Abstract
Bromodeoxyuridine (5-bromo-2′-deoxyuridine, BrdU) is a halogenated nucleotide of low toxicity commonly used to monitor DNA replication. It is considered a valuable tool for in vitro and in vivo studies, including the detection of the small population of neural stem cells (NSC) in the mammalian brain. Here, we show that NSC grown in self-renewing conditions in vitro, when exposed to BrdU, lose the expression of stem cell markers like Nestin, Sox2 and Pax6 and undergo glial differentiation, strongly up-regulating the astrocytic marker GFAP. The onset of GFAP expression in BrdU exposed NSC was paralleled by a reduced expression of key DNA methyltransferases (DNMT) and a rapid loss of global DNA CpG methylation, as we determined by our specially developed analytic assay. Remarkably, a known DNA demethylating compound, 5-aza-2′-deoxycytidine (Decitabine), had similar effect on demethylation and differentiation of NSC. Since our key findings apply also to NSC derived from murine forebrain, our observations strongly suggest more caution in BrdU uses in stem cells research. We also propose that BrdU and its related substances may also open new opportunities for differentiation therapy in oncology.
INTRODUCTION
Ongoing neurogenesis in the mammalian brain is thought to depend on neural stem cells (NSC), which reside in few specialized areas of the adult brain, such as the subventricular and subgranular zones (1). Extensive in vivo and in vitro studies involving NSC have shown that these cells are capable of differentiating into all major brain cell types: neurons and glia, namely astrocytes and oligodendrocytes (2). The process of differentiation is associated with the loss of neural stem cell markers, such as the intermediate filament Nestin and the transcription factors Sox2 and Pax6 (2). In this study, we employed NSC derived from mouse embryonic stem (ES) cells, which reliably recapitulate the features of brain NSC (2). These NSC represent a fast growing, self-renewing and homogenous cell line, not susceptible to spontaneous differentiation under normal, fetal calf serum (FCS)-free culture conditions. Controlled exposure of NSC to the growth factor-rich FCS results in a rapid induction of astrocyte-specific intermediate filament GFAP and astrocytic differentiation (2,3). This model system has therefore several advantages compared to neurosphere or ex vivo assays in which self-renewing NSC are continuously undergoing spontaneous differentiation, thus eventually becoming a minority of the total cell population.
Neural stem cells (NSC) were discovered as the only DNA-replicating cells of the mammalian brain, initially through the incorporation of radioactive 3H isotopes (4,5). Later on, bromodeoxyuridine (5-bromo-2′-deoxyuridine, BrdU), a halogen-containing thymidine analogue which can become incorporated into the replicating DNA and detected by immunochemical analysis using specific antibodies (6) has been exploited to detect DNA-synthesizing and therefore proliferatingcells in the brain (7–9), and has since then become the reagent of choice for these uses.
Despite the fact that BrdU was used clinically for assessment of glioma proliferation (10), its genotoxic side effects are also known. Already in the 1960s, BrdU was recognized to cause chromosomal constrictions (i.e. sites of very densely packed chromatin, similarly to centromere regions) in leukocyte cultures (11,12). Currently, BrdU is also employed as a radio-sensitizing agent in oncology, thus leading to generation of anti-proliferative DNA cross-links upon subsequently applied ionizing radiation (13). On its own, BrdU was reported to negatively affect the growth of cancer cells without affecting their viability (14,15). The tenets and caveats of BrdU as a genotoxic agent are known, yet it is still widely used in the neurobiological research studies due to its apparent low toxicity (16).
DNA methylation and demethylation is an important and tightly controlled process in stem cells (17,18). Changes in methylation status of cytosines in so-called CpG islands at gene regulatory regions determine the expression of differentiation-relevant genes (19). DNA methylation is ensured by DNA methyltransferase (DNMT) enzymes, which add methyl groups to CpG cytosines, either de novo (DNMT3a and DNMT3b) or in the newly replicated DNA in order to maintain existing CpG methylation patterns (DNMT1) (18). DNA demethylation is achieved either by exchange of methylated cytosines or by inhibited DNMT activity upon DNA replication (17) and can then stimulate the transcriptional activation of affected genes. Indeed, genome-wide DNA demethylation was shown to be associated with differentiation of neural stem and cancer cells (20,21). So far, BrdU’s possible effect on DNA methylation was rarely studied (22), and in context of stem cells, it has not been apparently addressed yet.
MATERIALS AND METHODS
Derivation of murine neural stem cell lines
Cell lines were derived from murine embryonic stem cells (ESC) based on protocols established by A. Smith’s laboratory (2,23). Initially, ESC were grown in feeder-free conditions on high-glucose DMEM, supplemented with 15% fetal calf serum (FCS, PAN Biotech), recombinant LIF supernatant, 0.1 mM β-mercaptoethanol, 2 mM l-glutamine (l-Gln), 100 U/ml penicillin and 100 μg/ml streptomycin (P/S). In the first step, ESC were seeded on gelatinized dishes in N2B27 culture medium (1:1 DMEM/F12 and Neurobasal medium (Invitrogen), supplemented with 0.5× N2 supplement (Invitrogen) and 0.5× B27 supplement (Invitrogen), 0.1 mM β-mercaptoethanol, l-Gln and P/S. Medium was changed daily without passaging for 7 days. In the second step, colonies were gently dislodged using Accutase (Sigma Aldrich), washed with PBS, gently resuspended in NSC culture medium (see below), then transferred at high density into culture flasks. There, after 2 days of culturing, formation of neurospheres could be observed. Neurospheres were disrupted by Accutase treatment and gentle pipeting and transferred to cell culture dishes in NSC culture medium. Adherently outgrowing NSC were expanded into stable cell lines through several passages (≥20) before being used for experiments.
Cell culture and treatments
Murine ES-derived NSC and murine forebrain NSC, kindly provided by Luciano Conti (2) were grown in Euromed-N cell culture medium (Euroclone), supplemented with 2 mM l-Gln and P/S, 1× N2 supplement (Invitrogen), 20 ng/ml murine EGF and FGF2 (ProSpec, Israel) at 5% CO2 and 37°C. In the case of forebrain NSC, cell culture dishes with coated with 0.2% porcine gelatine (Sigma Aldrich). BrdU, CldU and IdU (Sigma Aldrich) 100× stock solutions were prepared in H2O at 330 µM. 5-Aza-2′-deoxycytidine (5-aza, also known as Decitabine, Tocris Bioscience) was dissolved in water and applied at 0.5 µM final concentration. Astrocyte differentiation and culture medium was DMEM/F12 with l-Gln and P/S, supplemented with 10% FCS.
Gene expression analysis
Total RNA was extracted from live cells with Trizol reagent (Invitrogen), precipitated with isopropanol and ethanol and dissolved in DEPC-treated water (Invitrogen). One micrograms of total RNA [as quantified with NanoVue device, General Electric (GE)] was used for retrotranscription using VILO reverse transcription kit (Invitrogen) according to manufacturer’s instructions and without RNAse treatment. RT-minus reactions (without reverse transcriptase enzyme) were also prepared. Estimated 20 ng of cDNA in 25 µl reaction volume were analyzed in triplicate by quantitative RT-PCR amplification on a Light Cycler 480 system (Roche) using SYBR Green assay (QuantiFast SYBR Green PCR Kit, Qiagen) according to manufacturer’s instructions and for 40 cycles. CT-values were obtained by calculation of the second derivative using Light Cycler 480 software (Roche) and normalized among samples against a housekeeping gene β-2-microglobulin (B2M). RT-minus preparations proved to be negative. Forward and reverse primers (FP and RP) were designed with Roche UniversalProbe Library software against Mus musculus:
B2M: FP: CTGCAGAGTTAAGCATGCCAGTA; RP: TCACATGTCTCGATCCCAGTAGA
DNMT1: FP: CAGAGACTCCCGAGGACAGA; RP: TTTACGTGTCGTTTTTCGTCTC
DNMT3A: FP: ACACAGGGCCCGTTACTTCT; RP: TCACAGTGGATGCCAAAGG
DNMT3B: FP: GAATGCGCTGGGTACAGTG; RP: GCCACCAGTTTGTCAGCAG
Ki67: FP: GCTGTCCTCAAGACAATCATCA; RP: GGCGTTATCCCAGGAGACT
Nestin: FP: CTGCAGGCCACTGAAAAGTT; RP: TCTGACTCTGTAGACCCTGCTTC
Sox2: FP: TGCTGCCTCTTTAAGACTAGGG; RP: TCGGGCTCCAAACTTCTCT
Pax6: FP: GTTCCCTGTCCTGTGGACTC; RP: ACCGCCCTTGGTTAAAGTCT
GFAP: FP: TGGAGGAGGAGATCCAGTTC; RP: AGCTGCTCCCGGAGTTCT
Tuj1: FP: GCGCATCAGCGTATACTACAA; RP: CATGGTTCCAGGTTCCAAGT
MBP: FP: GGCACGCTTTCCAAAATCT; RP: CCATGGGAGATCCAGAGC
Flow cytometry
Fluorescence-activated cell sorting (FACS) acquisition and analysis were performed on BD FACScalibur using CellQuest software. For Hal-dU and 5-methyl cytosine (5me-C) assay, cells were fixed in 75% ethanol (1 h, 4°C). CpG epitope was made accessible by heating the cells in dH2O at 90°C for 5′. Cells were then washed with 1% bovine serum albumin (BSA) in PBS and stained with either mouse anti-BrdU antibody (preference for CldU, #347580, BD Biosciences), rat anti-BrdU (preference for IdU, #ab6326, Abcam) or sheep anti-5me-C antibody (#GTX21884, Genetex), followed by fluorophore-labeled secondary antibody (Invitrogen). For DNA content and apoptotic DNA fragmentation assay, ethanol-fixed cells were stained with propidium iodide (Sigma Aldrich), signal was measured on a linear or logarithmic scale, respectively. Cell cycle phase distribution was calculated using ModFit software.
Immunoblotting
Cells were lysed in NP40 lysis buffer (1% NP40, 50 mM Tris–Cl pH 8, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 1 mM NaF, 100 µM Na2VO4 and protease inhibitor cocktail (Roche). 40 µg of whole cell lysate in Lämmli loading buffer were resolved by SDS-PAGE, transferred to nitrocellulose membranes (Protran) using Biorad electrophoresis systems and probed with primary and secondary antibodies in 5% bovine serum albumin (BSA) and skimmed milk, respectively. Primary antibodies used were: Nestin (#611658, BD Biosciences), GFAP (#Z0334 Dako), Pax6 (#PRB-278P, Covance), Vinculin (#V4505, Sigma Aldrich).
Immunofluorescence microscopy
Cells cultured on glass cover slips were fixed in 4% para-formaldehyde (PFA) in PBS (10′ at room temperature), permeabilized with 0.2% Triton X100, blocked with 0.5% BSA and 0.2% gelatin in PBS, then probed with appropriate primary antibodies and Alexa-fluor 488- and 647-labeled secondary antibodies (Invitrogen). Primary antibodies used were: Nestin (#MAB353, Millipore), GFAP (#Z0334 Dako); Ki67 antibody (Alexa-fluor 488 labelled, #561165, BD Biosciences). For BrdU detection, incubation with DNAse I (NEB) was performed together with the primary anti-BrdU antibody (#347580, BD Biosciences) in order to retrieve the epitope. For 5me-C and BrdU detection, cells were fixed for 5′ in methanol and rehydrated with PBS. Epitopes were retrieved by 2 M HCl treatment for 30′ at 37°C. Residual HCl was inactivated with borax buffer (0.1 M Na2B4O7, pH 8.5), staining was performed as above, using anti-BrdU antibody and anti-5me-C antibody described above. DNA was counterstained with DAPI (Sigma Aldrich). Confocal images were obtained with a Leica TCS SP2 AOBS confocal laser microscope by sequential scanning and processed with Leica LAS AF Lite software; wide field images with an Olympus AX70 upright microscope and processed with ImageJ software.
RESULTS
BrdU induces cell cycle arrest in NSC
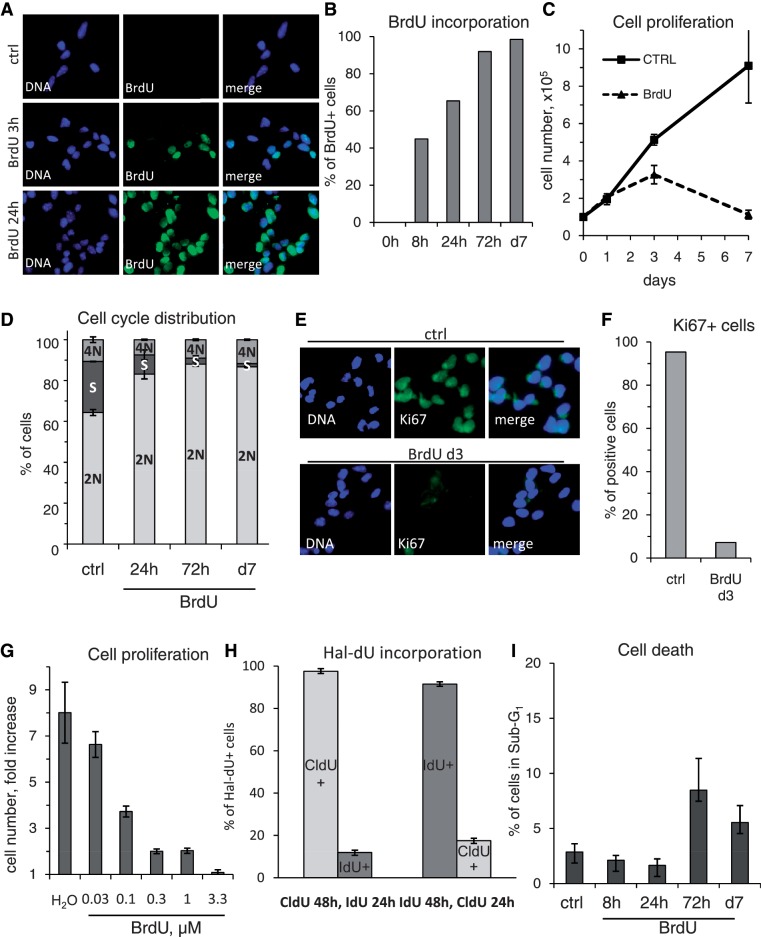
In our study, we employed embryonic stem cell-derived NSC (2), which serve as a reliable model for radial glia stem cells present in the subvetricular zone of the mammalian brain. These cells are self-renewing and undifferentiated over numerous passages (24), while tripotent in their capacity to differentiate in a controlled manner into glial cells (astrocytes and oligodendrocytes, Supplementary Figure S1A) and neurons (Supplementary Figure S1B). In order to address the impact of BrdU exposure on NSC physiology, we applied this reagent at a concentration of 3.3 µM, as commonly used for DNA labeling and cell proliferation studies. In all our BrdU exposure experiments, NSC were continuously cultured in their growth medium purposefully designed to maintain their self-renewal capacity (2), and were not intentionally subjected to any known differentiation stimuli. Immediately after its administration, BrdU became rapidly incorporated into the DNA of NSC and already after 8 h of exposure, nearly half of the cells became BrdU-positive, and at 72 h, practically, all cells showed a strong BrdU signal in their nuclei (Figure 1A and B). Strikingly though, our growth kinetics studies showed that the proliferation of BrdU-treated cells was markedly impaired when compared to control NSC which proliferated exponentially and in fact had to be passaged twice within the 7 days of the experiment (Figure 1D). By analyzing their cell cycle distribution, we observed that BrdU-treated cells progressively accumulated in 2 N state and the percentage of cells in S-phase decreased already after 24 h of BrdU exposure and even more so at day 3 (Figure 1D and Supplementary Figure S2A). Correspondingly, at the same time point, cells ceased to express Ki67, a widely used proliferation marker, as detected both by reduced gene expression and at the single cell level (Supplementary Figure S2B and Figure 1E and F). Both these sets of data indicate that BrdU-exposed NSC exit the cell cycle and arrest in the G1/G0 phase.
Figure 1.
BrdU is a potent cytostatic agent in NSC, with a rather moderate toxicity. (A) Wide-field microscopy analysis of BrdU incorporation kinetics in NSC. At T0 h, cell culture medium was supplemented with BrdU. Cells were fixed at given time points and analyzed by immunofluorescence using a BrdU-specific antibody. (B) Quantification of BrdU incorporation kinetics as representatively shown in (A). Cells displaying a clear nuclear BrdU signal were counted as positive. (C) Growth curve of NSC, treated with BrdU or H2O (ctrl). About 105 cells were seeded for both treatments in parallel in triplicate. At given time points, cells were detached and total cell number was calculated. Note: ctrl cells were split twice, on day 2 and 5 due to confluency, while BrdU treated cells never reached confluency. Error bars show SD. (D) Quantification of DNA content and cell cycle phase distribution in ctrl and BrdU-treated cells at different time points as analyzed by linear propidium iodide signal acquisition by flow cytometry and quantified using ModFit software. Error bars show SD of a triplicate experiment. (E) Wide-field microscopy analysis of immunofluorescence staining of control and BrdU-treated NSC using Alexa-fluor coupled Ki67 antibody. (F) Quantification of Ki67 positive cells in the control and BrdU-treated population as representatively shown in (E). (G) Titration of BrdU effect of cell proliferation. About 105 cells were seeded in medium supplemented with H2O or increasing range of BrdU (0.03–3.3 µM). On day 3, total cell number was assessed and the relative cell number increase versus cells seeded was calculated. Error bars show SD of a triplicate experiment. (H) Flow cytometrical analysis for BrdU-related compounds CldU and IdU which also induce cell cycle arrest in NSC. Cell were treated first with one of the substances for 48 h, then with the other for 24 h, fixed and stained with a mouse-anti-BrdU antibody [preference for CldU (25)] or rat-anti-BrdU antibody [preference for IdU (25)]. Note the strongly reduced degree of incorporation of either compound when added last. Error bars show SD of a triplicate experiment. (I) Flow cytometrical analysis of NSC for apoptosis-associated DNA fragmentation (Sub-G1). Time course study in triplicate of BrdU-treated NSC. Cells were scored for DNA content of less than 2 N, as detected with propidium iodide, measured on log10 scale. Error bars show SD. Note the rather low percentage of apoptotic cells at all time points.
We have next investigated at which concentrations BrdU’s cytostatic effect becomes apparent. Strikingly, within 3 days, as little as 100 nM BrdU was enough to reduce NSC proliferation by more than half, and 300 nM lead to an 8-fold decrease (Figure 1G). Furthermore, we discovered that also the BrdU-related halogenated deoxyuridine (Hal-dU) compounds, 5-chloro-2′-deoxyuridine (CldU) and 5-iodo-2′-deoxyuridine (IdU) displayed similar cytostatic effects on NSC (Figure 1H). We exploited this observation and the possibility to detect these two different Hal-dU by two specific antibodies (25), to quantify the DNA synthesis rates. Proliferating NSC were exposed to either compounds for 48 h and then switched to the other one for the following 24 h. Hal-dU incorporation into cells was detected by either of two BrdU-targeted antibodies from different species origin, of which one has a much stronger affinity to CldU over IdU and another has higher preference for IdU (25). Flow cytometrical analysis revealed that while nearly all cells incorporated the first compound in the initial 48 h, only a small percentage of cells took up the other Hal-dU compound supplied later. The order of addition of a Hal-dU compound was inconsequential (Figure 1H). In summary, we were able to detect a significant decrease in NSC proliferation already after 48 h of Hal-dU exposure. Also, the toxicity of CldU and IdU as determined by the analysis of Sub-G1 DNA fragmentation was low (data not shown), comparable to BrdU, with which we never detected more than 10% of cells displaying apoptotic DNA fragmentation (Figure 1I).
BrdU induces astrocytic differentiation in NSC
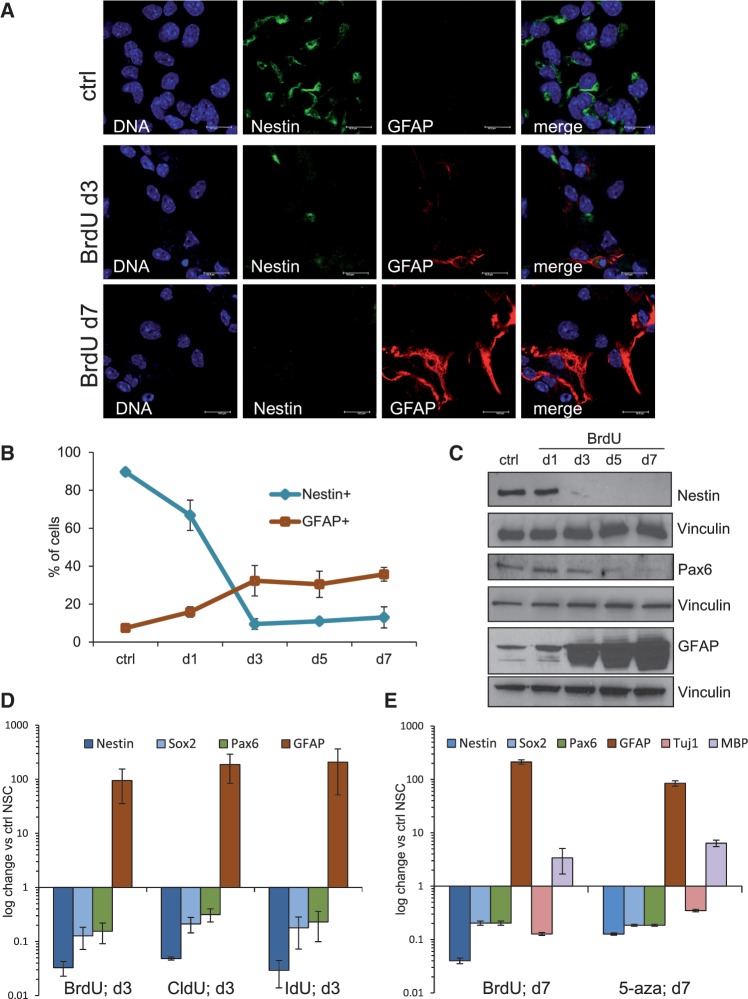
When we inspected with closer scrutiny the nature of the BrdU-induced cell cycle arrest, we detected signs consistent with astrocytic differentiation in these cells. Shortly after the beginning of BrdU treatment, we could observe characteristic changes in cellular morphology (i.e. increase in cell size and astrocyte-like stellate cell appearance), loss of Nestin (a known marker of NSC) and strong up-regulation of GFAP, a marker typical of astrocytes (Figure 2A). Hence, we performed a kinetics analysis of Nestin and GFAP expression by immunofluorescence (IF) and western blotting. Immunofluorescence quantification revealed that 72 h of BrdU exposure are sufficient to induce loss of Nestin expression in nearly 90% of NSC, while at the same time point approximately one-third of NSC has become GFAP positive; these values were roughly retained in the following 4 days of observation (Figure 2B). Also by western blotting, we observed an inverse correlation between the progressive loss of Nestin (and the NSC-specific transcriptional factor Pax6) and the concomitant increase of GFAP protein expression (Figure 2C). In addition, by analyzing by quantitative real time RT-PCR (qRT-PCR) the expression of the stem cell markers Nestin, Sox2 and Pax6, and the differentiation marker GFAP specific for astrocytes (2) at 72 h after BrdU treatment, we observed a strong reduction of Nestin, Sox2 and Pax6 gene expression and a 100-fold up-regulation of the astrocyte marker GFAP (Figure 2D). Consistent with these observations, the BrdU-related compounds CldU and IdU had a similar effect on the loss of stem cell markers and the induction of astrocytic differentiation (Figure 2D).
Figure 2.
NSC treated with BrdU rapidly lose the expression of stem cell markers and differentiate into astrocytes. (A) Confocal microscopy analysis of immunofluorescence staining of control and BrdU-treated NSC. GFAP-specific antibody (red) was used to visualize astrocytic differentiation. Note that while ctrl NSC are Nestin positive (green), the antibody against Nestin shows no signal in BrdU treated cells. Bar: 15 µm. (B) Immunofluorescence-based kinetics of Nestin and GFAP expression in BrdU-treated NSC. Cells were stained with specific antibodies and scored for the Nestin or GFAP signal positivity in triplicate for each time point. Blue line, Nestin positive cells; red line, GFAP positive cells. Error bars show SD of a triplicate experiment. (C) Western blot analysis of NSC for the kinetics of stem cell (Nestin, Pax6) and astrocyte (GFAP) relevant protein expression upon BrdU treatment. Membranes were normalized for vinculin. (D) Quantitative RT-PCR analysis of NSC treated with with BrdU and BrdU-related reagents CldU and IdU. Gene expression of stem cell markers (Nestin, Sox2, Pax6) and astrocyte differentiation marker (GFAP) was normalized against untreated NSC. β2-microglobulin (β2M) was used as housekeeping gene, error bars show SD. (E) Quantitative RT-PCR analysis of NSC treated with BrdU or 5-aza-2′-deoxycytidine (5-aza). Gene expression of stem cell markers (Nestin, Sox2 and Pax6) and differentiation markers (GFAP for astrocytes, Tuj1 for neurons and MBP for oligodendrocytes) was normalized against untreated NSC. β2M was used as housekeeping gene, error bars show SD.
Hal-dUs are modified nucleotides, which become incorporated into DNA during replication. Unexpectedly, however, the uridine residue of BrdU was reported not only to compete with thymidine during DNA replication but also with cytidine (26), thus becoming incorporated against guanine as well as adenine residues of the DNA (27,28). We have tested whether a moderate excess of thymidine or cytidine (10 µM, a triple concentration of that of BrdU) would impact on stem cell physiology. While BrdU lead to a strong drop or decrease in numbers of Nestin or GFAP positive cells, respectively, no such effect was observed with thymidine or cytidine (Supplementary Figure S3). However, modified cytidine analogues, such as cytosine arabinoside (Ara-C) and 5-aza-2′-deoxycytidine (5-aza, also known as Decitabine), were shown to induce differentiation in cancer cells (29) and embryonic stem cells (30,31) or stem cells from the umbilical cord (32), respectively. Ara-C proved highly toxic for NSC (data not shown), also 5-aza had a rather high toxicity in NSC, when compared to BrdU (Supplementary Figure S4A). However, we observed that NSC which survived 5-aza treatment arrested their cell cycle (Supplementary Figure S4B) and underwent glial differentiation. In agreement with this, 5-aza-treated cells showed a clear reduction in Nestin, Sox2 and Pax6 stem cell gene expression as detected by qRT-PCR and a significant up-regulation of GFAP, all to a level similar to BrdU-treated cells (Figure 2E). Furthermore, we detected some ectopic expression of the oligodendrocyte gene MBP, which is commonly absent in NSC, while neuronal marker Tuj1 was not up-regulated in these conditions (Figure 2E).
BrdU induces DNA demethylation
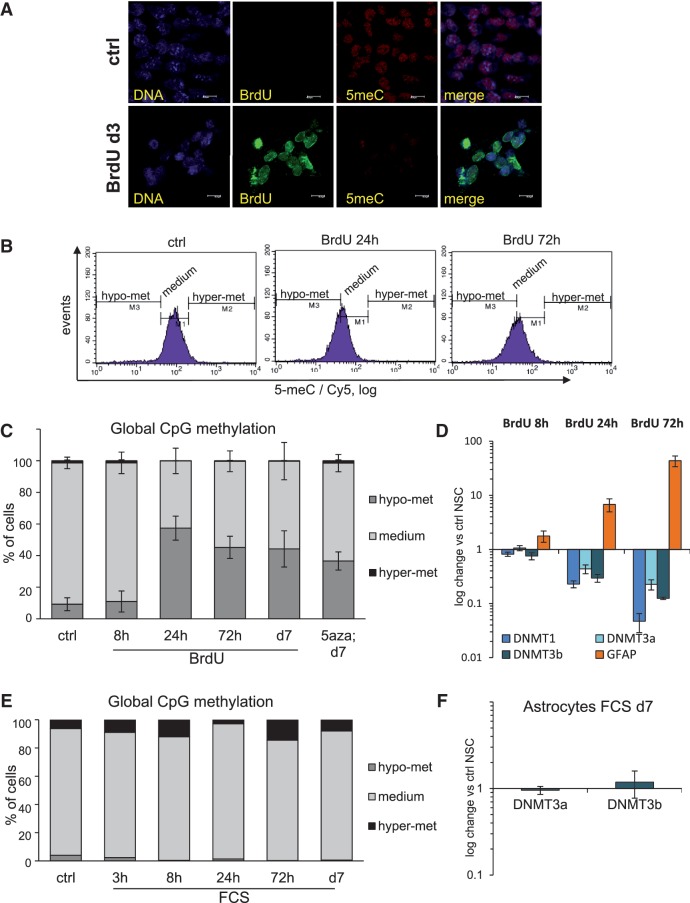
5-aza is primarily used in the cancer therapy for its ability to arrest tumour growth by inducing DNA demethylation (33); indeed, it was confirmed as an efficient DNA demethylating agent in neurosphere assays (21). Following these observations, we analyzed BrdU-exposed NSC for their global DNA methylation status. Strikingly, we detected a strong decrease of the signal generated by a specific antibody against 5-methyl-2′-deoxycytidine (5me-C) in BrdU-treated NSC as compared to control cells, indicating a comparatively reduced CpG methylation status (Figure 3A). In order to further address this effect more quantitatively, we set on to develop a new method for studying relative changes of global CpG DNA methylation, based on specific antibody against 5-methyl-2′-deoxycytidine (5me-C) and FACS. Here, 5me-C epitope is unmasked by hypotonic thermal denaturation of nuclear DNA, at temperatures exceeding the melting threshold of BrdU-substituted DNA (34). Also here we addressed the changes in global DNA methylation relatively to its status in untreated proliferating NSC. We hence arbitrarily set the gates for ‘medium’ DNA methylation so as to include most control untreated NSC, while cell nuclei outside this gate were considered as hypo- or hyper-methylated (Figure 3C). Remarkably, already 24 h after exposure to BrdU, more than half of the NSC population showed a DNA methylation profile below the medium gate, indicating a loss of global CpG methylation. This loss of DNA methylation persisted also at day 3 and even on the day 7 of BrdU treatment (Figure 3C and D). Remarkably, also 5-aza prompted a clear increase in the hypomethylated population of cells, thus confirming both its role as DNA demethylating agent also in a culture of homogenous self-renewing NSC and the reliability of the DNA methylation analytic assay we have developed (Figure 3D). Furthermore, also the other Hal-dUs, CldU and IdU, had a similar effect on the global DNA demethylation of NSC (Supplementary Figure S5A). Noteworthy, in all cases the hypomethylation gate corresponded to the negative control of cells stained with the secondary antibody alone (Supplementary Figure S5A), indicating that the loss of global DNA methylation may be nearly total for the cells in this gate.
Figure 3.
BrdU treatment leads to a loss of global CpG methylation in NSC. (A) Wide-field microscopy analysis of immunofluorescence staining of control and BrdU-treated NSC. BrdU was detected with a specific antibody (green, note the absence of nuclear signal in ctrl cells). Specific antibody for 5-methyl-2′-deoxycytidin (5me-C) was used to detect global DNA methylation at CpG sites (red). Representative images are shown. (B) Representative flow cytometrical analyses of NSC for the relative changes in global DNA methylation upon exposure to BrdU at 24 h and 72 h. Global DNA methylation at CpG islands was detected with same antibody as in (A). Cy5-coupled secondary antibody signal was measured on log10 scale. Gates were set to discriminate ‘medium’ methylated cells, while the population migrations out of this gate were classified as ‘hypo-’ or ‘hyper-methylated’. (C) Statistical quantification of the flow cytometrical data representatively shown in (B). The kinetics of BrdU treated NSC and NSC treated with 5-aza for 7 days are shown. Gates were set as described above; same gate set was used for all measurements of each experiment done in triplicate. Error bars show SD. (D) Expression kinetics of the main DNA methyltransferases (DNMT) and differentiation marker GFAP in BrdU treated NSC, as measured by qRT-PCR analysis and normalized against untreated NSC. β2M was used as housekeeping gene, error bars show SD. (E) Flow cytometrical analysis of NSC for the relative changes in global DNA methylation. Shown is the kinetics of serum (FCS) treated NSC in order to induce astrocytic differentiation. Acquisition and gating were set as in (D). (F) Quantitative RT-PCR analysis of expression of the 2 cell-cycle independent DNA methyltransferases (DNMT) in FCS-treated NSC, normalized against untreated NSC. β2M was used as housekeeping gene, error bars show SD.
Interestingly, in our FACS assay, the quantity of hypomethylated cells observed upon BrdU treatment was as high as in 5-aza-treated cells, used as positive control: this indicates the efficacy of BrdU as a DNA-demethylating agent. At the same time, BrdU proved significantly less toxic than 5-aza (Supplementary Figure S4A). We next aimed to determine whether the DNA methylation machinery is affected in BrdU-treated NSC, and we therefore examined the expression of the main DNA methyltrasferases (DNMTs): DNMT1 and the related enzymes DNMT3a and DNMT3b. Our qRT-PCR analyses revealed that all these three enzymes were unchanged after 8 h of BrdU exposure, but became progressively down-regulated at later time points (Figure 3E and Supplementary Figure S5B). This is insofar remarkable since only DNMT1 is known to be down-regulated in non-proliferating cells, while DNMT3a/3b expression is reportedly cell-cycle independent (18). Moreover, the observed DNMT down-regulation coincided in its rate and temporal progression with the increased expression of the differentiation marker GFAP (Figure 3E). Our data hence correlate the loss of global CpG methylation, the down-regulation of all three key DNMTs and the astrocytic differentiation in BrdU-treated NSC.
Both 5-aza (35) and BrdU (14) were reported to induce DNA damage and activate the DNA damage response [DDR, (36)] signaling in cancer cells. We have recently shown that NSC are fully DDR proficient in response to DNA double strand breaks (3). However, when we tested DDR activation in BrdU-exposed NSC, we could not detect any foci formation of 53BP1, γH2AX or any focal kinase activity of Ataxia telangiectasia mutated (ATM) neither after 24 h nor 3 days (Supplementary Figure S6A and S6B). In fact, ATM was found to be down-regulated at the transcriptional level (Supplementary Figure S6C), in agreement with our recent report on suppressed ATM expression and DDR activity in astrocytes (3). The lack of DDR activity in BrdU-exposed NSC can hence be interpreted as further supporting evidence of their differentiation into astrocytes.
It may be argued that the loss of global DNA methylation is a general feature associated with astrocytic differentiation or exit from cell cycle. If that is the case, demethylation induced by BrdU might be the indirect outcome of arrested cell cycle and/or differentiation. However, when we analyzed the levels of global DNA methylation of NSC exposed to FCS, a treatment known to lead to their rapid cell-cycle exit and terminal differentiation into astrocytes (2,3), we did not detect any enrichment for cells with hypomethylated DNA (Figure 3F). Of course, growth factor-stimulated astrocytic differentiation of NSC is still likely to be associated with promoter-specific DNA methylation changes (37), but our data indicate that this is distinct from the global DNA demethylation observed following BrdU. Moreover, astrocytes, unlike BrdU-exposed NSC, retain stable mRNA levels of the two cell cycle independent DNMT enzymes, DNMT3a and DNMT3b, when compared with proliferating NSC (Figure 3G). Therefore, we conclude that astrocytic differentiation or cell cycle exit per se does not require and is not intrinsically associated with decreased global methylation in NSC.
BrdU impact on forebrain NSC
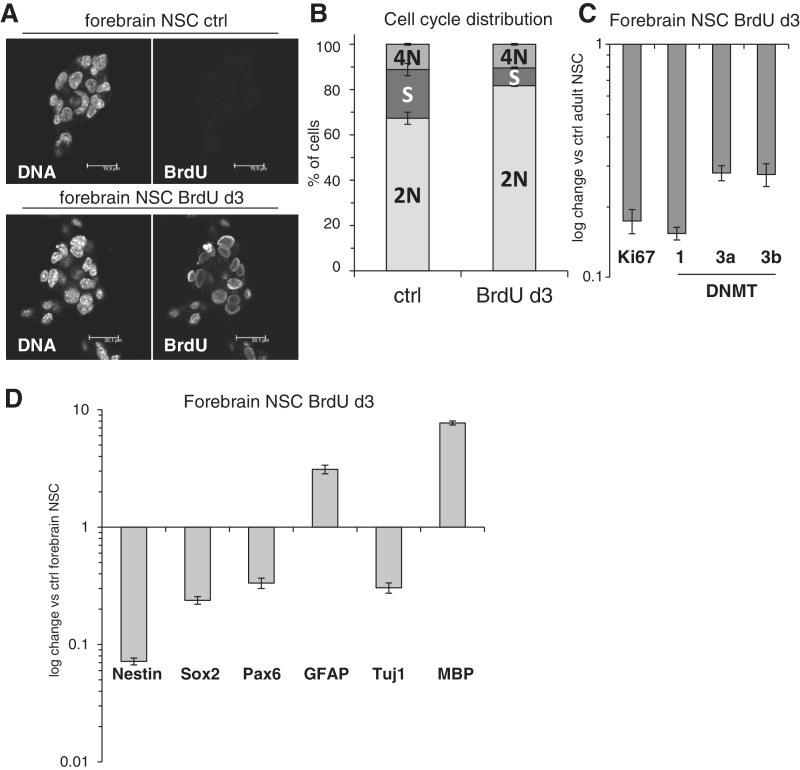
Finally, we were able to recapitulate our key observations in an ex vivo NSC model system. Here, we employed adherently growing murine NSC derived from the mouse forebrain, which in their characteristics and culture conditions widely resemble ES-derived NSC (2). Also these cells readily incorporated BrdU (Figure 4A) and at day 3 showed signs of cell cycle arrest, as determined by a decrease of the percentage of cells in S-phase and the Ki67 gene expression (Figure 4B and C). Remarkably, also in these forebrain NSC, the expression of DNMT1, 3a and 3b was down-regulated upon BrdU exposure, similarly to ES-derived NSC (Figure 4C). Consistent with the results described above, in BrdU-treated forebrain NSC, we detected reduced expression of stem cell markers Nestin, Sox2 and Pax6, while neuronal marker Tuj1 was not induced and, moreover, the glial genes GFAP and MBP were found up-regulated when compared to control forebrain NSC (Figure 4D). We were also able to detect some GFAP up-regulation on protein level (Supplementary Figure S7A) despite the fact that unlike ES-derived and fetal NSC, these forebrain NSC express detectable endogenous levels of GFAP (Supplementary Figure S7B) due to their particular origin from adult brain (1). In summary, also NSC derived from a mouse brain arrest proliferation, lose the expression of DNMTs and stem cell genes and show signs of glial gene induction when exposed to BrdU.
Figure 4.
Forebrain-derived NSC also undergo cell cycle arrest upon BrdU exposure and lose the expression of stem cell and DNMT genes. (A) Confocal microscopy analysis of BrdU incorporation in cultured forebrain NSC, immunofluorescence performed using a BrdU-specific antibody. Bar: 20 µm. (B) Quantification of DNA content and cell cycle phase distribution in ctrl and BrdU-treated forebrain NSC at day 3 as analyzed by linear propidium iodide signal acquisition by flow cytometry and quantified using ModFit software. Error bars show SD of a triplicate experiment. (C) Quantitative RT-PCR gene expression analysis of the proliferation marker Ki67 and the three DNMTs in BrdU exposed forebrain NSC, normalized against untreated forebrain NSC. β2M was used as housekeeping gene, error bars show SD. (D) Quantitative RT-PCR analysis of BrdU-treated forebrain NSC. Gene expression of stem cell markers (Nestin, Sox2 and Pax6) and differentiation markers (GFAP for astrocytes, Tuj1 for neurons and MBP for oligodendrocytes) was normalized against untreated forebrain NSC. β2M was used as housekeeping gene, error bars show SD.
DISCUSSION
In this study, we show that BrdU is readily incorporated into proliferating NSC, without activating canonical DNA-damage response signaling or provoking significant toxicity. However, already after 24 h of exposure of NSC to moderate levels of BrdU, NSC proliferative capacity diminishes and this is paralleled by widespread loss of global DNA CpG methylation. Ultimately, BrdU causes the loss of stem cell characteristics and the profound induction of glial, mainly astrocytic, differentiation. Here, the tendency to undergo glial, and not neural differentiation in the absence of appropriate cues may be explained by the so-called ‘glial identity of the neural stem cells’ (1), thus making an astrocytic differentiation the default choice for these cells. Furthermore, we show that this outcome is stable over time and that both commonly used BrdU-related halogenated deoxyuridines CldU and IdU had comparable effects on DNA methylation and astrocytic differentiation. Finally, we show that BrdU affects forebrain NSC in a very similar manner.
At this stage, it is unclear whether BrdU has a direct or an indirect cytostatic effect on NSC. The former may be brought about by inhibition of DNA polymerase activity and hence DNA replication by this thymidine analogue, while the latter may be caused by DNA demethylation. Of course, both direct and indirect effects may co-operate in arresting cell proliferation. Cytostatic effects of BrdU onto cell lines have been reported before (38). Several studies have demonstrated BrdU’s impact on proliferation rate and cellular senescence in cancer cells (14,15,39). Two recent studies on NSC have shown that also BrdU treatment of neurosphere cultures resulted in their cell cycle arrest, reduction of neurosphere size and cellular senescence (40,41). The latter study also demonstrated that BrdU-exposed neurospheres did not undergo neuronal or oligodendrocytical differentiation when stimulated to do so, while their capacity to undergo growth factor-stimulated astrocytic differentiation was even slightly increased (40). However, our study had the unique important distinguishing feature of employing a homogenous adherent population of tri-potent, self-renewing NSC (2), and, hence, is unaffected by the potentially confounding effects of differentiating cells commonly present in neurospheres. Due to this key advantage, we were able to detect two main changes triggered by BrdU in NSC: loss of global methylation and astrocytic differentiation.
Importantly, this astrocytic differentiation was achieved in an unmodified NSC culture medium (aside from BrdU addition), which stimulates proliferation and self-renewal and inhibits differentiation of NSC, without any additional exposure to any known differentiation-inducing growth factors or stimuli. Indeed, the certain delayed toxicity of BrdU did coincide with the onset of ectopic differentiation, which in turn may have affected the viability of the cells in their pro-proliferative and differentiation-suppressing culture medium. This culture medium conflict may explain why many of BrdU exposed cells remain in a kind of transition state, where their minimal cytoplasmic Nestin levels are not detectable anymore while their differentiation has not proceeded as far as to identify them as GFAP positive. Hence, a number of cells in our immunofluorescence assay were determined as Nestin negative but not GFAP positive. The same may apply to DNA methylation, since also our methylation detection assay may have its limits of sensitivity.
The mechanism by which BrdU affects DNA CpG methylation in a manner similar to 5-aza is presently unclear. Indeed, 5-aza is a modified cytosine, hence being chemically able to interfere in the CpG-methylation pathway (42), while BrdU is generally considered as a thymidine analogue. Interestingly, however, research performed in the 1980s has shown that BrdU competes with cytidine during DNA replication (26), and can be incorporated to a high extent against guanine residues, resulting in a U-G base mismatch, which can be then repaired back to C-G (27,28). Therefore, BrdU, just as 5-aza, can be incorporated in place of cytidine, thus leading to the loss of CpG methylation. The down-regulation of DNMTs may be a secondary effect of BrdU-induced chromatin changes, which however further promotes global CpG demethylation. Indeed, DNA demethylation can be only achieved by replacement of methylated cytosine by the DNA repair machinery or by inhibited activity of DNMT enzymes during the DNA replication process (17) and, in our experiments, differentiation did neatly correlate with the reduced expression of DNMTs. Also elsewhere, decreased DNMT activity and reduced DNA methylation were shown to be associated with NSC differentiation (21), while DNMT1 deficiency (43) or inhibition of global DNA methylation (30–32) were both reported to result in ectopic stem cell differentiation.
Finally, our study suggests a word of caution when using BrdU in stem cells studies and when interpreting the results deriving from its use, especially in living animals. Our data on forebrain NSC strongly implies that also in vivo BrdU may indeed perturb stem cell physiology in a similar way. Long-term BrdU administration to mice in order to detect NSC may hence lead to ectopic differentiation of NSC and to misinterpretation of data, e.g. by assuming the proliferation of differentiated cells. It is also possible that other somatic stem cell types may respond to prolonged BrdU exposure in a similar manner.
Bromodeoxyuridine (BrdU) is already used as a radio-sensitization drug in cancer treatment (13). At the same time, many approaches for cancer treatment aim at targeting DNA methylation (e.g. by 5-aza) (33), or inducing differentiation of so-called cancer stem cells, thus depriving tumours of their repopulating capacity and preventing relapse (44). Interestingly, while no DNA demethylating effect of BrdU was seen in rat hepatoma cells (22), the newly discovered BrdU substitute EdU was shown to have anti-proliferative and apoptotic effects on human glioblastoma cells (45), thus indicating that BrdU may affect differently various cell types. In any case, we suggest that BrdU and its related compounds should be re-evaluated as an anti-cancer agent on the basis of its unique potential to induce stem cell differentiation while showing low toxicity to the patient.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Figures 1–7, and Supplementary Reference [46].
FUNDING
L.S. is an EMBO and DFG (Deutsche Forschungsgesellschaft) fellow. F.d’A.d.F laboratory is supported by FIRC (Fondazione Italiana pr la Ricerca sul Cancro), AIRC (Associazione Italiana per la Ricerca sul Cancro), Cariplo Foundation (grant number 2010.0818), the European Community's 7th Framework Programme (FP7/2007-2013) under grant agreement n° 202230, acronym “GENINCA", HFSP (Human Frontier Science Program) and the EMBO Young Investigator Program. Funding for open access charge: EMBO Young Investigator Program.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Luciano Conti for the critical manuscript reading, L.C. and Elena Cattaneo for advice, discussions and sharing of protocols and reagents, Thomas Burgold and Giuseppe Testa for help with the establishing of NSC cultures, the IFOM-IEO Campus Imaging Facility for reagents and technical advice, Sara Tomassini for experimental help and all F.d'A.d.F. lab members for discussion and feedback throughout this work.
REFERENCES
- 1.Doetsch F. The glial identity of neural stem cells. Nat. Neurosci. 2003;6:1127–34. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- 2.Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider L, Fumagalli M, d'Adda di Fagagna F. Terminally differentiated astrocytes lack DNA damage response signaling and are radioresistant but retain DNA repair proficiency. Cell Death Differ. 2012;19:582–591. doi: 10.1038/cdd.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 1965;124:319–35. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 5.Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc. Natl Acad. Sci. USA. 1993;90:2074–7. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gratzner HG. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: a new reagent for detection of DNA replication. Science. 1982;218:474–5. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn A-M, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat. Med. 1998;4:1313–7. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn H, Dickinson-Anson H, Gage F. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J. Neurosci. 1996;16:2027–33. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowakowski RS, Lewin SB, Miller MW. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 1989;18:311–8. doi: 10.1007/BF01190834. [DOI] [PubMed] [Google Scholar]
- 10.Hoshino T, Nagashima T, Cho KG, Davis RL, Donegan J, Slusarz M, Wilson CB. Variability in the proliferative potential of human gliomas. J. Neuro-Oncol. 1989;7:137–143. doi: 10.1007/BF00165098. [DOI] [PubMed] [Google Scholar]
- 11.Palmer CG. 5-bromodeoxyuridine-induced constrictions in human chromosomes. Can. J. Genet. Cytol. 1970;12:816–830. doi: 10.1139/g70-106. [DOI] [PubMed] [Google Scholar]
- 12.Kaback MM, Saksela E, Mellman WJ. The effect of 5-bromodeoxyuridine on human chromosomes. Exp. Cell Res. 1964;34:182–186. doi: 10.1016/0014-4827(64)90193-4. [DOI] [PubMed] [Google Scholar]
- 13.Berry SE, Kinsella TJ. Targeting DNA mismatch repair for radiosensitization. Semin. Radiat. Oncol. 2001;11:300–315. doi: 10.1053/s1053-4296(01)80067-9. [DOI] [PubMed] [Google Scholar]
- 14.Masterson JC, O'Dea S. 5-Bromo-2-deoxyuridine activates DNA damage signalling responses and induces a senescence-like phenotype in p16-null lung cancer cells. Anticancer Drugs. 2007;18:1053–1068. doi: 10.1097/CAD.0b013e32825209f6. [DOI] [PubMed] [Google Scholar]
- 15.Levkoff LH, Marshall GP, II, Ross HH, Caldeira M, Reynolds BA, Cakiroglu M, Mariani CL, Streit WJ, Laywell ED. Bromodeoxyuridine inhibits cancer cell proliferation in vitro and in vivo. Neoplasia. 2008;10:804–816. doi: 10.1593/neo.08382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taupin P. BrdU immunohistochemistry for studying adult neurogenesis: paradigms, pitfalls, limitations, and validation. Brain Res. Rev. 2007;53:198–214. doi: 10.1016/j.brainresrev.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatada I, Morita S, Kimura M, Horii T, Yamashita R, Nakai K. Genome-wide demethylation during neural differentiation of P19 embryonal carcinoma cells. J. Hum. Genet. 2008;53:185–191. doi: 10.1007/s10038-007-0228-0. [DOI] [PubMed] [Google Scholar]
- 21.Singh RP, Shiue K, Schomberg D, Zhou FC. Cellular epigenetic modifications of neural stem cell differentiation. Cell. Transplant. 2009;18:1197–1211. doi: 10.3727/096368909X12483162197204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer J, Stellwagen RH, Roberts-Ems J, Riggs AD. 5-Methylcytosine content of rat hepatoma DNA substituted with bromodeoxyuridine. J. Biol. Chem. 1977;252:5509–5513. [PubMed] [Google Scholar]
- 23.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 24.Spiliotopoulos D, Goffredo D, Conti L, Di Febo F, Biella G, Toselli M, Cattaneo E. An optimized experimental strategy for efficient conversion of embryonic stem (ES)-derived mouse neural stem (NS) cells into a nearly homogeneous mature neuronal population. Neurobiol. Dis. 2009;34:320–331. doi: 10.1016/j.nbd.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Vega CJ, Peterson DA. Stem cell proliferative history in tissue revealed by temporal halogenated thymidine analog discrimination. Nat. Methods. 2005;2:167–169. doi: 10.1038/nmeth741. [DOI] [PubMed] [Google Scholar]
- 26.Ashman CR, Davidson RL. Bromodeoxyuridine mutagenesis in mammalian cells is related to deoxyribonucleotide pool imbalance. Mol. Cell Biol. 1981;1:254–260. doi: 10.1128/mcb.1.3.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasken RS, Goodman MF. The biochemical basis of 5-bromouracil-induced mutagenesis. Heteroduplex base mispairs involving bromouracil in G × C—-A × T and A × T—-G × C mutational pathways. J. Biol. Chem. 1984;259:11491–11495. [PubMed] [Google Scholar]
- 28.Kaufman ER. The role of deoxyribonucleotide metabolism in 5-bromo-2′-deoxyuridine mutagenesis in mammalian cells. Mutat. Res. 1988;200:149–155. doi: 10.1016/0027-5107(88)90077-2. [DOI] [PubMed] [Google Scholar]
- 29.Takagaki K, Katsuma S, Kaminishi Y, Horio T, Tanaka T, Ohgi T, Yano J. Role of Chk1 and Chk2 in Ara-C-induced differentiation of human leukemia K562 cells. Genes Cells. 2005;10:97–106. doi: 10.1111/j.1365-2443.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 30.Musch T, Öz Y, Lyko F, Breiling A. Nucleoside drugs induce cellular differentiation by caspase-dependent degradation of stem cell factors. PLoS One. 2010;5:e10726. doi: 10.1371/journal.pone.0010726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee S, Bacanamwo M. DNA methyltransferase inhibition induces mouse embryonic stem cell differentiation into endothelial cells. Exp. Cell Res. 2010;316:172–180. doi: 10.1016/j.yexcr.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.So A-Y, Jung J-W, Lee S, Kim H-S, Kang K-S. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS One. 2011;6:e19503. doi: 10.1371/journal.pone.0019503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Issa J-PJ. DNA methylation as a therapeutic target in cancer. Clin. Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 34.David J, Gordon JS, Rutter WJ. Increased thermal stability of chromatin containing 5-bromodeoxyuridine-substituted DNA. Proc. Natl Acad. Sci. USA. 1974;71:2808–2812. doi: 10.1073/pnas.71.7.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 37.Namihira M, Kohyama J, Abematsu M, Nakashima K. Epigenetic mechanisms regulating fate specification of neural stem cells. Philos Trans R Soc Lond B Biol Sci. 2008;363:2099–2109. doi: 10.1098/rstb.2008.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriko M, Nakabayashi K, Suzuki T, Kaul SC, Ogino H, Fujii M, Mitsui Y, Ayusawa D. 5-Bromodeoxyuridine induces senescence-like phenomena in mammalian cells regardless of cell type or species. J. Biochem. 1999;126:1052–1059. doi: 10.1093/oxfordjournals.jbchem.a022549. [DOI] [PubMed] [Google Scholar]
- 39.Minagawa S, Nakabayashi K, Fujii M, Scherer SW, Ayusawa D. Early BrdU-responsive genes constitute a novel class of senescence-associated genes in human cells. Exp. Cell Res. 2005;304:552–558. doi: 10.1016/j.yexcr.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 40.Lehner B, Sandner B, Marschallinger J, Lehner C, Furtner T, Couillard-Despres S, Rivera F, Brockhoff G, Bauer H-C, Weidner N, et al. The dark side of BrdU in neural stem cell biology: detrimental effects on cell cycle, differentiation and survival. Cell Tissue Res. 2011;345:313–328. doi: 10.1007/s00441-011-1213-7. [DOI] [PubMed] [Google Scholar]
- 41.Ross HH, Levkoff LH, Marshall GP, Caldeira M, Steindler DA, Reynolds BA, Laywell E. Bromodeoxyuridine induces senescence in neural stem and progenitor cells. Stem Cells. 2008;26:3218–3227. doi: 10.1634/stemcells.2008-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-Deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou B-BS, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 45.Ross H, Rahman M, Levkoff L, Millette S, Martin-Carreras T, Dunbar E, Reynolds B, Laywell E. Ethynyldeoxyuridine (EdU) suppresses in vitro population expansion and in vivo tumor progression of human glioblastoma cells. J. Neurooncol. 2011;105:485–498. doi: 10.1007/s11060-011-0621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glaser T, Pollard SM, Smith A, Brustle O. Tripotential differentiation of adherently expandable neural stem (NS) cells. PLoS One. 2007;2:e298. doi: 10.1371/journal.pone.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.