8361–8367. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC Protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 15.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 16.Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta A, Mehra P, Deshmukh A, Dar A, Mitra P, Roy N, Dhar SK. Functional dissection of the catalytic carboxyl-terminal domain of Origin Recognition Complex subunit 1 (PfORC1) of the human malaria parasite Plasmodium falciparum. Eukaryot. Cell. 2009;8:1341–1351. doi: 10.1128/EC.00170-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta A, Mehra P, Dhar SK. Plasmodium falciparum origin recognition complex subunit 5: functional characterization and role in DNA replication foci formation. Mol. Microbiol. 2008;68:646–665. doi: 10.1111/j.1365-2958.2008.06316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueiredo LM, Pirrit LA, Scherf A. Genomic organization and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 2000;106:169–174. doi: 10.1016/s0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- 20.Vernick KD, McCutchan TF. The sequence and structure of a Plasmodium telomere. Mol. Biochem. Parasitol. 1998;28:85–94. doi: 10.1016/0166-6851(88)90055-2. [DOI] [PubMed] [Google Scholar]
- 21.Dolan SA, Herrfeldt JA, Wellems TE. Restriction polymorphism and fingerprint patterns from an interspersed respective element of Plasmodium falciparum DNA. Mol. Biochem. Parasitol. 1993;61:137–142. doi: 10.1016/0166-6851(93)90166-u. [DOI] [PubMed] [Google Scholar]
- 22.Wasserman M, Contreas J, Pinilla G, Rojas MO, Paez A, Caminos E. Plasmodium falciparum: characterization of a 0.7-kbp, moderately repetitive sequence. Exp. Parasitol. 1995;81:165–171. doi: 10.1006/expr.1995.1105. [DOI] [PubMed] [Google Scholar]
- 23.Ouendo P, Goman M, Mackay M, Langsley G, Walliker D, Scaife J. Characterization of a repetitive DNA sequence from the malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 1986;18:89–101. doi: 10.1016/0166-6851(86)90053-8. [DOI] [PubMed] [Google Scholar]
- 24.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 27.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 29.Kyes S, Horrocks P, Newbold C. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- 30.Mancio-Silva L, Roias-Meza AP, Vargas M, Scherf A, Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell. Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Toledo K, Roias-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, Martinez-Cavillo S, Scherf A, Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amava AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, et al. Plasmodium falciparum Heterochromatin Protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;6:e1000784. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, Duffy MF, Silva LM, Ivens A, Speed TP, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;14:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 35.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 36.Crabb BS, Rug M, Gilberger TW, Thompson JK, Triglia T, Maier AG, Cowman AF. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 2004;270:263–276. doi: 10.1385/1-59259-793-9:263. [DOI] [PubMed] [Google Scholar]
- 37.Fiddock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl Acad. Sci. USA. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dar A, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA Gyrase genes: unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Eukaryot. Cell. 2007;6:398–412. doi: 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prusty D, Mehra P, Srivastava S, Shivange AV, Gupta A, Roy N, Dhar SK. Nicotinamide inhibits Plasmodium falciparum Sir2 activity in vitro and parasite growth. FEMS Microbiol. Lett. 2008;282:266–272. doi: 10.1111/j.1574-6968.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 40.Gatton ML, Prters JM, Gresty K, Fowler EV, Chen N, Cheng Q. Detection sensitivity and quantitation of Plasmodium falciparum var gene transcripts by real-time RT-PCR in comparision with conventional RT-PCR. Am. J. Trop. Med. Hyg. 2006;75:211–218. [PMC free article] [PubMed] [Google Scholar]
- 41.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 42.Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinson C, Myakishev M, Acharya, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gentz R, Rauscher FJ, III, Abate C, Curran T. Parallel association of Fos and jun leucine zippers juxtaposes DNA-binding domains. Science. 1989;243:1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- 45.Benjamin JM, Torke SJ, Demeler B, McGarry TJ. Geminin has dimerization, Cdt1-binding, and destruction domains that are required for biological activity. J. Biol. Chem. 2004;279:45957–45968. doi: 10.1074/jbc.M407726200. [DOI] [PubMed] [Google Scholar]
- 46.Jenuwein T, Muller R. Structure-function analysis of fos protein: a single amino acid change activates the immortalizing potential of v-fos. Cell. 1987;48:647–657. doi: 10.1016/0092-8674(87)90243-1. [DOI] [PubMed] [Google Scholar]
- 47.Hickman MA, Rusche LN. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc. Natl Acad. Sci. USA. 2010;107:19384–19389. doi: 10.1073/pnas.1006436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.