Abstract
Trypanosoma brucei undergoes an essential process of mitochondrial uridine insertion and deletion RNA editing catalyzed by a 20S editosome. The multiprotein mitochondrial RNA-binding complex 1 (MRB1) is emerging as an equally essential component of the trypanosome RNA editing machinery, with additional functions in gRNA and mRNA stabilization. The distinct and overlapping protein compositions of reported MRB1 complexes and diverse MRB1 functions suggest that the complex is composed of subcomplexes with RNA-dependent and independent interactions. To determine the architecture of the MRB1 complex, we performed a comprehensive yeast two-hybrid analysis of 31 reported MRB1 proteins. We also used in vivo analyses of tagged MRB1 components to confirm direct and RNA-mediated interactions. Here, we show that MRB1 contains a core complex comprised of six proteins and maintained by numerous direct interactions. The MRB1 core associates with multiple subcomplexes and proteins through RNA-enhanced or RNA-dependent interactions. These findings provide a framework for interpretation of previous functional studies and suggest that MRB1 is a dynamic complex that coordinates various aspects of mitochondrial gene regulation.
INTRODUCTION
The Order Kinetoplastida includes numerous human and animal pathogens such as Trypanosoma brucei, T. cruzi and Leishmania spp., which are the causative agents of African sleeping sickness and nagana, Chagas’ disease and several forms of leishmaniasis, respectively. The kinetoplastids are named for their unique mitochondrial DNA, called the kinetoplast, which is comprised of a catenated network of approximately 50 maxicircles and thousands of minicircles (1). Maxicircles encode two mitochondrial rRNAs and 18 proteins, the majority of which are components of the respiratory complexes. Of 18 mitochondrial mRNAs, 12 undergo post-transcriptional modification through an exceptional process of RNA editing entailing specific uridine (U) insertion and deletion that can double the size of the primary transcript. U insertion/deletion editing is essential for creation of translatable open reading frames in kinetoplastid mitochondria (2,3). Sequence information governing the sites and numbers of uridines to be inserted and deleted is provided by the mostly (with two exceptions) minicircle-encoded guide RNAs (gRNAs). RNA editing is catalyzed by the multiprotein RNA editing core complex (RECC), also known as the editosome. Pre-mRNA and cognate gRNA form an anchor duplex, with the sites to be edited located upstream of the anchor duplex. The central region of the gRNA then acts as the template to direct the editing by the RECC, in a series of reactions including endonucleolytic cleavage of the mRNA, U insertion or deletion, and RNA ligation. Some mRNAs are minimally edited, while a number of mRNAs are extensively edited (pan-edited) and require dozens of gRNAs to act sequentially in the 3′- to 5′-direction along the mRNA to produce the fully edited translatable mRNAs (2,3). In addition to the RECC, a number of editing accessory factors are required for efficient editing. These accessory proteins can modulate editing through substrate production, substrate delivery, and editing processivity, as well as associated gene regulatory processes including RNA turnover. For example, MRP1/2 and RBP16 are essential for editing of a subset of mRNAs, a function that may involve their RNA–RNA annealing properties (4–7). The helicase REH1 facilitates RNA editing progression when multiple gRNAs are involved (8). Another protein, p22, transiently interacts with the editosome and is specifically required for editing of cytochromes oxidase subunit II (9).
A number of other accessory proteins have been characterized, and these were either subsequently or simultaneously identified as components of an ill-defined RNA editing accessory complex. Three groups independently identified this multiprotein accessory complex by immunopurification and mass spectrometry of tagged gRNA-associated proteins, GAP1 and GAP2. This complex was named the mitochondrial RNA-binding complex 1 (MRB1) in T. brucei and gRNA-binding complex (GRBC) in Leishmania major (10–12). This MRB1/GRBC complex, hereafter called MRB1, has an imprecisely defined composition because the initial and subsequent MRB1 purifications identified both common and distinct components (10–14). MRB1 complex proteins characterized to date are essential for growth and affect RNA editing directly or indirectly at multiple steps including initiation (MRB3010, TbRGG2), 3′- to 5′-editing progression (TbRGG2), gRNA stabilization (GAP1/2 and REH2), as well as undefined steps of RNA editing (Tb11.02.5390 and Tb927.6.1680) (12,14–17). Additionally, some MRB1 pulldowns contain components of the MERS1 (mitochondrial edited mRNA stability) and kPAP1 (mRNA polyadenylation) complexes (12,17). The kPAP1 complex protein PPR1, also known as KPAF1, stimulates kPAP1 and RET1 post-editing 3′ A/U long tail addition, which marks the transcripts for translation (18). Thus, the MRB1 complex may play a central role in coordinating RNA editing, stability, polyadenylation and translation.
MRB1 pulldowns consistently identify a common set of proteins; however, copurification of other proteins varies depending on the component of MRB1 that is tagged and the laboratory (10–14). This variability suggests that the MRB1 complex is composed of subcomplexes that have different temporal and physical associations. Characterization of the MRB1 complex is further complicated by the RNA-dependent associations that are likely taking place. To understand the functional roles of this complex, we must first determine the physical associations that make up the MRB1 complex and its component subcomplexes and proteins. To this end, we undertook a large scale yeast two-hybrid analysis to characterize direct protein–protein interactions in the MRB1 complex. Additional affinity purifications of MRB1 components in the presence and absence of nuclease treatment, followed by immunoblots and mass spectrometric analysis, were used to confirm associations in vivo, and determine their RNA dependence. We identify a core MRB1 complex containing six proteins, GAP1, GAP2, MRB3010, MRB5390 (Tb11.02.5390), MRB8620 (Tb11.01.8620) and MRB11870 (Tb927.10.11870). The MRB1 core complex interacts with additional subcomplexes and proteins directly or in a manner enhanced by the presence of RNA. Yet other proteins associate with the MRB1 complex in a completely RNA-dependent manner. This work reveals the basic structural architecture of the MRB1 complex, which is essential in understanding its functions and their spatial and temporal organization. Overall, our results suggest a model in which the MRB1 core complex participates in multifaceted dynamic and RNA-dependent associations that coordinate the numerous roles of the MRB1 complex in mitochondrial RNA biogenesis and editing.
MATERIALS AND METHODS
Yeast two-hybrid screen
The complete open reading frames of 31 putative MRB1 genes were PCR amplified from either T. brucei procyclic form (PF) strains 29–13 genomic DNA or cDNA using Pfx (Invitrogen) or Phusion (Finnzymes) polymerase with the primers listed in the Supplementary Materials and Methods. PCR products were cloned into the yeast two-hybrid Gal4 activation domain (AD) vector pGADT7 and into the Gal4-binding domain (BD) vectors pAS2-1 or pGBKT7 (Clontech). Binary combinations of the plasmids (1 µg each) were cotransformed into the Saccharomyces cerevisiae strain PJ69-4 A using the lithium acetate method. Cotransformed yeast was plated to synthetically defined (SD) media lacking leucine (–leu) and tryptophan (–trp), and incubated for 3 days at 30°C. Next, 5–10 colonies of cotransformed yeast were inoculated onto SD (−leu/−trp) media, to select for the two cotransformed plasmids, and onto SD (−leu/−trp) also lacking histidine (−his), to select for protein–protein interaction. SD (−leu/−trp/−his) plates were supplemented with 1, 2, 3.5 and 5 mM 3-amino-1,2,4-triazole (3-AT), which inhibits PJ69-4 A yeast growth due to leaky expression of the HIS gene. The inoculated plates were incubated 3 days at 30°C. The entire procedure was repeated for each binary combination yielding growth.
Tagged cell line construction (plasmid construction, cell culture, transfection)
For PTP tagging, The C-terminal 400 bp of MRB6070 were cloned into the ApaI and NotI sites of pC-PTP (19) that was previously modified to contain the puromycin resistance gene (pC-PTP-PURO) (20), and the resulting pC-PTP-PURO-MRB6070 was digested at the unique MRB6070 cut site BoxI. The C-terminal 569 bp of MRB5390 were cloned into pC-PTP-PURO ApaI and NotI sites, and the resulting pC-PTP-PURO-MRB5390 was linearized at the unique BsgI site. The C-terminal 583 bp of MRB10130 were cloned into pC-PTP-PURO HindIII and NotI sites and the resulting pC-PTP-PURO-MRB10130 was linearized at the unique NcoI site. The C-terminal 518 bp of MRB11870 were cloned into pC-PTP-PURO using the ApaI and NotI sites, and the resulting pC-PTP-PURO-MRB11870 was linearized at the unique MfeI site. All plasmids were subsequently transfected into procyclic form T. brucei strain 29–13. To construct an RNAi vector for TbRGG2, a 350-nt DNA fragment corresponding to the TbRGG2 3′-UTR was cloned into the BamHI and HindIII sites of p2T7-177 vector. An amount of 50 µg of NotI linearized p2T7-177-TbRGG2 3′-UTR was transfected by electroporation into 29–13 cells, and transformants were selected with phleomycin (2.5 µg/ml). Growth effects of TbRGG2-3′-UTR RNAi were monitored for at least 10 days in the absence or presence of 2.5 µg/ml tetracycline and protein downregulation was verified by immunoblotting. To construct the addback vector for myc-TbRGG2, the entire ORF of TbRGG2 was amplified and cloned into Zero blunt-end vector (Invitrogen). This TbRGG2 fragment was excised and cloned into HindIII and XbaI sites of p2Myc-100 vector (21). An amount of 50 µg of NotI linearized p2Myc-TbRGG2 was transfected into TbRGG2-3′-UTR RNAi cells. Transformants were selected with blasticidin (20 µg/ml) and downregulation of the endogenous TbRGG2 as well as expression of the exogenous myc-TbRGG2 was confirmed by immunoblotting. All primers used in this study are listed in Supplementary Table S1.
PTP purification and immunoprecipitation
Tandem affinity purification was carried out using 5 × 1010 procyclic cells containing the PTP-tagged proteins as described previously (19,22) except for minor modifications. Prior to binding the IgG Sepharose 6 Fast Flow column the supernatant was split in half. One-half was incubated with 50 U of the RNase inhibitor SuperaseIn (Ambion). The other half was treated with a nuclease cocktail containing RNase A (0.1 U/µl), RNase T1 (0.1 U/µl), RNase H (0.01 U/µl), RNase 1 (0.1 U/µl), RNase V1 (0.002 U/µl), DNase 1 (0.002 U/µl) and micrococcal nuclease (0.25 U/µl) (Fermentas) for 60 min on ice. The TEV eluates from the first step of the tandem affinity purification were analyzed by western blot. The tandem affinity purified eluates from the anti-protein C columns were analyzed by mass spectrometry. Myc affinity purification of TbRGG2 was carried out under similar conditions to the published PTP and TAP purification schemes. Mitochondrial extract from Day 4 TbRGG2-2myc overexpression PF cells was incubated with anti-myc polyclonal antibody (ICL laboratories) cross-linked to protein A sepharose beads (GE Healthcare) for 2 h at 4° in the presence of the SuperaseIn inhibitor and Complete Protease inhibitor (Roche). The flow through was collected and the column washed extensively with PBST (phosphate buffered saline with 0.1% NP-40). The antibody-antigen complex was disrupted using elutions of 100 mM glycine (pH 2.5). The eluted complexes were neutralized using 1 M Tris buffer (pH 8.7) and analyzed using western blotting.
Glycerol gradient
Mitochondria were enriched from 1 × 1010 PTP-MRB3010 RNAi procyclic cells as described previously (11). The enriched mitochondria were lysed in 1 ml of the lysis buffer (10 mM Tris at pH 7.2, 10 mM MgCl2, 100 mM KCl, 1 mM DTT, 1 mg/ml pepstatin, 2 mg/ml leupeptin) with 1% Triton X-100 and treated with the nuclease cocktail described above for 60 min at 4°C. The lysate was cleared and loaded on a 11-ml 10–30% glycerol gradient and the gradient centrifuged at 32 000 rpm in a Beckman SW41 rotor for 16 h at 4°C. Twelve 0.5-ml fractions were then collected from the top and sedimentation of MRB proteins was analyzed by western blot.
Western blots
Proteins were transferred onto nitrocellulose membrane and probed with polyclonal antibodies against GAP1 (14), GAP2 (17), TbRGG2 (23) and KREPA6 (24) (a kind gift from Ken Stuart, Seattle BioMed) described previously. Polyclonal antibodies were produced against recombinant MRB11870 and the oligopeptides CNLSNETTSDLKGKENSEESQ (MRB6070) and CGNGPKDGTTIHSGPGGREK (MRB8170) (Bethyl Laboratories). Anti-MRB11870 antibodies were further affinity purified against recombinant MRB11870.
Mass spectrometry
LC–MS/MS analysis was performed by Dr Yuko Ogata at the Fred Hutchinson Cancer Research Center using LTQ mass spectrometer (Thermo Fisher Scientific). The LC system configured in a vented format (25) consisted of a fused-silica nanospray needle packed in-house with Magic C18 AQ 100 A reverse-phase media (Michrom Bioresources Inc.) and a trap containing Magic C18 AQ 200 A reverse-phase media. The peptide samples were loaded onto the column and chromatographic separation was performed using a two-mobile-phase solvent system consisting of 0.1% formic acid in water (A) and 0.1% acetic acid in acetonitrile (B). The mass spectrometer operated in a data-dependent MS/MS mode over the m/z range of 400–1800. For each cycle, the five most abundant ions from each MS scan were selected for MS/MS analysis using 35% normalized collision energy. Selected ions were dynamically excluded for 45 s. For data analysis, raw MS/MS data were submitted to the Computational Proteomics Analysis System (CPAS), a web-based system built on the LabKey Server (26) and searched using the X! Tandem search engine (27) against T. brucei protein database v. 4.0 (ftp://ftp.sanger.ac.uk/pub/databases/T.brucei_sequences/T.brucei_genome_v4/), which included additional common contaminants such as human keratin. The search output files were analyzed and validated by ProteinProphet (28). Proteins and peptides with a probability scores of ≥0.9 were accepted.
RESULTS
Identifying direct protein–protein interactions in the MRB1 complex by yeast two-hybrid analysis
The MRB1 complex has been ascribed multiple functions and overlapping protein compositions (10–14,16,17). However, these functional analyses of individual MRB1 complex components are difficult to interpret without an understanding of the RNA-dependent and RNA-independent protein–protein interactions and subcomplexes that comprise the large MRB1 complex. To gain an understanding of the architecture of this complex, or possibly a consortium of subcomplexes, we began by performing a comprehensive yeast two-hybrid screen to identify direct protein–protein interactions. Here, we included 26 components reported in the three original MRB1 (also known as GRBC) purifications (10–12) and Tb11.01.0880, which was subsequently isolated with the REH2 helicase (13) and MRB3010 (14). In addition, some components of the kPAP1 polyadenylation and MERS1 RNA stability complexes were also associated with the MRB1 complex (12), prompting us to include four additional proteins reported to be components of the kPAP1 and/or MERS1 (but not MRB1) complexes. A complete list of the 31 proteins tested by yeast two-hybrid analysis is shown in Table 1. Throughout the manuscript, we refer to MRB1 components with the last four or five digits of their TriTrypDB designation, preceded by MRB, or by previously reported descriptive names. Unnamed proteins that are kPAP1 or MERS1 components are only listed by the last four digits of their TriTrypDB accession number.
Table 1.
Trypanosoma brucei mitochondrial MRB1 and MRB1-associated proteins analyzed in the comprehensive yeast two-hybrid screen
| Tb Name | GeneDB # | Other Name or # | Complex(es) | Predicted Size (kDa) | LmjF homolog |
|---|---|---|---|---|---|
| MRB3010 | Tb927.5.3010 | MRB1 (10–14)a,b,c,d,e | 57 | LmjF08.1170 | |
| MRB5390 | Tb11.02.5390 | MRB1 (10–14)a,b,c,d,e | 120 | LmjF28.0340 | |
| MRB4160f | Tb927.4.4160 | MRB1 (10–14)a,b,c,d,e | 100g | LmjF31.0640 | |
| MRB8620 | Tb11.01.8620 | MRB1 (10–14)a,b,c,d,e | 53g | LmjF32.3180 | |
| MRB8170f | Tb927.8.8170 | MRB1 (10–14)a,b,c,d,e | 100 | LmjF31.0640 | |
| TbRGG2 | Tb927.10.10830 | Tb10.406.0050, RGGm | MRB1 (10,11,13)a,b,d | 32 | LmjF33.0260 |
| GAP1 | Tb927.2.3800 | GRBC2 | MRB1 (10–14)a,b,c,d,e | 55 | LmjF33.2730 |
| GAP2 | Tb927.7.2570 | GRBC1 | MRB1 (10–12,14)a,b,c,e | 52 | LmjF22.0650 |
| Helicase | Tb927.4.1500 | Hel1500, REH2 | MRB1 (10–13)a,b,d,e | 241 | LmjF34.3230 |
| MRB11870 | Tb927.10.11870 | Tb10.389.1910 | MRB1 (11–14)b,c,d,e | 34 | LmjF33.1250 |
| MERS1 | Tb11.01.7290 | NUDIX Hydrolase | MERS1 (12)e MRB1 (11)b | 44 | LmjF32.2440 |
| MRB1820 | Tb927.3.1820 | MRB1 (11)b | 25g | LmjF25.1740 | |
| MRB1860 | Tb927.2.1860 | MRB1 (10,13,14)a,c,d | 96 | LmjF33.1730 | |
| MRB6070 | Tb927.2.6070 | MRB1 (10)a | 31g | ||
| MRB800 | Tb927.7.800 | MRB1 (10,13,14)a,c,d | 60 | LmjF26.1140 | |
| MRB10130 | Tb927.10.10130 | Tb10.6k15.0150 | MRB1 (10,14)a,c | 61 | LmjF36.4770 |
| MRB8180h | Tb927.8.8180 | Tb927.4.4150 h | MRB1 (10,12,14)a,c,e | 103 | LmjF31.0630 |
| MRB1680 | Tb927.6.1680 | MRB1 (10)a | 58g | LmjF30.0260 | |
| MRB1590 | Tb927.3.1590 | MRB1 (10,11)a,b | 72 | LmjF25.1540 | |
| MRB0880 | Tb11.01.0880 | MRB1 (10,13,14)a,c,d | 18 | LmjF28.1810 | |
| kPAP1 | Tb11.02.5820 | kPAP (12)e | 58 | LmjF28.0780 | |
| 7510 | Tb11.01.7510 | kPAP (12)e | 86 | LmjF32.2670 | |
| 0024 | Tb11.47.0024 | kPAP (12)e MERS1 (12)e | 98 | LmjF27.0630 | |
| PPR5 | Tb927.10.380 | Tb10.70.7960 | MRB1 (12)e | 39 | LmjF21.1620 |
| 3180 | Tb11.02.3180 | MRB1 (12)e kPAP (12)e MERS1 (12)e | 94 | LmjF24.0830 | |
| PPR1 | Tb927.2.3180 | kPAF1 | MRB1 (12)e kPAP (12)e | 114 | LmjF18.0010 |
| 0130 | Tb11.02.0130 | MRB1 (12)e kPAP (12)e MERS1 (12)e | 84 | LmjF33.2510 | |
| 3900 | Tb927.10.6850 | Tb10.6k15.3900 | MRB1 (12)e kPAP (12)e MERS1 (12)e | 36 | LmjF36.2310 |
| 5120 | Tb11.02.5120 | MERS1 (12)e | 104 | LmjF28.0040 | |
| MRB2140 | Tb927.6.2140 | Hydratase | MRB1 (11,13)b,d | 28 | LmjF30.0705 |
| TbRGG1 | Tb927.6.2230 | RGG1 | MRB1 (11)b | 88g | LmjF30.0780 |
MRB1 and MRB1-associated proteins identified by multiple groups in five manuscripts:
a,b,c,dProteins are identified as components of indicated complex if present after RNase treatment.
eProteins are identified as components of indicated complex if at least three peptides were identified and greater than half remained after RNase treatment.
fBoth MRB8180/MRB4150 and MRB8170/MRB4160 are the result of a chromosomal duplication in T. brucei (but not L. major), and therefore these two proteins have the same single L. major homolog.
gSome of the predicted sizes for the T. brucei proteins are different from those listed on TriTrypDB because of alternative initiation site utilization.
hMRB8180/MRB4150 are 99% identical and thus are considered identical for the purpose of this study.
Tb, T. brucei; LmjF, L. major.
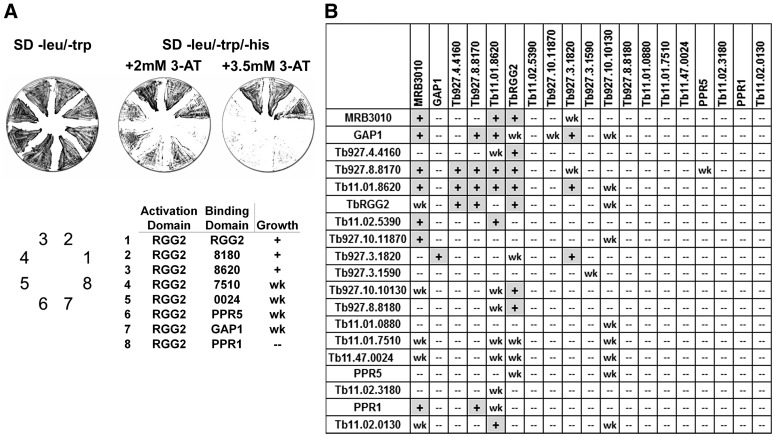
For yeast two-hybrid analysis, genes encoding each of the 31 proteins were cloned into bait and prey vectors, and the resulting plasmids were cotransformed into yeast to screen for all 961 possible binary combinations. Selection was carried out on SD (−leu/−trp/−his) plates containing 1, 2, 3.5 or 5 mM 3-AT to eliminate background growth and establish increasingly stringent growth conditions. We categorized each interaction as strong or weak based on reproducible growth patterns as follows. Strong interactions were defined as those for which cotransformants grew robustly on plates containing 3.5 mM 3-AT, and weak interactions were defined as those for which cotransformants grew on plates containing 2 mM 3-AT, but not at higher 3-AT concentrations. An example of the selection and scoring scheme is shown in Figure 1A, where plasmid cotransformation was confirmed by growth on SD (−leu/−trp) and protein–protein interaction was screened on SD (−leu/−trp/−his) supplemented with 3-AT inhibitor. Interactions between TbRGG2-activation domain (AD) and MRB8180-BD, MRB8620-BD and TbRGG2-BD were scored as strong [shown as (+) in Figure 1] based on their robust growth at 3.5 mM 3-AT. Interactions between TbRGG2-AD and GAP1-BD, 7510-BD and PPR5-BD (latter two are members of the kPAP1 complex proteins), and 0024-BD (member of the kPAP1 and MERS1 complexes) were scored as weak [shown as (wk) in Figure 1], based on growth at 2 mM, but not 3.5 mM 3-AT. The TbRGG2-AD/PPR1-BD interaction was scored as negative due to the absence of growth on 2 mM 3-AT. Using this scoring system, we designated each of the 961 binary interactions as strong, weak, or negative. In Figure 1B, we show a summary of the interactions only for those proteins that exhibited at least one interaction. Of the 31 proteins tested, 19 displayed interactions in at least one direction. The 12 proteins that showed no interactions are GAP2 (Tb927.7.2570), REH2 (Tb927.4.1500), MERS1 (Tb11.01.7290), Tb927.2.1860, Tb927.2.6070, Tb927.7.800, Tb927.6.1680, kPAP1 (Tb11.02.5820), Tb927.10.6850, Tb11.02.5120, Tb927.6.2140 and TbRGG1 (Tb927.6.2230). The absence of interactions in the yeast two-hybrid screen may reflect an RNA-dependent association between a given protein and the MRB1 complex. However, we cannot rule out that poor protein expression or the presence of the AD or BD tag precluded interactions in this assay.
Figure 1.
Yeast two-hybrid analysis of direct interactions in the MRB1 complex. (A) Representative plates showing yeast cotransformed with bait (BD) and prey (activation domain). Yeast cells were grown on synthetically defined (SD) media (−leu/−trp) plates to select for cotransformants and subsequently grown on SD (−leu/−trp/−his) plates with 2 or 3.5 mM 3-AT to select for bait-prey interaction. Growth on 2 mM 3-AT but not 3.5 mM 3-AT was scored as a weak (wk) interaction, and growth on both 2 and 3.5 mM 3-AT was scored as a strong (+) interaction. (B) Summary of yeast two-hybrid results for all proteins showing at least one interaction (19 out of 31). The prey proteins are on the x-axis and the bait proteins are on the y-axis.
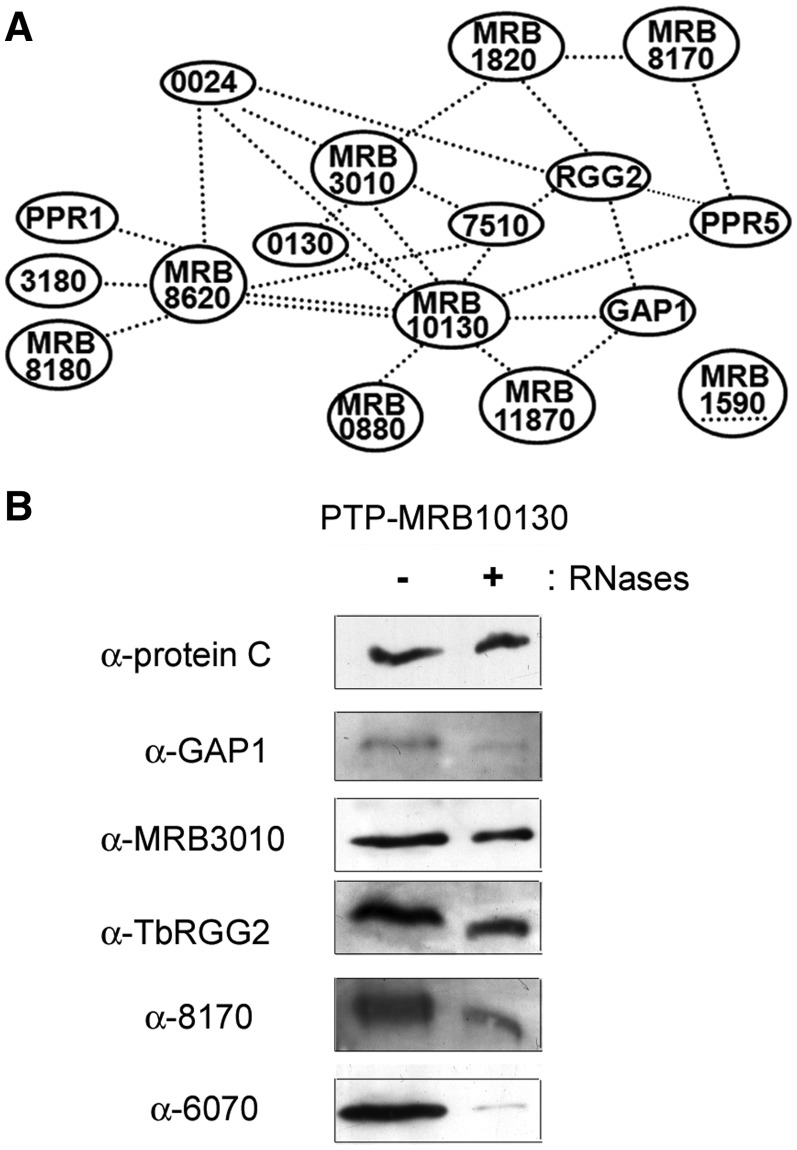
In total, we observed 62 protein–protein interactions, 31 strong and 31 weak. Strikingly, a small number of proteins are responsible for the majority of interactions. The proteins MRB3010, GAP1, TbRGG2, MRB8620, MRB8170 and MRB4160 are involved in 30 of 31 strong interactions and 24 of 31 weak interactions. Notably, the protein MRB10130 is involved in many of the weak interactions (Figures 1B and 3). These results suggest these proteins are important binding partners in the MRB1 complex.
Figure 3.
MRB10130 mediates multiple protein–protein interactions in the MRB1 complex. (A) Schematic representation of all protein–protein pairs exhibiting only weak interactions in the yeast two-hybrid screen. Each black dashed line represents a weak interaction in one direction and an underline represents self-interaction. (B) Purification of PTP-tagged MRB10130 from cell extracts that were either nuclease treated (+ RNases) or left untreated (− RNases). TEV protease eluates of IgG Sepharose 6 Fast Flow columns were analyzed by immunoblot as described in Figure 2.
MRB1 contains a core subcomplex
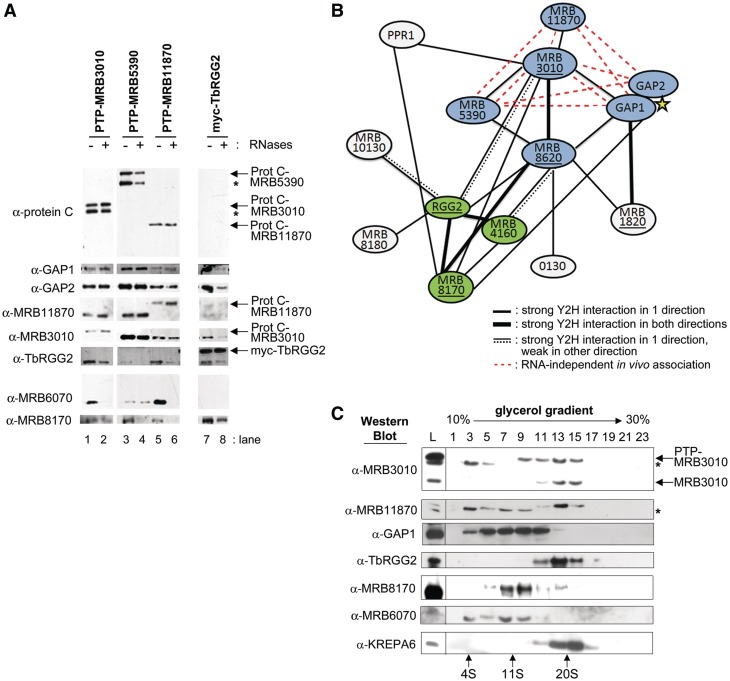
We next set out to confirm a subset of the strong protein–protein interactions identified by the yeast two-hybrid screen using in vivo pull downs, followed by western blot, glycerol gradient, and mass spectrometry analysis. We generated PF T. brucei cell lines in which MRB5390 and MRB11870 were tagged at one endogenous locus with a C-terminal PTP (Protein A-TEV cleavage-Protein C) tag (19). We also utilized PTP-MRB3010 cell lines, which were previously described (14). Initially, we employed all available antibodies to detect proteins that copurify with the PTP-tagged proteins by western blot. Extracts were either incubated with RNase inhibitor (− RNases) or were pretreated with a cocktail containing RNases A, T1, V1, H and 1, DNase 1, and micrococcal nuclease (+ RNases). Untreated and nuclease treated extracts were subjected to IgG Sepharose chromatography and TEV protease cleavage, a protocol that avoids RNases that often contaminate proteins subjected to a second affinity chromatography step (29). Comparison of copurifying proteins from both RNase inhibited and nuclease treated extracts allows us to distinguish RNA-dependent, RNA-enhanced and RNA-independent interactions. Western blot analysis of proteins associated with PTP-MRB3010, MRB5390 and MRB11870 revealed numerous RNA-independent protein–protein interactions, several of which confirmed direct interactions detected by yeast two-hybrid screen. Affinity purified PTP-MRB3010 associated in vivo with GAP1 and MRB11870 in an RNA-independent manner (Figure 2A, lanes 1 and 2), consistent with the direct MRB3010-GAP1 and MRB3010-MRB11870 interactions identified by yeast two-hybrid assay (Figures 1B and 2B). The latter interaction was corroborated by the reciprocal RNA-independent copurification of MRB3010 with PTP-MRB11870 (Figure 2A, lanes 5 and 6). Western blot analysis of affinity purified PTP-MRB5390 demonstrated an RNA-independent association between this protein and MRB3010 (Figure 2A, lanes 3 and 4), again supporting the direct interaction identified by yeast two-hybrid assay.
Figure 2.
The MRB1 core complex and TbRGG2 subcomplex have RNA-independent and RNA-enhanced interactions. (A) Purification of MRB1 complexes from extracts of cells expressing PTP- or myc-tagged MRB1 components that were either nuclease treated (+ RNases) or left untreated (− RNases). Proteins were eluted from IgG Sepharose 6 Fast Flow columns by TEV protease cleavage or immunoprecipitated with anti-myc antibody and electrophoresed on SDS–PAGE gels, followed by immunoblot analysis using an antibody specific to protein C (to detect the PTP-tagged proteins) or antibodies specific to MRB1 complex proteins. Asterisks indicate breakdown products. (B) Schematic representation of all yeast two-hybrid (Y2H) strong interactions. Thin black lines represent a strong interaction in one direction, thick black lines represent strong interactions in both directions, a thin black line with a dashed line represents a strong interaction in one direction and a weak interaction in the other direction, and an underline represents self-interaction. The yellow star is used to indicate that the GAP1 and GAP2 interaction is based on previous work (12,17), not yeast two-hybrid results. The MRB1 core complex is shown in blue and the TbRGG2 subcomplex is shown in green. Red dashed lines represent RNA-independent interactions between core proteins identified in in vivo purifications in (A). (C) Glycerol gradient sedimentation of MRB components after RNase treatment. Mitochondrial extract from PF PTPMRB3010 cells was treated with RNases and fractionated on a 10–30% glycerol gradient. Alternate gradient fractions were electrophoresed on SDS–PAGE and immunoblotted with the indicated antibodies. The mitochondrial extract loaded on the gradient is shown on the left (abbreviated as L). Asterisks indicate breakdown products or a nonspecific band.
Unexpectedly, western blot analysis of nuclease treated extracts also revealed numerous RNA-independent interactions between GAP1, GAP2, MRB3010, MRB5390 and MRB11870 that were not detected by the yeast two-hybrid assay. For example, we observed RNA-independent associations between GAP2 and PTP-MRB3010, PTP-MRB5390 and PTP-MRB11870, although GAP2 showed no two-hybrid interactions (Figure 2A, lanes 1–6). Likewise, PTP-MRB5390 and PTP-MRB11870 associated with GAP1 in an RNA-independent manner even though these proteins lacked direct interactions by yeast two-hybrid analysis (Figure 2A, lanes 3–6). These results suggest that the in vivo interactions between MRB5390, MRB11870 and GAP2 are indirect and mediated by other proteins such as MRB3010. The diagram in Figure 2B summarizes all strong yeast two-hybrid interactions, which are shown with solid black lines, and incorporates RNA-independent associations identified by in vivo tagged protein pulldowns as shown by dashed red lines. Collectively, the yeast two-hybrid screening and in vivo pulldown analyses support the presence of an RNA-independent subcomplex containing at least MRB5390, MRB3010, MRB11870, GAP1 and GAP2, which entails numerous direct and indirect interactions between these proteins (Figure 2B, shown in blue). The existence of such a subcomplex is consistent with all previous MRB1 complex pulldowns, the majority of which contained these five proteins [summarized in (14)]. We were not able to obtain an antibody against MRB8620 to test its copurification with the subcomplex proteins by western blot. However, yeast two-hybrid results demonstrating direct interactions between MRB8620 and MRB5390, MRB3010 and GAP1 (Figures 1B and 2B) in combination with mass spectrometric analyses of MRB1 complex components performed by our laboratories and others are consistent with MRB8620 constituting a sixth member of the subcomplex (Table 2) (10–14).
Table 2.
Proteins associated with RNase-treated MRB1 components were identified by LC–MS/MS
 |
 |
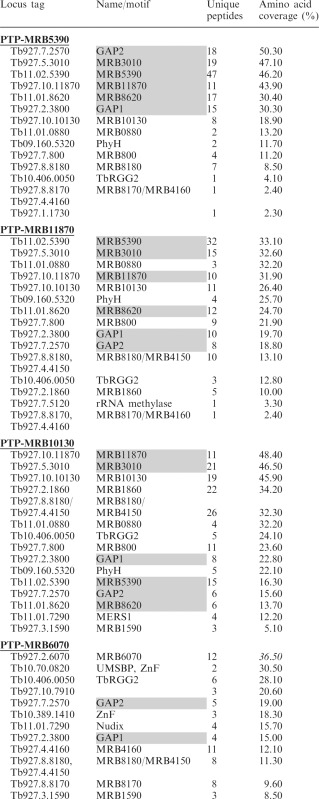
Mass spectrometric analysis of proteins identified in tandem affinity purifications of nuclease treated extracts from PF cells containing endogenously PTP-tagged MRB1 complex components; MRB5390, MRB11870, MRB10130 and MRB6070. The numbers of unique peptides and amino acid coverage for each protein are shown. Proteins highlighted in gray are components of the MRB1 core complex.
To further confirm the core complex composition and identify additional RNA-independent interactions within the MRB1 complex, we performed mass spectrometric analysis of proteins that associate with PTP-tagged MRB5390 and MRB11870 in an RNA-independent manner. PTP-MRB5390 and PTP-MRB11870 were isolated from nuclease treated extracts by tandem affinity purification via sequential IgG Sepharose chromatography, TEV cleavage and mAb anti-Protein C chromatography. PTP pulldowns contained likely contaminants including characterized nuclear and cytoplasmic proteins, and these additional proteins are listed in Supplementary Table S2. Of the 14 unlikely contaminant proteins identified in PTP-MRB5390 purifications, the six proteins with the highest amino acid sequence coverage were PTP-MRB5390 itself along with GAP1, GAP2, MRB3010, MRB8620 and MRB11870 (Table 2). Thus, the proposed core subcomplex components make up the six proteins with the highest coverage. These results are consistent with previous mass spectrometry results for PTP-MRB3010, in which these six proteins were also isolated with the highest amino acid coverage (14). In MRB11870 purifications, the six core subcomplex components were present, comprising 6 of the 10 proteins with the highest amino acid coverage (Table 2), again supporting RNA-independent interactions between these proteins. Mass spectrometry also revealed nuclease-resistant interactions between both PTP-MRB5390 and PTP-MRB11870 and reported MRB1 components TbRGG2, MRB8170/4160, MRB8180, MRB10130 and MRB800. In addition, PTP-MRB5390 copurified with MRB0880, while PTP-MRB11870 copurified with MRB1860, possibly reflecting specific or higher affinity interactions. Taken together, our yeast two-hybrid and in vivo copurification results indicate that a particle comprising MRB5390, MRB3010, MRB11870, MRB8620, GAP1 and GAP2 constitutes an RNA-independent core subcomplex of the larger MRB1 complex.
Next we wanted to determine if the core proteins function only within the core subcomplex, or if they have additional functions outside of the core. We fractionated mitochondrial lysates from nuclease treated PTP-MRB3010 cells on 10–30% glycerol gradients, and analyzed MRB component sedimentation by immunoblot. PTP-MRB3010 sediments in fractions 9–15 and endogenous MRB3010 appears in fractions 11–15, which corresponds to a size of <20 S as compared to the 20 S editosome marker, KREPA6 (Figure 2C, bottom) (14). A breakdown product of PTP-MRB3010 is also evident in fractions 3–5 (Figure 2C, top, asterisk) (14). MRB11870 cosediments with both endogenous and PTP-tagged MRB3010 primarily in fractions 11–15 (Figure 2C). Together with the pull down results in Figure 2B, this suggests that MRB3010 and MRB11870 exist primarily, if not exclusively, in the MRB1 core subcomplex. In contrast, GAP1 (and therefore its heterotetramer partner GAP2) sediments broadly in fractions 3–13 after nuclease treatment (Figure 2C). The small degree of cosedimentation of the GAP proteins with other core complex components suggests GAP1 and GAP2 have additional functions outside of the core subcomplex.
TbRGG2 exhibits RNA-enhanced interactions with core subcomplex and other proteins
TbRGG2 is an RNA-binding protein that impacts both initiation and 3′- to 5′-progression of RNA editing (15,23). In the yeast two-hybrid screen, TbRGG2 displayed numerous strong interactions with both the core subcomplex, as well as with MRB8170, MRB4160, MRB8180 and MRB10130 (Figures 1B and 2B). Regarding interactions with the core, in the yeast two-hybrid screen TbRGG2 bound strongly both MRB3010 and MRB8620 (Figure 1B). In the glycerol gradient fractionation of nuclease treated mitochondrial lysates TbRGG2 cosedimented in fractions 13–17 with the core proteins MRB3010 and MRB11870 (Figure 2C). To further examine interactions between TbRGG2 and the above-described MRB1 core in vivo, we performed anti-TbRGG2 western blots of affinity purified MRB3010, MRB5390 and MRB11870. PTP-MRB3010 pulled down TbRGG2 in both untreated and nuclease treated extracts, although the signal in nuclease treated extracts was markedly diminished (Figure 2A, lanes 1 and 2). Thus, we define the MRB3010–TbRGG2 interaction as RNA-enhanced. Notably, PTP-MRB11870 also pulled down TbRGG2 in an RNA-enhanced manner (Figure 2A, lanes 5 and 6), although these proteins did not interact in the yeast two-hybrid screen. This presumably reflects an indirect MRB11870–TbRGG2 interaction mediated through MRB3010. The core protein PTP-MRB5390 also pulled down TbRGG2 (Figure 2A, lanes 3 and 4), although the interaction appears to be weaker than that observed between TbRGG2 and other core components. In addition, the MRB5390-TbRGG2 interaction may be RNA-dependent, although the apparent absence of signal in the nuclease treated sample may reflect the overall weak signal. It is possible that different core components interact with TbRGG2 somewhat differently, or the PTP tag on MRB5390 may perturb its interaction with TbRGG2. Nevertheless, this interaction is likely indirect as MRB5390 and TbRGG2 did not interact in the yeast two-hybrid screen. The RNA-enhanced nature of the in vivo interactions between TbRGG2 and core components is in striking contrast to the RNA-independent interactions observed between the core components themselves, and further supports the composition of the core. Together, these results suggest that TbRGG2 interacts with the core subcomplex (through MRB3010 and perhaps MRB8620, as suggested by yeast two-hybrid results), but it is not part of the core.
To confirm the numerous TbRGG2 interactions detected in the yeast two-hybrid screen, we generated a PF T. brucei line overexpressing exogenous myc-tagged TbRGG2 in a 3′-UTR-based knockdown background, such that the tagged protein constitutes a large percentage of mitochondrial TbRGG2. Initially, we generated a PTP-tagged TbRGG2 cell line, but the cells had severely altered growth rates, likely a result of improper function of TbRGG2 with the larger tag. Immunopurification of myc-TbRGG2 was carried out using untreated or nuclease treated cell extracts, as previously described for the PTP-tagged proteins. Interestingly, myc-TbRGG2 coprecipitated endogenous TbRGG2 in an RNA-enhanced manner (Figure 2A, lanes 7 and 8). This result suggests that multiple TbRGG2 proteins, or TbRGG2-containing complexes, can bind the same RNA or associated RNAs. The core subcomplex proteins GAP1, GAP2 and MRB3010 also demonstrate RNA-enhanced associations with myc-TbRGG2 (Figure 2A, lanes 7 and 8), consistent with the reciprocal PTP-MRB3010 pulldown described above. The signal for MRB11870 in myc-TbRGG2 immunoprecipitates is low, and it is difficult to discriminate between an RNA-enhanced or RNA-dependent association. These data further support an RNA-enhanced interaction between TbRGG2 and the core subcomplex.
In addition to its interactions with the core subcomplex, TbRGG2 also exhibited strong yeast two-hybrid interactions with MRB8180, MRB10130, MRB4160 and MRB8170, with the latter two displaying strong interactions in both directions. The result of gene duplication unique to T. brucei, MRB4160 and MRB8170 display >80% homology and show a number of the same interactions in yeast two-hybrid screens, including the core protein MRB8620, TbRGG2 and MRB8170 itself (Figure 1B). In addition, MRB8170 interacts with PPR1 and the core proteins MRB3010 and GAP1 (Figures 1B and 2B). Interestingly, the existence of a subcomplex comprising TbRGG2, MRB8170 and MRB4160 (green in Figure 2B) was suggested by Madina et al. (29) based on the isolation of these three proteins with the endonuclease mRNP1, a non-MRB1 complex protein. The TbRGG2-MRB8170 interaction is supported by western blot analysis of myc-TbRGG2 pulldown, which shows an RNA-enhanced TbRGG2-MRB8170 interaction, with a large amount remaining after nuclease treatment (Figure 2A, lanes 7 and 8). Indeed, the diminished signal upon nuclease treatment may reflect in part RNA-dependent interactions between myc-TbRGG2 and TbRGG2–MRB8170–MRB4160 complexes formed with the endogenous TbRGG2 (Figure 2A, compare α-TbRGG2 and α-8170 in myc-TbRGG2 pull down lane).
Interactions between MRB8170 and the MRB1 core subcomplex were probed by anti-MRB8170 western blot analysis of PTP-MRB3010, PTP-MRB5390 and PTP-MRB11870 purifications. The latter two proteins exhibit an RNA-dependent association with MRB8170, while PTP-MRB3010 demonstrated an RNA-enhanced interaction with MRB8170 (Figure 2A, lanes 1–4). These results are consistent with MRB8170 (and perhaps MRB4160 and TbRGG2 in a subcomplex with it) associating with the core, but not comprising part of the core complex. Similar to TbRGG2, yeast two-hybrid results indicate that the interaction of MRB8170 with the core may be mediated by MRB3010 and MRB8620 (Figures 1B and 2B). Collectively, the yeast two-hybrid screen and in vivo pulldown results suggest a model in which TbRGG2, MRB8170, MRB4160 comprise a subcomplex with primarily RNA-enhanced interactions with the core subcomplex. In glycerol gradients, MRB8170 partially cosedimented with the core proteins MRB3010 and MRB11870 and TbRGG2 in fractions 11 and 13; however, MRB8170 peaked in fractions 7 and 9 (Figure 2C). These results suggest that MRB8170 engages in dynamic interactions with the core and TbRGG2.
ARM/HEAT repeat protein MRB10130 mediates multiple protein–protein interactions
In addition to the 31 strong interactions, yeast two-hybrid analysis identified 31 weak interactions amongst the MRB1 complex proteins (Figure 1B, diagrammed in Figure 3A). MRB10130 is involved in 11 of these interactions, demonstrating weak association with nine other proteins, including numerous core components (GAP1, MRB3010, MRB11870, MRB8620), TbRGG2, MRB0880 [identified through its interactions with the REH2 RNA helicase (13) and MRB3010 (14)], PPR5 [a component of both MRB1 and kPAP1 complexes (12)], MRB0130 [also a MERS1 complex component (12)] and 0024 [a component of both kPAP1 and MERS1 complexes (12)]. The observation that MRB10130 mediates numerous, diverse interactions is particularly striking because analysis using the (PS)2-v2 protein structure prediction server predicts that MRB10130 is almost entirely composed of ARM/HEAT repeat units, which often act as a protein–protein interaction platforms (30,31). To determine if these weak interactions involving MRB10130 are relevant in vivo, we generated PF T. brucei cells expressing PTP-tagged MRB10130 and performed pulldowns as described above. Mass spectrometric analysis of PTP-MRB10130 that had been tandem affinity purified from nuclease treated cell extracts identified a number of proteins that associated with MRB10130 in the yeast two-hybrid analysis (Table 2). This included TbRGG2, the only protein with which MRB10130 showed strong interaction, as well as 5 of the 9 proteins with which this protein exhibited weak two-hybrid interactions (MRB11870, MRB3010, GAP1, MRB8620 and MRB0880). PPR5, MRB0130, 7510 and 0024 were not identified in the pulldowns by mass spectrometry, yet they interacted with MRB10130 in two-hybrid assays. Whether these four proteins have transient in vivo interactions with MRB10130 that are lost in the two-step purification, or are simply false positives, has yet to be determined. However, it is notable that we did identify MERS1 by mass spectrometry of MRB10130 purifications, which could reflect an interaction mediated by MRB0130 and/or 0024. Western blot analysis of TEV eluates from PTP-MRB10130 purifications reveals RNA-enhanced interactions between this protein and MRB3010, GAP1, MRB8170 and TbRGG2 (Figure 3B). These pulldowns thus support the in vivo relevance of the weak yeast two-hybrid interactions with both the MRB1 core and the TbRGG2 subcomplex. Collectively, the yeast two-hybrid and in vivo pulldown results suggest a role for MRB10130 in mediating numerous protein–protein interactions in the MRB1 complex, and possibly between MRB1 and other RNA-modifying complexes.
RNA-dependent associations in the MRB1 complex
As stated above, 12 of the 31 proteins tested by yeast two-hybrid analysis exhibited no interactions in this assay, although most have been identified in more than one MRB1 complex pulldown. This suggests that interaction of at least some of these proteins with the MRB1 core is entirely RNA-mediated. To test the RNA-dependent associations of a protein lacking direct interactions by yeast two-hybrid screen, we generated antibodies against MRB6070 (10,13), and analyzed its presence in untreated and nuclease treated MRB1 component pulldowns. MRB6070 was present in pulldowns of core components PTP-MRB3010 and PTP-MRB11870, as well as PTP-MRB10130 pulldowns, when extracts were untreated. However, MRB6070 was essentially absent if the same pulldowns were performed after nuclease treatment (Figure 2A, lanes 1 and 2, 5 and 6; Figure 3B). Western blots of TEV elutions from PTP-MRB5390 pulldowns showed a very weak association with MRB6070 in the presence or absence of RNase treatment; however, no MRB6070 peptides were identified in the mass spectrometric results (Figure 2A, lanes 3 and 4). This is similar to the relatively weak interaction between MRB5390 and TbRGG2, again suggesting that the tag on MRB5390 may affect some of its in vivo interactions. These results show that core proteins, MRB3010 and MRB11870, as well as MRB10130 associate with MRB6070 in a strictly RNA-dependent manner. Consistent with these results, MRB6070 fractionated on glycerol gradients in complexes of ≤11 S, smaller than the core proteins MRB3010 and MRB11870 (Figure 2C).
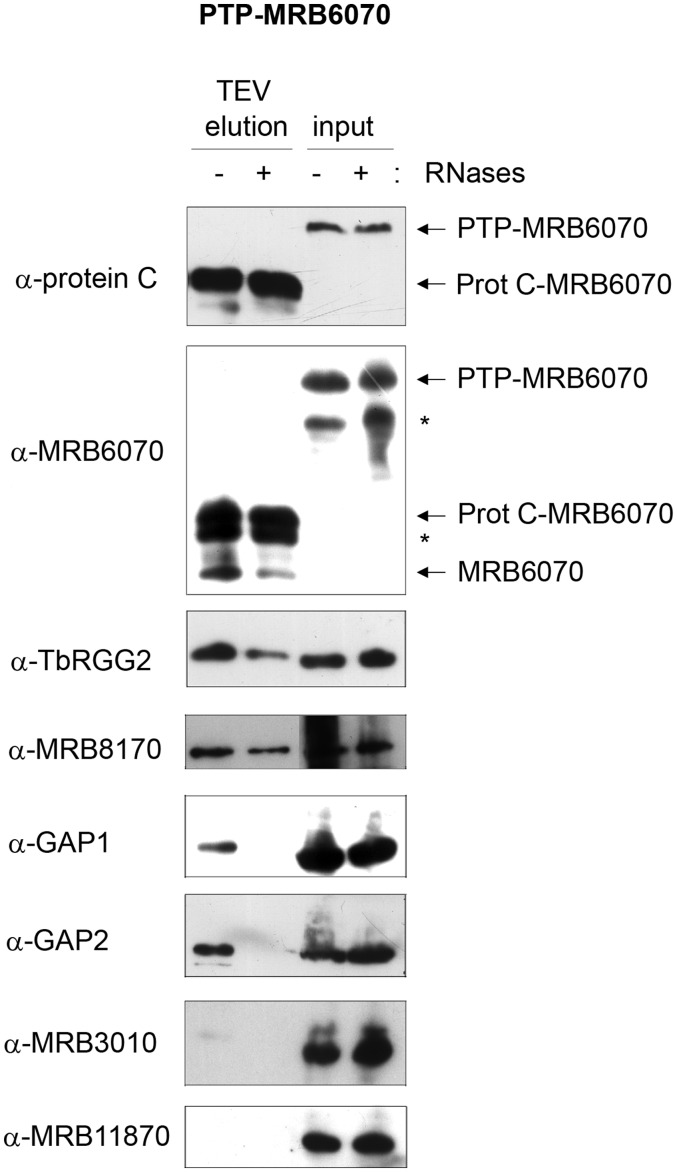
To further examine the interactions involving MRB6070 in the MRB1 complex, we generated a PF cell line expressing PTP-tagged MRB6070 and examined eluates of tandem affinity purification by western blot and mass spectrometry. Western blots of TEV elutions reveal RNA-dependent interactions with core subcomplex GAP1 and GAP2, RNA-enhanced interactions with TbRGG2, MRB8170 and endogenous MRB6070, but no interactions with the core subcomplex proteins MRB3010 and MRB11870 (Figure 4). Consistent with these results, mass spectrometry of the anti-protein C elution from RNase-treated PTP-MRB6070 extracts shows higher coverage for GAP1, GAP2, TbRGG2 and MRB8170, while MRB3010 and MRB11870 were identified by only 1 and 2 unique peptides, respectively (Table 2). The apparent preferential association of GAP1 and GAP2 with PTP-MRB6070 as compared to other core components supports our conclusion that the GAP1/2 heterotetramer can engage in interactions apart from the core subcomplex. MRB6070 also pulled down MRB8180 and MRB4150, which may be a TbRGG2-interacting protein (as suggested by yeast two-hybrid screen), and MRB4160 which is believed to be in a subcomplex with MRB8170 and TbRGG2 [Figure 1 and (29)]. Additional MRB1 components MRB1590 and MERS1, which only rarely copurify with the MRB1 complex are here pulled down with MRB6070 (Table 2). Mass spectrometry also identified a number of additional proteins that are not members of the MRB1 complex copurifying with PTP-MRB6070; most of these proteins contain zinc finger (ZnF) motifs, including the universal minicircle-sequence binding protein (UMSBP). These results are consistent with a model in which MRB6070 interacts with the MRB1 complex through association with a common RNA. It is likely that a number of the other proteins that only purify with a subset of the MRB1 pulldowns also have RNA-dependent associations with the MRB1 complex.
Figure 4.
MRB6070 exhibits RNA-dependent interactions with MRB1 complex proteins. Purification of PTP-tagged MRB6070 from cell extracts that were either nuclease treated (+ RNases) or left untreated (− RNases). Both the cell extracts (input) and the eluates from TEV protease cleavage off IgG Sepharose columns (TEV elution) were analyzed by immunoblot as described in Figure 2. Asterisks indicate breakdown products.
DISCUSSION
MRB1 is a multifunctional complex with reported roles in numerous aspects of mitochondrial gene expression, including RNA editing and differing reported protein compositions (10–17). In this study, we address the architecture of the MRB1 complex by identifying the direct and indirect (protein- and RNA-mediated) interactions between its component proteins to provide a roadmap for understanding the function of the MRB1 complex and constituent subcomplexes and proteins. We identified a six-protein core subcomplex, containing GAP1, GAP2, MRB3010, MRB5390, MRB8620 and MRB11870 that interacts with other subcomplexes and proteins through RNA-enhanced and RNA-dependent interactions. Yeast two-hybrid and immunoblot analyses of affinity purified MRB1 complex proteins confirmed numerous direct and indirect interactions between the core complex proteins, which are independent of RNA. These results validate the consistent copurification of these six proteins in MRB1 pulldowns from various groups (10–14). The core complex also has numerous and likely dynamic interactions with other subcomplexes and proteins involved in various processes including RNA processing, polyadenylation, stabilization and editing (Figure 5). What is unclear at this point is whether the core is a structural coordinator for all of the functions associated with the MRB1 complex, whether it has a distinct function of its own, or if the core is responsible for both of these activities.
Figure 5.
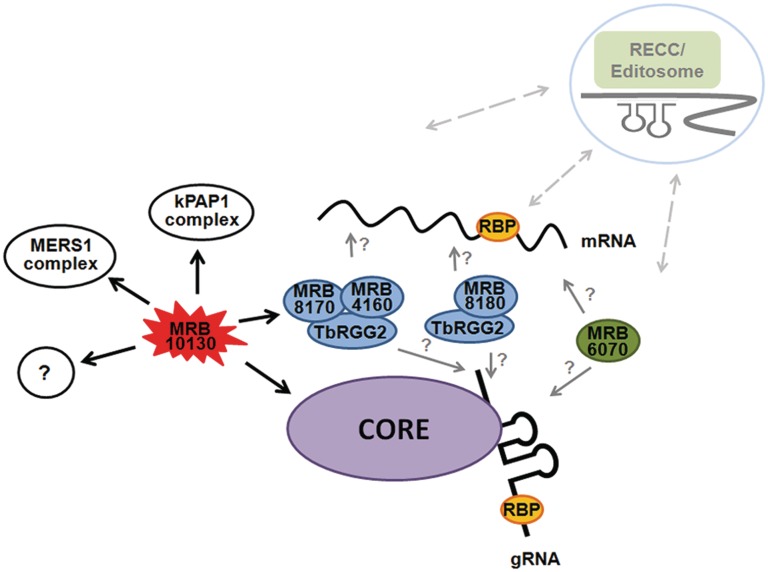
Model for MRB1 complex interactions. The core complex binds gRNA through GAP1/2. The TbRGG2 subcomplexes interact with the core and RNA. MRB6070 associates with the MRB1 complex in an RNA-dependent manner. The gray arrows and question marks indicate remaining questions regarding whether TbRGG2 subcomplexes and MRB6070 bind gRNA, mRNA, or both. MRB10130 binds the core, TbRGG2 subcomplex, MERS1 complex and kPAP1 complex and may coordinate association of these proteins. Because repression of numerous MRB1 components affects RNA editing, the MRB1 complex presumably interacts transiently with the RECC, as suggested by several studies.
Although the function of the core complex as a whole is not known, some of its component proteins have been characterized. Knockdown experiments show that both MRB5390 and MRB3010 affect RNA editing of most transcripts, with MRB3010 having a role at an early step in the editing process (14,16). Knockdowns of GAP1 and GAP2 primarily cause destabilization of gRNAs, which was not seen with the MRB3010 knockdowns (12,14,17). Thus, while these proteins are all components of the core MRB1 complex, they appear to affect different steps of the editing process. This may in part be a result of using knockdowns to analyze protein function. With this method, not only do we observe the in vivo effect of the direct loss of the protein’s activity, but if the protein has binding partners we may also see the effect of the loss of these interactions. Therefore, knocking down the protein results in phenotypes summarizing compromised function of some or all of the proteins it touches, highlighting the critical nature of the structural analysis undertaken in this study.
GAP1 and GAP2 form an α2/β2 heterotetramer that binds gRNAs in vitro and in vivo (12,32), and is likely responsible for mediating the interaction of the core complex with gRNAs. The inhibition of editing at an early stage by MRB3010 knockdowns suggests that one function of the core may be to recruit GAP1/2-bound gRNA to the editing complex. However, it appears that GAP1/2 can also exist as a subcomplex apart from the core as evidenced by their enriched purification in the MRB6070 pulldown separate from the rest of the core (Table 2) as well as sedimentation separate from other core complex components (Figure 2C). Perhaps GAP1/2 exists as a heterotetramer when it initially binds the gRNA and then brings it to the core complex, although additional core proteins could facilitate GAP1/2-gRNA interaction. Interestingly, although GAP1 and GAP2 have 31% sequence identity and 48% sequence similarity, only GAP1 displayed interactions in the yeast two-hybrid screen. Although false negative results occur in yeast two-hybrid screens, our results may indicate that GAP1 is responsible for all protein interactions of the heterotetramer.
A number of other proteins demonstrated interaction with the core complex in our study, including TbRGG2. Indeed, apart from the core complex proteins, TbRGG2 and its interacting proteins are responsible for most of the remaining strong interactions by yeast two-hybrid screen. TbRGG2 and the paralogs MRB8170 and MRB4160 exhibited multiple interactions with each other and, thus, may exist as a subcomplex, although we cannot distinguish between multiple binary complexes or a single subcomplex containing these proteins from our results. Support for a TbRGG2 subcomplex, or subcomplexes, comes from the recent report that only TbRGG2, MRB8170 and MRB4160 copurified with the endoribonuclease mRNP1, which is involved in gRNA processing (29). Together, these data suggest these three proteins exist together apart from the rest of the MRB1 complex. Interestingly, the TbRGG2 subcomplex(es) containing MRB8170 and MRB4160 may not be the only subcomplex carrying TbRGG2. Mass spectrometry results from tandem affinity purifications of MRB10130 (a protein that has a strong interaction with TbRGG2 by yeast two-hybrid) revealed high amino acid coverage for TbRGG2; however, MRB8170 and MRB4160 were not present (Table 2). Rather, in these MRB10130 pulldowns, another protein that had a strong interaction with TbRGG2 in the yeast two-hybrid screen, MRB8180, exhibited high amino acid coverage comparable to that of TbRGG2 (Table 2). Additionally, TbRGG2 and MRB8170 had overlapping but distinct sedimentation patterns (Figure 2C). Thus, TbRGG2 may have multiple subcomplex associations within the MRB1 complex. The complexity of the TbRGG2 interactions in the MRB1 complex adds to the difficulty in determining the role of TbRGG2 in RNA editing. It is known that TbRGG2 is a multifunctional protein that affects gRNA utilization and editing processivity (15). However, the specific functions of the TbRGG2 subcomplex(es) or its binding partners including MRB8170, MRB4160, MRB8180 and MRB10130 are unknown. Although the TbRGG2 subcomplex(es) containing MRB8170 and MRB4160 was found associated with mRPN1, an endonuclease involved in gRNA processing, RNAi results for TbRGG2 suggest that gRNA processing is not one of its primary functions (15). TbRGG2 has two putative RNA-BDs, and in vitro binds RNA (23), so we postulate that TbRGG2 is one of the proteins responsible for MRB1 complex binding to RNA, possibly facilitating gRNA or mRNA association and utilization during editing. Where and when the different TbRGG2-binding partners play a role has yet to be determined, and we speculate that there may be temporal differences in these TbRGG2 associations.
The results discussed thus far suggest the occurrence of dynamic associations within the MRB1 complex, particularly with the core. Likely there are proteins whose purpose is to coordinate these interactions and processes, and an attractive candidate for this role is MRB10130. This protein is predicted on TriTrypDB to consist mostly of an ARM repeat Superfamily SSF48371 domain, and modeling analysis reveals that the majority of the protein forms HEAT repeats. ARM Superfamily proteins (which includes HEAT motif-containing proteins), often act as organizers of protein complexes because the superhelix of α-helices and hydrophobic core formed by tandem ARM repeat units creates a versatile platform for interactions with numerous partners (33–37). In fact, MRB10130 has numerous yeast two-hybrid interacting partners, all of which display weak interactions except for TbRGG2. Immunopurification experiments demonstrate clear association of MRB10130 with the core complex, TbRGG2 subcomplex(es), other MRB proteins and the MERS1 Nudix hydrolase. Could MRB10130 be facilitating binding of specific proteins with the core or the TbRGG2 subcomplex (Figure 5)? Could MRB10130 be coordinating association of the core and TbRGG2 subcomplex with each other? Does MRB10130 play a role in coordinating association of the kPAP1 and MERS1 complexes with the core? Further analysis is necessary to determine if MRB10130 is acting as an MRB1 complex organizer and how cooperative and/or competitive protein binding to MRB10130 plays a role.
Finally, a number of proteins previously identified in MRB1 pulldowns were included in our yeast two-hybrid screen, but exhibited no direct interactions in this assay. In some cases, this may be the result of false negatives; however, we anticipate that a number of these proteins have only RNA-dependent or transient interactions with the MRB1 complex. Likely, numerous RNA-binding proteins interact with gRNAs and mRNAs and through this associate with the MRB1 complex. Whether these proteins are essential for some of the functions carried out by the MRB1 complex has not been addressed. In this study, we identified an RNA-dependent component of the MRB1 complex, termed MRB6070. MRB6070 is predicted to contain 5–6 repeats of a Ran-binding protein 2 type ZnF domain which can act as either a protein recognition motif or a single-stranded RNA-binding motif (38). The fact that the yeast two-hybrid screen revealed no direct protein interactions with MRB6070, yet in pulldown experiments this protein consistently displayed RNA-dependent and RNA-enhanced interactions with other MRB1 proteins, suggests the ZnF motifs in MRB6070 function in RNA binding. Western blot analysis provided additional evidence for RNA-dependent association of MRB6070 with the MRB1 complex (Figures 2A and 3B), yet it remains unknown if MRB6070 is interacting through mRNA, gRNA, or both (Figure 5). When MRB6070 was tandem affinity purified, mass spectrometry of associated proteins revealed primarily the RNA-binding proteins of the MRB1 complex (GAP1, GAP2, TbRGG2). The copurification of TbRGG2 and MRB8170 with PTP-MRB6070 after RNase treatment suggests these proteins are in close proximity on the same RNA, and thus the interactions are partially resistant to RNAse treatment (Figure 4), although we cannot exclude a direct interaction undetected by yeast two-hybrid analysis. The tandem affinity purification of PTP-MRB6070 also identified a number of other likely nucleic acid-binding proteins that were not previously identified as part of the MRB1 complex, including the UMSBP. The association of MRB6070 with UMSBP may be a result of either MRB6070 binding to nascent minicircle-encoded RNA, DNA-binding activity of MRB6070 and resultant interaction with UMSBP on the minicircle, or a role for UMSBP outside of DNA replication in which it interacts with MRB6070.
In conclusion, the present study provides significant insight into the direct and indirect interactions within the MRB1 complex. Our results suggest a dynamic complex comprised of subcomplexes and RNA-binding proteins that are likely subject to temporal and spatial regulation as they carry out numerous functions in mitochondrial gene regulation. The interaction studies described here illuminate potential functions of the MRB1 complex components, such as a direct role for the core in gRNA recruitment during RNA editing and a role for MRB10130 in MRB1 complex organization. They further provide a framework for the future interpretation of functional studies of proteins that comprise this enigmatic complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1 and S2.
FUNDING
National Institutes of Health grants (RO1 AI061580 and RO1 AI077520 to LKR, F32 AI07718501 to J.C.F.); the Grant Agency of the Czech Republic (204/09/1667 to J.L.); the Praemium Academiae award to J.L.; and RNPnet FP7 program (289007 to J.L.). Funding for open access charge: NIH (grant NIAID AI077520).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Achim Schnaufer and Fred Ponticelli for advice on yeast two-hybrid screens and Dr Ken Stuart for the gift of anti-KREPA6 antibodies.
REFERENCES
- 1.Liu T, Bundschuh R. Model for codon position bias in RNA editing. Phys. Rev. Lett. 2005;95:088101. doi: 10.1103/PhysRevLett.95.088101. [DOI] [PubMed] [Google Scholar]
- 2.Lukeš J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 3.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Fisk JC, Presnyak V, Ammerman ML, Read LK. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol. Cell Biol. 2009;29:5214–5225. doi: 10.1128/MCB.00520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumacher MA, Karamooz E, Zíková A, Trantírek L, Lukeš J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 6.Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ammerman ML, Fisk JC, Read LK. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA. 2008;14:1069–1080. doi: 10.1261/rna.982908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F, Herrera J, Zhou S, Maslov DA, Simpson L. Trypanosome REH1 is an RNA helicase involved with the 3'-5' polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc. Natl Acad. Sci. USA. 2011;108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprehe M, Fisk JC, McEvoy SM, Read LK, Schumacher MA. Structure of the Trypanosoma brucei p22 protein, a cytochrome oxidase subunit II-specific RNA-editing accessory factor. J. Biol. Chem. 2010;285:18899–18908. doi: 10.1074/jbc.M109.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panigrahi AK, Zíková A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Hashimi H, Zíková A, Panigrahi AK, Stuart KD, Lukeš J. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J. Biol. Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammerman ML, Hashimi H, Novotná L, Čičová Z, McEvoy SM, Lukeš J, Read LK. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA. 2011;17:865–877. doi: 10.1261/rna.2446311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA. 2010;16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acestor N, Panigrahi AK, Carnes J, Zíková A, Stuart KD. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimi H, Čičová Z, Novotná L, Wen YZ, Lukeš J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol. Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimanski B, Nguyen TN, Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisk JC, Sayegh J, Zurita-Lopez C, Menon S, Presnyak V, Clarke SG, Read LK. A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 2009;284:11590–11600. doi: 10.1074/jbc.M807279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulah CC, Pelletier M, Read LK. Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA. 2006;12:1545–1555. doi: 10.1261/rna.90106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunzl A, Schimanski B. Tandem affinity purification of proteins. Curr. Protoc. Protein Sci. 2009 doi: 10.1002/0471140864.ps1919s55. Chapter 19, Unit 19 19. [DOI] [PubMed] [Google Scholar]
- 23.Fisk JC, Ammerman ML, Presnyak V, Read LK. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal. Chem. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 26.Rauch A, Bellew M, Eng J, Fitzgibbon M, Holzman T, Hussey P, Igra M, Maclean B, Lin CW, Detter A, et al. Computational Proteomics Analysis System (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments. J. Proteome Res. 2006;5:112–121. doi: 10.1021/pr0503533. [DOI] [PubMed] [Google Scholar]
- 27.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 29.Madina BR, Kuppan G, Vashisht AA, Liang YH, Downey KM, Wohlschlegel JA, Ji X, Sze SH, Sacchettini JC, Read LK, et al. Guide RNA biogenesis involves a novel RNase III family endoribonuclease in Trypanosoma brucei. RNA. 2011;17:1821–1830. doi: 10.1261/rna.2815911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 32.Aphasizheva I, Aphasizhev R. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol. Cell Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giesecke A, Stewart M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J. Biol. Chem. 2010;285:17628–17635. doi: 10.1074/jbc.M110.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J. Cell Sci. 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 35.Zhao G, Li G, Schindelin H, Lennarz WJ. An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc. Natl Acad. Sci. USA. 2009;106:16197–16202. doi: 10.1073/pnas.0908321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol. Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Nardozzi J, Wenta N, Yasuhara N, Vinkemeier U, Cingolani G. Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5. J. Mol. Biol. 2010;402:83–100. doi: 10.1016/j.jmb.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen CD, Mansfield RE, Leung W, Vaz PM, Loughlin FE, Grant RP, Mackay JP. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J. Mol. Biol. 2011;407:273–283. doi: 10.1016/j.jmb.2010.12.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.