Abstract
Chemically modified siRNAs are expected to have resistance toward nuclease degradation and good thermal stability in duplex formation for in vivo applications. We have recently found that 2′-OMe-4′-thioRNA, a hybrid chemical modification based on 2′-OMeRNA and 4′-thioRNA, has high hybridization affinity for complementary RNA and significant resistance toward degradation in human plasma. These results prompted us to develop chemically modified siRNAs using 2′-OMe-4′-thioribonucleosides for therapeutic application. Effective modification patterns were screened with a luciferase reporter assay. The best modification pattern of siRNA, which conferred duration of the gene-silencing effect without loss of RNAi activity, was identified. Quantification of the remaining siRNA in HeLa-luc cells using a Heat-in-Triton (HIT) qRT–PCR revealed that the intracellular stability of the siRNA modified with 2′-OMe-4′-thioribonucleosides contributed significantly to the duration of its RNAi activity.
INTRODUCTION
Since its discovery in 1998 (1), RNAi has rapidly become a standard method for the investigation of gene function in various eukaryotic cells. However, long dsRNA (>30 nt) has the potential to elicit severe innate immune response when introduced in mammalian cells. A few years after the discovery of RNAi, Elbashir et al. reported that exogenously introduced siRNAs, 21- to 23-mers in length, could achieve sequence-specific gene-silencing in mammalian cell lines without sequence nonspecific silencing arising from interferon (IFN) response (2). Owing to its potency, target specificity and ability to silence virtually any gene, this procedure applying siRNAs has been used not only as a genetic tool but also as an aid in identifying new drug candidates. However, despite these advantages, questions concerning poor nuclease resistance in biological fluids and off-target effects (3), among others, still need to be addressed. In addition, recent studies have shown that even a standard 21-mer siRNA has the potential to induce severe innate immune response in vivo (4). In order to overcome these problems, chemical modifications on the sugar, the phosphate backbone or the nucleobase moiety of siRNAs have been attempted (5). A variety of chemically modified nucleosides, such as 2′-O-methyl (2′-OMe) (6–8) and 2′-fluoro (2′-F) (9–11) substitutions, have been used to improve the nuclease and thermal stability of siRNA. These chemical modifications, in particular the use of 2′-OMe derivatives, have also been shown to prevent innate immune response. Recently, Judge et al. reported that incorporation of as few as two 2′-OMe guanosine and uridine residues in a highly immunostimulatory siRNA sequence is sufficient to abrogate siRNA-mediated IFN and inflammatory cytokine induction without disrupting its gene-silencing effect in human peripheral blood mononuclear cells and in mice in vivo (12).
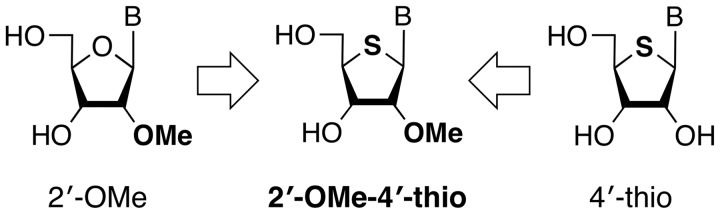
On the other hand, our group (13,14) and others (15,16) have studied siRNAs modified with 4′-thionucleoside derivatives. We have recently published a detailed study on the modification pattern–siRNA activity relationship of siRNAs modified with 4′-thioribonucleosides against the photinus luciferase and renilla luciferase genes in three different mammalian cell lines. siRNAs modified with 4′-thioribonucleosides were well tolerated in both the sense and antisense strands (13). From our search for the best modification pattern, the modified siRNAs with four 4′-thioribonucleosides in both ends of the sense strand and four residues at the 3′-end of the antisense strand showed the most potent RNAi activity against the two target luciferase genes in NIH/3T3, HeLa and MIA PaCa-2 cell lines (14). These successful results prompted us to develop a new siRNA molecule possessing a hybrid type of chemically modified nucleoside based on 2′-OMe and 4′-thioribonucleosides (Figure 1).
Figure 1.
Structure of 2′-OMe-4′-thioribonucleoside.
We have already developed a synthesis of 2′-OMe-4′-thioribonucleoside and examined the physical and physiological properties of fully 2′-OMe-4′-thioribonucleoside modified RNAs, that is, 2′-OMe-4′-thioRNAs (17). In its hybridization property, 2′-OMe-4′-thioRNA preferred RNA as a complementary partner rather than DNA in duplex formation. In addition, an investigation of their stability revealed that 2′-OMe-4′-thioRNA was extremely stable in 50% human plasma (t1/2 = 1631 min) compared with 2′-OMeRNA (t1/2 = 451 min), 2′-FRNA (t1/2 = 53 min) and 4′-thioRNA (t1/2 = 187 min). From these results, we expected 2′-OMe-4′-thioribonucleoside modified siRNA to be thermally stable in duplex formation and quite stable in biological fluids as well as in cellular compartments, which make them ideally suitable for therapeutic applications. In this study, we investigated the potency and duration of the gene-silencing activity of siRNAs modified with 2′-OMe-4′-thioribonucleosides.
MATERIALS AND METHODS
Oligonucleotides
The chemically modified oligonucleotides used in this study were synthesized on an Applied Biosystem 3400 DNA synthesizer as described previously (17). Briefly, each of phosphoramidite units was used at concentration of 0.1 M in dry acetonitrile, and the coupling time was extended to 10 min for each step. After cleavage and de-protection, RNAs were purified by reversed-phase high performance liquid chromatography (C18). The analytical data of the synthesized oligonucleotides are summarized in Supplementary Tables S1 and S2. Unmodified (UM) oligonucleotides were purchased from Hokkaido System Science Co., Ltd. For preparation of duplexes, sense- and antisense-strand oligonucleotides (150 µM each) were mixed together in an annealing buffer (100 mM potassium acetate, 30 mM HEPES–KOH at pH 7.4, 2 mM magnesium acetate) and incubated for 3 min at 90°C then cooled gradually to room temperature. The siRNAs used in this study consisted of a 21-nt with a 2-nt overhang at the 3′-end of each strand (luc siRNA: sense 5′-GCGCUGCUGGUGCCAACCCtt-3′, antisense 5′-GGGUUGGCACCAGCAGCGCtt-3′; scrambled siRNA: sense 5′-ACCGACUAGGGGCGUCGCGtt-3′, antisense 5′-CGCGACGCCCCUAGUCGGUtt-3′).
Cell culture
HeLa cells stably expressing luciferase (HeLa-luc) were cultured at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 µg/ml streptomycin and 0.4 mg/ml G418, as described previously (18). Cells were regularly passaged to maintain exponential growth.
Preparation of multifunctional envelop-type nano device
A Multifunctional Envelop-type Nano Device (MEND) was used as a transfection reagent and was prepared as described previously (19). SiRNA (0.1 mg/ml) was mixed with STR-R8 (0.1 mg/ml at a nitrogen/phosphate ratio of 1.05) in 250 µl of HEPES buffer (pH 7.4). A lipid film was formed by evaporation of the chloroform solution containing 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and cholesterol (125 nmol total lipids in a 3:4:3 molar ratio). The lipid films for the modified MENDs were prepared by evaporation. The resulting lipid film was then incubated for 10 min at room temperature to hydrate the lipids. The lipid film was sonicated for ∼1 min in a bath-type sonicator, followed by sonication for 10 min with a probe sonicator. The siRNA/STR-R8 complex was applied to the lipid film. The average diameter and the ζ-potential of the condensed siRNA and MENDs were determined using a Zetasizer Nano ZS ZEN3600 (MALVERN Instrument, Worchestershire, UK).
Luciferase reporter assay
HeLa-luc cells were seeded in a 24-well plate (4 × 104 cells/500 µl per well) in DMEM at 70% confluency for 24 h prior to transfection. siRNAs formulated with MEND were applied to each well in a total volume of 250 µl Opti-MEM I (Invitrogen). After 3 h of incubation at 37°C, 750 µl of culture medium containing 10% serum was added to the cells, followed by incubation at 37°C. For the time-course experiment, multiple parallel cultures were maintained for 9 days by serial passage on alternate days without additional treatment. The cells were washed with 0.5 ml PBS and lysed with reporter lysis buffer (Promega). Luciferase activity in the cell lysate was measured using luminometer (Luminescencer-PSN, ATTO). The protein concentration was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). Luciferase activity was expressed as relative light units (RLU) per milligram of protein. Data points represent the ratio of the average luciferase signal intensity from triplicate wells receiving anti-luciferase siRNA and scrambled siRNA. The scrambled siRNA used in this study had no effect on luciferase expression level by comparison with untreated cells. All experiments were performed in triplicate and data shows mean values from at least three assays.
Pharmacokinetic data analysis of time-course experiment
According to the report of Takahashi et al. (20), the area under the curve of the inhibitory effect (AUCIE) and the mean response time of the inhibitory effect (MRTIE) values were calculated using following equations: AUCIE = ∫ RGEIdt; MRTIE = ∫ t·RGEIdt / ∫ RGEIdt, where RGEI represents the ratio of inhibition of gene expression.
Stability of siRNAs in 50% mouse serum
Each sense strand of siRNA labeled with 32P at the 5′-end (5 pmol) was mixed with the corresponding unlabeled sense strand (20 pmol) and antisense strand (30 pmol). The labeled siRNA duplex was prepared by annealing in HEPES buffer (pH 7.4) containing 100 mM potassium acetate and 2 mM magnesium acetate. The siRNA was incubated in HEPES buffer (pH 7.4, 20 µl) containing 50% mouse serum at 37°C. At appropriate periods, 2 µl aliquots of the reaction mixture were added to 8 µl of 50 mM EDTA, and the mixtures were frozen immediately in liquid nitrogen. After the final sample was taken, all the samples were then dissolved on ice and extracted with phenol and chloroform. The extracts were mixed with the loading buffer (1× TBE, 10% glycerol, 0.05% bromophenol blue, 0.05% xylene cyanol) and analyzed by electrophoresis on native 15% PAGE. Radioactive densities of the remaining full-length siRNAs were visualized and measured by a Bio-imaging analyzer (Bas 2500, Fuji Co., Ltd).
Quantification of siRNA in HeLa-luc cells
SiRNAs formulated with MEND (120 nM) were transfected into HeLa-luc cells. At the indicated days, cells were trypsinized and pelleted by centrifugation. The cell pellets were resuspended in 0.25% Triton X-100 in phosphate-buffered saline (PBS, 500 µl) and heated at 95°C for 10 min, then immediately cooled on ice for 10 min. The lysates were centrifuged at 20 000g for 20 min at 4°C and the supernatants were put into a new tube and kept at −80°C. Reverse transcription reactions were performed using a TaqMan MicroRNA RT kit (Applied Biosystems). The lysates were heated at 95°C for 10 min, and aliquots of each hot sample were directly added into the RT reaction (30 min, 16°C, 30 min, 42°C, 5 min, 85°C). Quantitative PCR reactions were performed using TaqMan PCR kit (TaqMan probe, forward primer, reverse primer and TaqMan Universal PCR Master Mix) on MX3005P QPCR System (Agilent Technologies). All experiments were performed in triplicate and data shows mean values from at least three assays.
RESULTS AND DISCUSSION
Optimization of modification patterns
We first optimized modification patterns of siRNA directed against firefly luciferase. In our previous study, we found that modified siRNAs, which have four residues of the 4′-thioribonucleoside unit on both ends of the sense strand and four 4′-thioribonucleosides on the 3′-end of the antisense strand, or four residues on both ends of the sense strand and two residues on both ends of the antisense strand (modification similar to SM1 and SM2 in Table 1), exhibited potent RNAi activity. Incorporation of more 4′-thioribonucleosides in the siRNAs resulted in a decrease in RNAi activity. Therefore, we decided to prepare SM1 and SM2, which have similar modification patterns as those of the best for 4′-thioribonucleoside modified siRNAs. We also prepared SM3–SM6 (Table 1), whose antisense strands were UM. Since it is well known that nuclease-resistant chemical modifications at the RISC cleavage site of an siRNA sense strand (between positions 9 and 10) impaired its gene-silencing activity (13,21), we avoided introducing the modifications adjacent to this cleavage site.
Table 1.
Sequence and modification pattern of siRNAs against firefly luciferase gene (GL3)
| siRNA sequences | |
|---|---|
| UM | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| SMl | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| SM2 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| SM3 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| SM4 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| SM5 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| SM6 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| M6 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
| S6 | 5′-GCGCUGCUGGUGCCAACCCtt-3′ |
| 3′-ttCGCGACGACCACGGUUGGG-5′ | |
Upper case letters represent ribonucleotides; lower case letters represent 2′-deoxyribonucleotides; underlined bold italics are 2′-OMe-4′-thioribonucleotides; underlined are 2′-OMe; 4′-thioribonucleotides are in italics.
Each siRNA was transfected into HeLa cells, which constitutively express the luciferase gene (HeLa-luc cells), at a 120 nM concentration using MEND (19), which was developed for use as an efficient non-viral system for the delivery of nucleic acids. The average diameters and ζ-potentials of the MENDs containing the siRNAs used in this study were ∼120 nm and −30 mV, respectively, and there were no obvious differences in the size and potential of the MEND containing either UM or modified siRNAs. Since the transfection efficiency of siRNA into the cell may also influence the duration of activity, we examined the transfection efficiency of the MEND and found that it was comparable to that of the commercially available Lipofectamine® 2000 (the results are shown in the Supplementary Figure S1). We also confirmed that the transfection of both UM and chemically modified siRNA seemed to have no effect on cell viability and cell division rate.
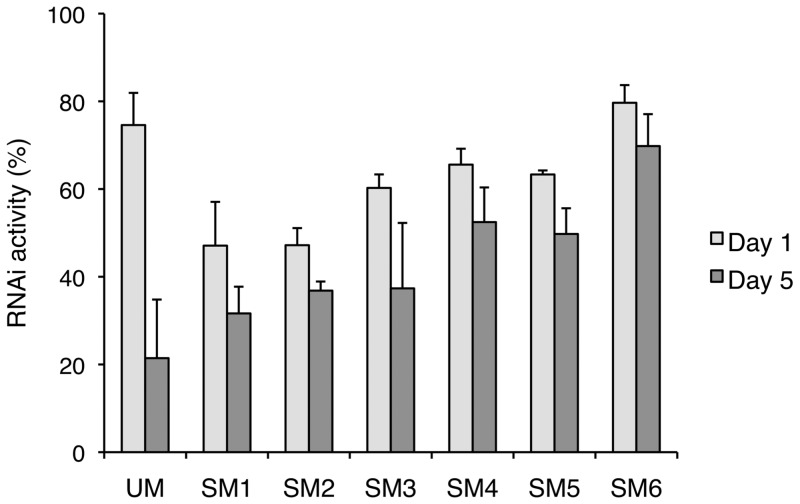
To assess whether the modifications of siRNA with 2′-OMe-4′-thioribonucleosides affected RNAi activity in HeLa-luc cells, we carried out luciferase reporter assays at designated hours after transfection of siRNAs, and the resulting RNAi activities are shown in Figure 2. The UM siRNA showed high potency and suppressed the luciferase gene expression by 74% 1 day after the transfection (Day 1). Unlike the modification by 4′-thioribonucleosides, the modification by 2′-OMe-4′-thioribonucleosides on both the sense and antisense strand (SM1 and SM2) resulted in reduction of RNAi activity. When the antisense strand was unmodified, the RNAi activity was restored by >60% (SM3–6). Thus, SM3–SM5, which have four consecutive 2′-OMe-4′-thioribonucleosides at the 5′-end of the sense strand but a different number of consecutive residues at the 3′-end of the sense strand (4, 2 and 0 residues, respectively), all showed similar RNAi activity (60–65%). Among the siRNAs examined, the best potency was observed in SM6, which has three consecutive 2′-OMe-4′-thioribonucleosides at both ends of the sense strand, with a higher RNAi activity (79%) than that of UM (74%).
Figure 2.
Relative potency of siRNAs variously modified with 2′-OMe-4′-thioribonucleosides. siRNAs were transfected with MEND into HeLa-luc cells stably expressing luciferase at 120 nM concentrations at Day 0 and luciferase reporter assays were performed at Day 1 and Day 5.
It is worth noting that at Day 5 all the modified siRNAs had higher RNAi activity than UM. In particular, SM6 still had higher RNAi activity at Day 5, only a 10% decrease from Day 1, whereas UM showed less than one-third of the Day 1 RNAi activity at Day 5. The results indicated that the siRNAs modified with 2′-OMe-4′-thioribonucleosides enhanced the duration of gene-silencing activity.
Detailed study of the duration of RNAi activity
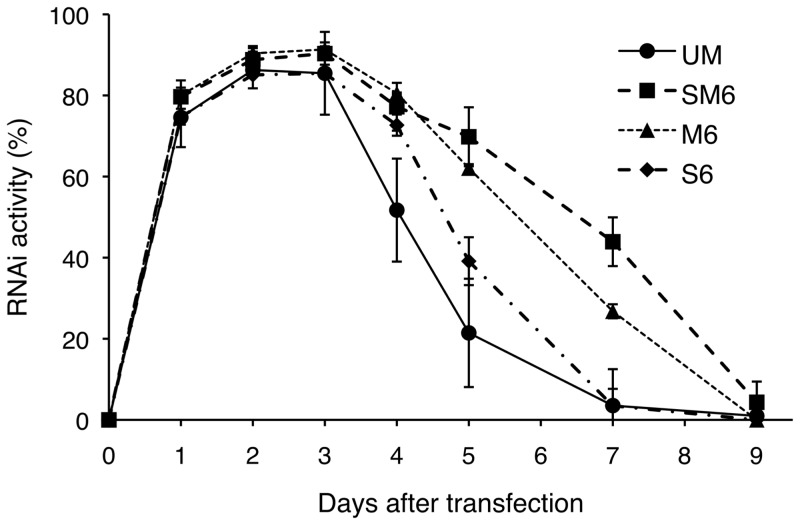
As shown in Figure 2, SM6 appeared to be the most promising candidate for activity and duration of gene-silencing; therefore, we carried out more detailed time-course experiments using SM6. For comparison, we also synthesized siRNAs modified with 2′-OMe (M6) and 4′-thioribonucleoside (S6) with the same modification pattern as SM6, and evaluated the duration of RNAi activity along with that of UM. As shown in Figure 3, effective suppression of luciferase expression was observed in the cells transfected with either unmodified or three modified siRNAs (SM6, M6 and S6) up to Day 3 with 70–90% RNAi activity. Drastic changes in their RNAi activities were observed after Day 3. Thus, UM showed a significant decrease in its RNAi activity at Day 4, while all other modified siRNAs still maintained >70% of their RNAi activity. Then, significant differences in the duration of RNAi activity among the three modified siRNAs were observed after Day 5. Although S6 still had comparable RNAi activity compared with that of the other two modified siRNAs up to Day 4, the activity declined sharply in the following days and almost completely disappeared at Day 7, just as it did with UM. Meanwhile, SM6 and M6 showed extended duration of RNAi activity; even at Day 7, these siRNAs still showed 44% and 27% RNAi activity, respectively. These results suggested that 2′-O-methylation appeared to be the major contributing factor in the duration of the RNAi activity of SM6. Thus, the rank order of the duration of RNAi activity was SM6 > M6 > S6 > UM siRNA.
Figure 3.
Time-course of RNAi activity of UM and modified (SM6, M6 and S6) siRNAs. siRNAs were transfected with MEND into HeLa-luc cells stably expressing luciferase at 120 nM concentrations at Day 0, and luciferase reporter assays were performed at indicated days.
To examine whether the optimized modification pattern can be applicable to other sequences and targets, we chose an endogenous cancer-related gene, Raf-1. The siRNA against Raf-1 mRNA (Raf-SM6) having the same modification pattern as SM6 was synthesized and evaluated its potency comparing UM siRNA (Raf-UM) (sequences of Raf-1 siRNA are shown in Supplementary Table S3). The siRNAs were transfected into human prostate cancer PC-3 cells at 1, 10, 25 nM concentrations, and the relative Raf-1 mRNA levels were assessed by real-time RT–PCR at 24 h after transfection (Supplementary Figure S2). Raf-SM6 efficiently reduced Raf-1 mRNA in a dose-dependent manner with an IC50 value of 3.4 nM, which was more potent than that of Raf-UM (an IC50 value of 4.7 nM). We also examined the duration of Raf-1 mRNA silencing by Raf-SM6 compared with Raf-UM. The siRNAs were transfected into PC-3 cells at 100 nM concentrations and measured Raf-1 mRNA levels by real-time RT–PCR at Day 2 and Day 6. As a result, Raf-UM and Raf-SM6 showed similar potency and effectively silenced Raf-1 mRNA at Day 2 (Supplementary Figure S3). As was the case with the results of luciferase reporter assay described above, Raf-SM6 remained higher silencing activity compared with Raf-UM at Day 6. These results suggested that our optimized modification pattern by 2′-OMe-4′-thioribonucleoside modified siRNA could be compatible to other siRNA sequences and targets.
We next carried out dose response experiments using these siRNAs. All siRNAs exhibited RNAi activity in a dose-dependent manner (dose response curves are shown in the Supplementary Figure S4) with the IC50 values summarized in Table 2. At Day 2, no difference was observed among the siRNAs studied here with an IC50 values of 22.6–29.2 nM. In contrast, a significant difference was observed in their RNAi activity on Day 6, where the IC50 value for UM was 597 nM, while that of SM6 was 118 nM, indicating that SM6 was 5-fold more potent than UM. The RNAi activity of S6 increased moderately with the IC50 value of 555 nM, which was similar to that of UM. The IC50 value of M6 (252 nM) was between SM6 and S6.
Table 2.
IC50 values and pharmacokinetic data (AUCIE and MRTIE) of siRNAs
| IC50 (nM) |
||||
|---|---|---|---|---|
| Day 2 | Day 6 | AUCIE | MRTIE | |
| UM | 26.6 | 597 | 3.37 | 2.66 |
| SM6 | 22.6 | 118 | 4.99 | 3.61 |
| M6 | 26.3 | 252 | 4.70 | 3.41 |
| S6 | 29.2 | 555 | 3.61 | 2.81 |
We also evaluated the time-course activity quantitatively from a kinetic viewpoint. We used the pharmacokinetic parameters, that is, the area under the curve of the inhibitory effect (AUCIE) as an index of the total intensity of RNAi activity and the mean response time of the inhibitory effect (MRTIE) as an index of its duration, which were proposed by Takahashi et al. (20). These parameters can be calculated by simple numerical integration using time-course experimental data. Table 2 summarizes the AUCIE and MRTIE values of the siRNAs studied here. Both AUCIE and MRTIE values of all three modified siRNAs were higher than those of the UM siRNA. The AUCIE values of UM and SM6 were calculated to be 3.37 and 4.99, respectively, indicating that, in terms of total RNAi potency, SM6 was 1.53-fold more potent than UM, and 1.1- and 1.44-fold more potent than M6 and S6, respectively. The MRTIE value of SM6 provided the numerical confirmation of the duration of its RNAi activity. Thus, SM6 had the highest value compared to the UM siRNA, M6 and S6. Together, these parameters provided confirmation that the 2′-OMe-4′-thioribonucleoside modification improved not only its RNAi activity but also its duration.
Bartlett and Davis examined the duration of RNAi mediated gene-silencing activity in vitro and reported that the activity of UM siRNA lasted less than 1 week at ∼100 nM siRNA concentrations in rapidly dividing cell lines (22). They also observed that, despite its nuclease stability, chemically modified siRNA (siSTABLEv2, Dharmacon) did not show any significant increase in the duration of RNAi activity relative to the UM siRNA in HeLa cells (23). They suggested that stabilization of siRNA against nuclease degradation by chemical modification is important in preventing siRNA degradation in the extracellular environment, such as the bloodstream. However, once the siRNA has reached the cytosol of the target cells, it does not offer any advantages over UM siRNAs with respect to the duration of RNAi activity. Likewise, Collingwood et al. demonstrated the duration of the gene-silencing activity using a 2′-OMe modified Dicer substrate siRNA (a duplex composed of a 25-mer sense strand and a 27-mer antisense strand) compared with its UM duplex. As a result, no significant difference in duration of silencing was observed between the UM and 2′-OMe modified duplexes (24). In contrast to these reports, the 2′-OMe-4′-thioribonucleoside modified siRNA (SM6) provided a substantial improvement in the duration of RNAi activity. As described above, 2′-OMe-4′-thioRNAs showed significant stability against nuclease degradation; therefore, we hypothesized that the duration effect of the 2′-OMe-4′-thioribonucleoside modified siRNAs might arise from the intracellular stability of the duplex.
Intracellular nuclease stability of the siRNA leads to long-term RNAi activity
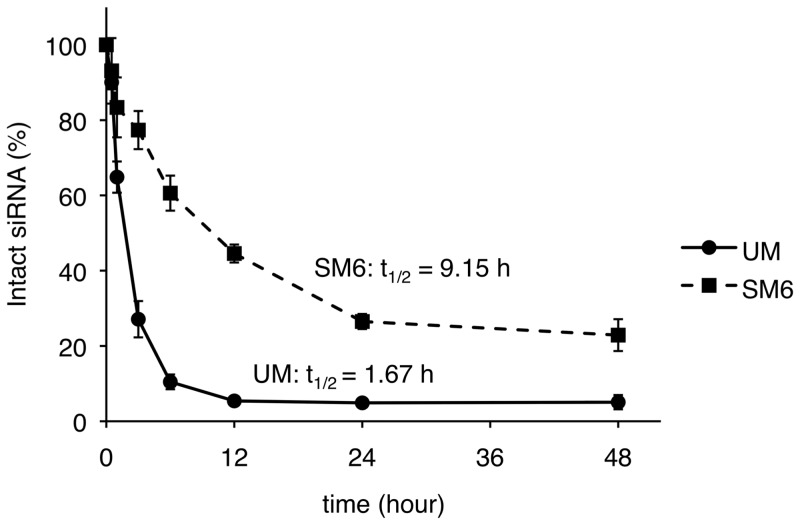
Thus far, a wide variety of chemical modifications have been reported to improve the thermal stabilization of siRNA duplexes and their resistance toward nuclease degradation. Among them, the 2′-OMe modification is well known to increase nuclease resistance without interrupting RNAi activity (6–8). On the other hand, we and other groups have found that 4′-thioribonucleoside modified RNAs have high resistance toward nuclease degradation (15,25). Furthermore, we noticed that 4′-thioribonucleoside modified siRNAs showed durable RNAi activity compared with the corresponding UM siRNAs (13). Therefore, we next carried out a comparison of nuclease stability of SM6 and the UM siRNA in 50% freshly prepared mouse serum (Figure 4, the results of PAGE are presented in the Supplementary Figure S5). SM6 showed a 5.5-fold higher stability with a half-life of 9.15 h compared with that of UM, which gave a half-life of 1.67 h. Thus, these results indicated that the 2′-OMe-4′-thioribonucleoside modifications caused the increase in siRNA stabilization in mouse serum.
Figure 4.
Stability of siRNAs in 50% mouse serum.
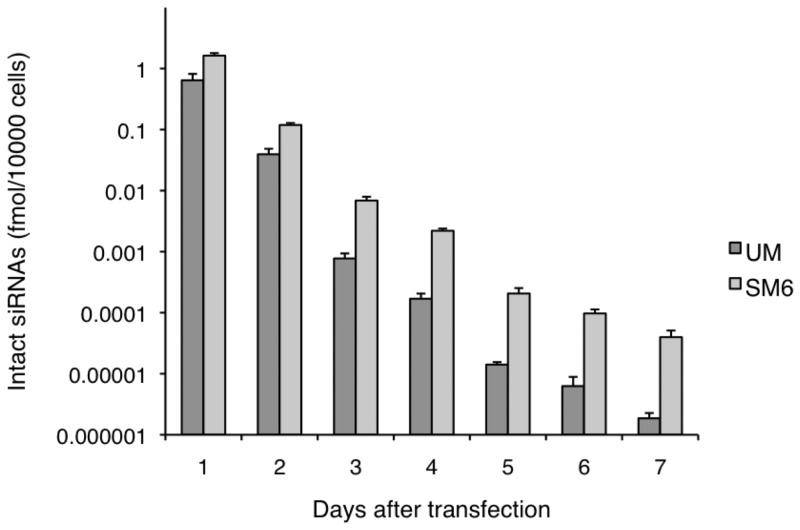
In order to determine whether the 2′-OMe-4′-thioribonucleoside modified siRNA actually remained intact which caused the persistent RNAi activity in cells, we next set out to quantify the siRNA in the cells. Recently, Landesman et al. developed a novel method for quantitating siRNA from cell lines, tissues and plasma (26). The method, Heating-in-Triton quantitative reverse transcription PCR (HIT qRT–PCR), is based on the stem-loop RT–PCR technique and allows a highly sensitive quantification of chemically modified siRNAs. By using this method, we estimated the amount of transfected siRNAs in HeLa-luc cells. Either 120 nM of SM6 or UM was transfected into HeLa-luc cells, and siRNA concentrations were measured by monitoring the amplification of the antisense strand of each siRNA by qRT–PCR over 7 days (Figure 5). The amount of SM6 detected at Day 1 was 2.5 times larger than that of UM siRNA (1.63 fmol/10 000 cells and 0.64 fmol/10 000 cells, respectively), even though there was little difference in RNAi activity measured by luciferase reporter assay described above between SM6 and UM siRNA. The differences became larger as time passed and more than 20 times the amount of SM6 molecules remained in the HeLa-luc cells compared to the UM siRNA on Day 7. We thus concluded that the intracellular stability of the modified siRNA caused by its nuclease resistance property contributed significantly to the duration of the RNAi activity of the 2′-OMe-4′-thioribonucleoside modified siRNA.
Figure 5.
Quantification of intracellular siRNAs using HIT qRT–PCR.
In addition to the intracellular stability of siRNA, several other factors can influence the duration of RNAi activity, including rate of cell division (22), siRNA transfection efficiency and half-life of the target protein. In order to better understand the duration of RNAi activity, we should keep in mind these factors. Chemical modification of siRNA is thought to be an easy way to alter the property of siRNA and improve the activity and duration of gene-silencing. With this study, we have successfully demonstrated the importance of intracellular stability of siRNA for duration of RNAi activity.
CONCLUSION
We have evaluated the effect of chemical modification of siRNA using 2′-OMe-4′-thioribonucleosides on RNAi activity. Optimization of both the number and position of 2′-OMe-4′-thioribonucleoside incorporation afforded modified siRNAs with more potent and persistent RNAi activity, where the modified siRNAs have three residues each on both ends of the sense strand, compared with the UM siRNAs. Further investigation of the duration of RNAi activity revealed that the 2′-OMe-4′-thioribonucleoside modified siRNAs showed the most persistent RNAi activity compared with 2′-OMe, 4′-thioribonucleoside modified and UM siRNAs. The dose-response experiments provided further confirmation of this finding, as well as the rank order of the duration of their RNAi activity, which is 2′-OMe-4′-thioribonucleoside > 2′-OMe > 4′-thioribonucleoside > UM.
Thus far, a number of reports have demonstrated the long-term RNAi activity of chemically modified siRNA both in vitro (9,10,23,27) and in vivo (23,28). However, the advantages of nuclease-resistant siRNAs inside the cell cytoplasm remain less well defined. Our studies revealed that the nuclease resistance of siRNA is a dominant factor in prolonging gene-silencing activity. Therefore, the remarkable nuclease resistance of the 2′-OMe-4′-thioribonucleosde modification facilitates the intracellular stability of siRNA leading to the duration of RNAi activity.
From a therapeutic viewpoint, this has important implications. For instance in patients requiring repeated administration of drugs, it may be beneficial to silence a target gene for longer time, since long-term treatment often produces unintended toxicity. Thus, prolonging RNAi activity is a major consideration for the therapeutic application of siRNA. To this aim, we have demonstrated here the potential advantage and significance of the 2′-OMe-4′-thioribonucleoside modification of siRNA, which leads to long-lasting RNAi activity. Overall, our data provide valuable insights into the therapeutic application of chemically modified siRNA.
Many questions still remain to be answered before this technology can be successfully applied to therapeutic use, such as unintended off-target effects, activation of innate immune response and target delivery of siRNA. Further study to address these concerns and problems not only in vitro but also in vivo using 2′-OMe-4′-thioribonucleoside modified siRNAs are currently under way. Details of these results will be published elsewhere.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–5.
FUNDING
Funding for open access charge: Grant-in-Aid for Scientific Research from the Japan Society for Promotion of Science (18109001 and 23249008).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Ms Y. Misawa (Hokkaido University) for technical assistance.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotide. 2009;19:89–101. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 5.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–931. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 7.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann T, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 9.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manoharan M, Akinc A, Pandey RK, Qin J, Hadwiger P, John M, Mills K, Charisse K, Maier MA, Nechev L, et al. Unique gene-silencing and structural properties of 2’-fluoro-modified siRNAs. Angew. Chem. Int. Ed. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Judge AD, Bola G, Lee ACH, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Hoshika S, Minakawa N, Kamiya H, Harashima H, Matsuda A. RNA interference induced by siRNAs modified with 4’-thioribonucleosides in cultured mammalian cells. FEBS Lett. 2005;579:3115–3118. doi: 10.1016/j.febslet.2005.04.073. [DOI] [PubMed] [Google Scholar]
- 14.Hoshika S, Minakawa N, Shionoya A, Imada K, Ogawa N, Matsuda A. Study of modification pattern–RNAi activity relationships by using siRNAs modified with 4’-thioribonucleosides. ChemBioChem. 2007;8:2133–2138. doi: 10.1002/cbic.200700342. [DOI] [PubMed] [Google Scholar]
- 15.Dande P, Prakash TP, Sioufi N, Gaus H, Jarres R, Berdeja A, Swayze EE, Griffey RH, Bhat B. Improving RNA interference in mammalian cells by 4’-thio-modified small interfering RNA (siRNA): Effect on siRNA activity and nuclease stability when used in combination with 2’-O-alkyl modifications. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- 16.Watts JK, Choubdar N, Sadalapure K, Robert F, Wahba AS, Pelletier J, Pinto BM, Damha MJ. 2’-Fluoro-4’-thioarabino-modified oligonucleotides: conformational switches linked to siRNA activity. Nucleic Acids Res. 2007;35:1441–1551. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi M, Minakawa N, Matsuda A. Synthesis and characterization of 2’-modified-4’-thioRNA: a comprehensive comparison of nuclease stability. Nucleic Acids Res. 2009;37:1353–1362. doi: 10.1093/nar/gkn1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai Y, Hatakeyama H, Sato Y, Akita H, Takayama K, Kobayashi S, Futaki S, Harashima H. Endosomal escape and the knockdown efficiency of liposomal-siRNA by the fusogenic peptide shGALA. Biomaterials. 2011;32:5733–5742. doi: 10.1016/j.biomaterials.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 19.Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R, Masuda T, Danev R, Nagayama K, Kogure K, et al. Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process. Biomaterials. 2009;30:2940–2949. doi: 10.1016/j.biomaterials.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y, Yamaoka K, Mishikawa M, Takakura Y. Moment analysis for kinetics of gene silencing by RNA interference. Biotechnol. Bioeng. 2006;93:816–819. doi: 10.1002/bit.20718. [DOI] [PubMed] [Google Scholar]
- 21.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live- animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartlett DW, Davis ME. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Nucleic Acids Res. 2007;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collingwood MA, Rose SD, Huang L, Hillier C, Amarzguioui M, Wiiger MT, Soifer HS, Rossi JJ, Behlke MA. Chemical modification patterns compatible with high potency Dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18:177–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshika S, Minakawa N, Matsuda A. Synthesis and physical and physiological properties of 4’-thioRNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landesman Y, Svrzikapa N, Cognetta A, III, Zhang X, Bettencourt BR, Kuchimanchi S, Dufault K, Shaikh S, Gioia M, Akinc A, et al. In vivo quantification of formulated and chemically modified small interfering RNA by heating-in-Triton quantitative reverse transcription polymerase chain reaction (HIT qRT-PCR) Silence. 2010;1:16. doi: 10.1186/1758-907X-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variation and stabilizing modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakarama T, Charrise K, Ndungo EM, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotech. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.