Abstract
Archaeal viruses have been the subject of recent interest due to the diversity discovered in their virion architectures. Recently, a new group of haloarchaeal pleomorphic viruses has been discovered. It is distinctive in terms of the virion morphology and different genome types (ssDNA/dsDNA) harboured by rather closely related representatives. To date there are seven isolated viruses belonging to this group. Most of these share a cluster of five conserved genes, two of which encode major structural proteins. Putative proviruses and proviral remnants containing homologues of the conserved gene cluster were also identified suggesting a long-standing relationship of these viruses with their hosts. Comparative genomic analysis revealed three different ways of the genome organization, which possibly reflect different replication strategies employed by these viruses. The dsDNA genomes of two of these viruses were shown to contain single-strand interruptions. Further studies on one of the genomes suggested that the interruptions are located along the genome in a sequence-specific manner and exhibit polarity in distribution.
INTRODUCTION
Archaeal viruses described to date comprise a relatively small group of prokaryotic viruses (1). Despite the small number, it is apparent that they exhibit a variety of morphologies (1,2). Among only 34 crenarchaeal viruses described such unique virion morphologies as bottle- and droplet-shaped, ovoid or bacilliform can be encountered (1,2). Although most of the ∼50 euryarchaeal viruses described resemble bacteriophages (3,4), new types of viruses have also been discovered (3,5,6). This diversity of archaeal viruses is encompassed in ten families and a few unclassified groups (1).
As more archaeal viruses are isolated, characterized and sequenced, comparative genomic analysis becomes a frequently used tool giving insights into the diversity and evolution of these viruses (4,7–10). Studies on fuselloviruses, lipothrixviruses, tailed archaeal viruses and proviruses showed that, similarly to bacteriophages, these viruses have mosaic genomes composed of variable and conserved regions, parts of which are derived from a shared pool of genes (4,8–12). The gene pool of archaeal viruses largely comprises open reading frames (ORFs) having significant similarity only to ORFs of other archaeal viruses or cells, and ORFans (13,14), genes that lack homologues in other genomes (15). Lately, many structural and functional studies have been undertaken in attempts to characterize proteins encoded by archaeal viruses (16–18). As a result, novel protein folds and viral proteins self-assembling into structures with rare symmetry were discovered (17,18).
In a recent study, 40 new haloarchaeal viruses were isolated and further studied (3). Here we focus on a group of haloarchaeal pleomorphic viruses, which includes two earlier described members Halorubrum pleomorphic virus 1 (HRPV-1) and Haloarcula hispanica pleomorphic virus 1 (HHPV-1) (6,19,20) and four new members Halorubrum sp. pleomorphic viruses 2, 3 and 6 (HRPV-2, HRPV3 and HRPV-6) (3,21) and Halogeometricum sp. pleomorphic virus 1 (HGPV-1) (3). These pleomorphic viruses have a narrow host range and, except for HHPV-1, all infect a host that originates from the same sample as the virus (3). Analyses of the pleomorphic virions showed that the HRPV-1 and HHPV-1 viral particles consist of a genome enveloped in a lipid membrane with two major structural proteins (6,20,22). The smaller major structural protein, VP3, is predicted to contain several transmembrane (TM) helices, and in HRPV-1 it was shown to be membrane-associated (22). The larger major structural protein, VP4, is N-terminally processed, exposed to the viral surface and anchored to the membrane with a C-terminal TM domain (6,20,22,23). Thus, the VP4 proteins are predicted to be involved in receptor binding and fusion of the host and viral membranes (6,20,22,23). In HRPV-1, a third structural protein, VP8, which is a putative P-loop NTPase, has been identified (6). Both HRPV-1 and HHPV-1 contain a predicted ORF encoding a putative rolling circle replication initiation protein (RCR Rep) (6,20,24).
HRPV-1 has a 7048-nucleotide (nt)-long circular single-stranded DNA (ssDNA) genome, whereas the HHPV-1 genome is a 8082-bp long circular double-stranded DNA (dsDNA) molecule (6,20). Despite different genome types, these two viruses share a set of homologous proteins and their genomes are collinear (20). Both viruses share a conserved cluster of two protein-coding genes and four predicted ORFs (6,20). Putative proviral elements containing an entire set of the HRPV-1 and HHPV-1 homologues have been identified in the sequenced genomes of Haloferax volcanii DS2 and Halomicrobium mukohataei DSM 12286, as well as in Haloferax lucentense plasmid pHK2 (20,23,25). Some homologues were found in haloarchaeal virus His2 (6,20). His2 virus has a linear 16 067-bp long dsDNA genome with inverted terminal repeats and with terminal proteins attached to 5′ termini (5,26).
Here we report the genome sequences of the four new haloarchaeal pleomorphic viruses HRPV-2, HRPV-3, HRPV-6 and HGPV-1. The genomes of HRPV-3 and HGPV-1 are shown to be circular dsDNA molecules with localized single-strand interruptions. We present evidence for the discontinuous nature of HRPV-3 and HGPV-1 genomes and describe the interruptions in the HRPV-3 genome in more detail. Comparative genomic analysis of the expanded set of haloarchaeal pleomorphic viruses and related putative proviral regions confirms the presence of the conserved cluster of identified and putative genes also found in the linear dsDNA genome of His2, a previously described haloarchaeal virus (5). In parallel study (21), His2, which is currently classified as a spindle-shaped virus (5), is suggested to belong to the group of haloarchaeal pleomorphic viruses on the basis of the virion architecture. The comparative genomic analysis reported in this work supports this proposal and further shows that these pleomorphic viruses can be divided into three subgroups according to their gene content and genome organization.
MATERIALS AND METHODS
Strains, culture conditions and stock solutions
Viruses and host strains used in this study are listed in Supplementary Table S1. For the modified growth medium (MGM) (27) used for the culture of all the host strains and viruses, the 30% artificial saltwater stock solution (SW) was diluted to 23% (MGM broth), 20% (MGM plates) or 18% (top layer agar) as described in the online resource—The HaloHandbook (http://www.haloarchaea.com/resources/halohandbook/index.html). Agar concentration of 1.4% for plates and 0.4% for the top layer agar was used. Viruses were propagated and ‘2×’ purified from the lysates according to the protocol described in Atanasova and others (3).
Purification and analysis of the genomes
Viral genomic DNA was obtained from 2× purified virus preparation by the phenol–chloroform extraction method as described previously (20). Shortly, 2× purified viruses were diluted 1:10 or 1:20 in solution containing final concentrations of 50 mM Tris-HCl (pH 7.5), 1% (w/v) SDS, 10 mM EDTA (pH 8.0) and 100–200 mM NaCl. Proteinase K (0.5 mg/ml) was added and the solution was incubated at +55 °C for 20 min. The solution was extracted with phenol and phenol–chloroform. DNA was precipitated from the final water phase with two volumes of ethanol in the presence of 0.2 M NaCl. The genomes were digested with DNase I (1 U of RQ1 RNase-Free DNase/1 µg of genomic nucleic acid; Promega), Exonuclease I (20 U/1 µg of genome; Fermentas) and RNase A (4 µg/1 µg of genome; Promega). Mung Bean Nuclease (MBN, Promega) digests were done in 10 µl reactions for 30 min at +37 °C using 0.025 U, 0.5 U or 5 U of per 1 µg of DNA. Phage φX174 ssDNA genome and its dsDNA replicative form RFII (New England Biolabs) were used as controls. HRPV-3 and HGPV-1 genomic DNA were incubated with Sulfolobus DNA polymerase IV (2 U/1 µg of DNA; New England Biolabs) in a reaction containing 1× enzyme buffer and 0.4 mM dNTP. Reactions were carried out at +37 °C for 3 h. Ligation was done with T4 DNA ligase (Fermentas) as described by manufacturer.
DNA manipulations
For cloning of the MBN-resistant fragments, MBN digested DNA of HRPV-3 was purified using QIAquick PCR purification kit (Qiagen) and ligated into pJET1.2/blunt vectors utilizing a CloneJETTM PCR Cloning Kit (Fermentas). Ligation mixtures were transformed to competent E. coli DH5α cells, and plasmid-containing colonies were selected on LB-plates supplemented with ampicillin (final concentration 150 µg/ml). Transformed colonies were screened for the presence of inserts by colony PCR using pJET1.2 forward and reverse primers (Fermentas). Selected plasmids were purified with QIAprep Spin Miniprep Kit (Qiagen), and the inserts were sequenced using pJET1.2 forward and reverse primers (Fermentas).
For the localization of the discontinuities in the HRPV-3 and HGPV-1 genomes, the free 3′ hydroxyl ends of genomic DNA were labelled with DIG-11-dUTP by terminal deoxynucleotidyl transferase (TdT, Fermentas). Tailing reactions for 0.5–1 µg of genomic DNA were carried out in 1× TdT buffer containing 0.25 mM CoCl2, 0.02 mM DIG-11-dUTP, 0.1 mM dATP and 15 U of TdT (Fermentas). Reactions were incubated at +37 °C for 40 min. After the tailing, DNA was concentrated in Microcon centrifugal filters with Ultracel® YM-100 membrane (Millipore) and digested with restriction enzymes. The digested fragments were separated in 1% agarose/1×TBE gel and transferred to nylon membrane (GE Osmonics). The DIG-11-dUTPs incorporated in genomic fragments were detected using anti-digoxigenin-AP conjugate (Roche) and CDP-Star substrate (Roche) as described by the manufacturer.
Sequencing and annotation of the genomes
The obtained viral genomic DNA was amplified using a GenomePhi kit (GE Healthcare) and used to make barcoded (10 bp) fragment libraries for 454 sequencing on a Genome Sequencer FLX Titanium (Roche/454 Life Science). The reads were assembled using the gsAssembler (Roche/454 Life Science). The coverage of the different genomes was 17× for HRPV-2, 14× for HRPV-3, 300× for HRPV-6 and 300× for HGPV-1. The obtained assembly was verified with PCR products that for HRPV-2, HRPV-3 and HGPV-1 covered the whole genome. The final genome sequences were obtained by sequencing these PCR products. For HRPV-6, several Sanger sequencing runs confirmed the initial assembly. The primers used for PCR and sequencing are listed in Supplementary Table S2. For the verification of the discontinuous regions, genomic DNA was used in sequencing reactions with primers, which are also listed in Supplementary Table S2. All sequencing reactions were carried out by using BigDye Termination Chemistry v. 3.1 and analysed on an ABI 3130 (Applied Biosystems). Sequencing reactions were purified with CleanSeq (Agencourt) using a Beckman NXp robot (Beckman Coulter). Sequences were assembled and edited using GAP4. The sequences were submitted to GenBank and can be found under the accession numbers JN882264 (HRPV-2), JN882265 (HRPV-3), JN882266 (HRPV-6) and JN882267 (HGPV-1).
The genome sequences were annotated using DNAMaster (http://cobamide2.bio.pitt.edu, version 5.22.2), pDRAW32 and GeneMark.hmm for Prokaryotes (version 2.8). Homologous nucleotide and protein sequences were searched for using BLASTN and BLASTP tools, respectively, (28) available at the National Center for Biotechnology Information (NCBI). Manually refined annotations were based on GeneMark.hmm coding potential analysis, the obtained protein chemistry data and colinearity with previously annotated pleomorphic virus genomes (6,20). The properties of identified proteins and the products of putative ORFs were analysed using expert protein analysis system (EXPASY) proteomics tools. Isoelectric points and molecular masses were determined using Compute pI/MW tool (29). The signal sequences for VP4-like proteins were predicted using Signal P (version 3.0) (30) and TatFind Servers (31). Conserved protein signature sequences were determined using InterProScan (32). Putative trans-membrane regions of proteins were additionally determined using combined predictions by TMHMM tool (33) and TMPred tool (34). Coiled-coil regions were predicted using Coil tool (35). Multiple sequence alignments were generated using T-Coffee (36), Muscle (37) and Praline (38). Conserved DNA motifs in aligned nucleotide sequences (T-Coffee) were visualized with WebLogo (39).
Phylogenetic analyses
Pairwise identity percentages between amino acid sequences of identified or putative proteins were determined using EMBOSS Needle tool at EMBL-EBI. Protein sequences were aligned with T-Coffee program, and conserved blocks for the phylogenetic analysis were selected using Gblocks (40). The phylogeny was reconstructed by maximum likelihood method using phyml algorithm (41). Resulting trees were visualized in Phylodendron (http://iubio.bio.indiana.edu/soft/molbio/java/apps/trees/).
HPLC-DAD-MS analysis of DNA
DNA extracted from HRPV-3 and HGPV-1 was dissolved in 20 mM NH4OAc pH 5.3 and incubated for 5 h at 37°C in the presence of 3 U nuclease P1 (Roche Diagnostics, Mannheim, Germany) per 100 µg DNA. Snake venom phosphodiesterase (Worthington, Lakewood, USA) was then added to a concentration of 0.06 U per 100 µg DNA, and the mixture was incubated at 37°C for another 2 h. Finally to convert the resulting mixture of mononucleotides to free nucleosides, 1/10 vol of 10 × SAP buffer (Fermentas, St. Leon-Roth, Germany) was added, followed by 3/20 vol of H2O, and ¼ vol of Shrimp Alkaline Phosphatase (SAP stock at 1 U/µl; from Fermentas, St. Leon-Roth, Germany). The mixture was incubated for 1 h at 37°C. Additionally a commercial oligomer containing deoxy-5-methyl-cytidine was digested and used as reference sample (rGrUrCrA[m5C]rGrCrGrGrGrArGrArCrCrGrGrGrGrUrUrCrGrArUrUrCrCrCrCrGrArCrGrGrGrGrArGrCrCrA, Iba, Göttingen, Germany).
The digested DNA was analysed on an Agilent 1260 series equipped with a diode array detector (DAD) and Triple Quadrupol mass spectrometer Agilent 6460. A Synergy Fusion RP column (4 µm particle size, 80 Å pore size, 250 mm length and 2 mm inner diameter) from Phenomenex (Aschaffenburg, Germany) with a guard column was used at 35°C. The solvents consisted of 5 mM ammonium acetate buffer adjusted to pH 5.3 using acetic acid (solvent A) and pure acetonitrile (solvent B). The elution started with 100% solvent A followed by a linear gradient to 20% solvent B at 10 min. Initial conditions were regenerated by rinsing with 100% solvent A for 7 min. The flow rate was 0.5 ml/min.
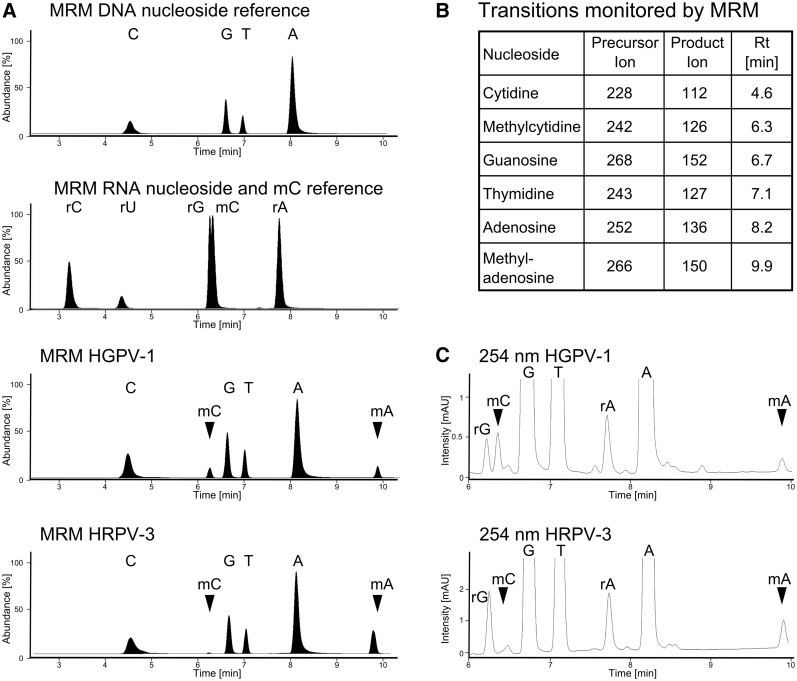
The effluent from the column was first measured photometrical at 254 nm by the DAD for later quantification followed by the mass spectrometer equipped with an electrospray ion source (Agilent Jet Stream). ESI parameters were as follows: gas temperature 300°C, Gas flow 5 l/min, Nebulizer pressure 35 psi, Sheath gas temperature 350°C, Sheath gas flow 12 l/min, capillary voltage 3500 V. The MS was operated in positive ion mode monitoring multiple fragmentation reactions (MRM mode) at previously optimized conditions. The transitions and retention times used for identification of nucleosides can be found in Figure 4B.
Figure 4.
LC-MS/MS analysis of HGPV-1 and HRPV-3 digests. (A) MRM chromatograms of a DNA digest, RNA digest including 5-deoxymethylcytidine, HGPV-1 DNA digest and HRPV-3 DNA digest. (B) Reactions monitored by MRM and retention times of nucleosides. (C) UV traces at 254 nm of HGPV-1 DNA digest and HRPV-3 DNA digest used for semi-quantitative calculations.
Quantification of modified nucleosides
For quantification of detected modified nucleosides, the UV traces at 254 nm monitored by the DAD were used. According to (42), the differences of extinction coefficients between methylated and unmethlyated nucleobases are negligible; hence, identical coefficients were used for quantification of C versus mC and A versus mA, respectively. The area under the absorption peak (AA) of each nucleoside was used for calculation of relative amounts using the formula %mC = AA(mC)/(AA(mC)+AA(C)×100% for mC and %mA = AA(mA)/(AA(mA)+AA(A)×100% for mA, respectively.
Protein analyses
The protein composition of the virions was analysed by 16% SDS-polyacrylamide gel electrophoresis (PAGE). The N-terminal sequences of the structural proteins were determined in the Protein Chemistry Core Laboratory, Institute of Biotechnology, University of Helsinki, as described previously (43).
For the analysis of glycan modifications, 5 µg of virion proteins were separated in 16% SDS-PAGE and stained using the Pro-Q Emerald 300 Glycoprotein Gel Stain Kit (Invitrogen) as described by the manufacturer.
RESULTS
In addition to the haloarchaeal pleomorphic viruses HRPV-1 and HHPV-1 reported before (6,20), four new pleomorphic viruses HRPV-2, HRPV-3, HRPV-6 and HGPV-1 were isolated from diverse geographical locations (3,21). The characterization of their life cycles and particle architectures by both biochemical dissociation and cryo-EM is described elsewhere (21). The N-terminal sequences of the structural proteins identified in this study are listed in Table 1. The virions were shown to contain two major structural proteins homologous to VP3 and VP4 proteins in HRPV-1 virus (6,21), the genome of which is used as a reference in this article (Figure 1). The names of the homologues in different viruses are shown in Table 1, and they will be referred to as VP3-like and VP4-like proteins further in the text. Other HRPV-1 protein homologues will be referred to in a similar manner (VP8-like proteins, ORF6-like and ORF7-like products). Besides VP3-like and VP4-like proteins, HGPV-1 contains one additional major structural protein (21). All major structural proteins were analysed for glycan modifications using in-gel staining. Among the major structural proteins of the new viral isolates, no glycan modifications were detected (data not shown).
Table 1.
N-terminal sequences of the major structural proteins determined in this study
| Virus | VP3-like protein | VP4-like protein |
|---|---|---|
| HRPV-2 | (VP4) ASSYRNSMGS … | (VP5) IAPLVGVGLA … |
| HRPV-3 | (VP1) ATSKLSGFAG … | (VP2) LAPLIAGXFL … |
| HRPV-6 | (VP4) ASSYRNS … | (VP5) IAPLVGYA … |
| HGPV-1 | n.d. | (VP4) EFVNCDLS … |
n.d. – not determined.
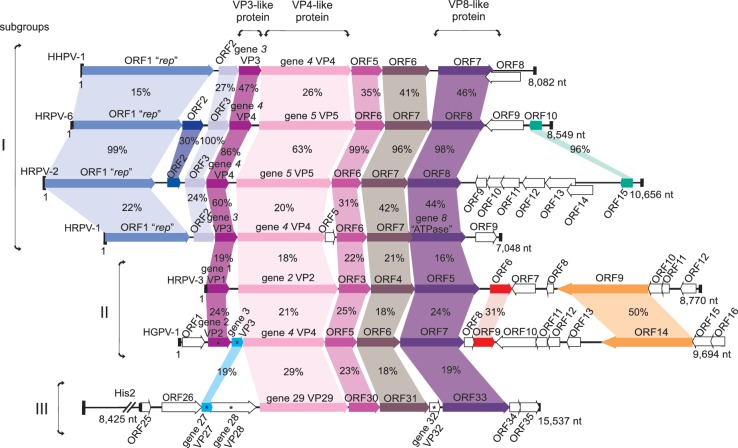
Figure 1.
Schematic genomic alignment of the haloarchaeal pleomorphic viruses HHPV-1, HRPV-6, HRPV-2, HRPV-1, HRPV-3 and HGPV-1 as linear representation and His2 (5) (GenBank accession no AF191797). HRPV-1 genome is used as a reference. Genes encoding VP3-like, VP4-like and VP8-like protein homologues are marked below the alignment of the genomes. Genes encoding structural proteins of HGPV-1 and His2 determined by M.K. Pietilä et al. (21) are denoted by asterisk. Percentages of identical amino acids in homologous putative polypeptides and proteins are indicated. Subgroups to which viruses were assigned according to genome organization are indicated on the right.
Genomic features
The genome types (DNA/RNA, single-stranded/double-stranded) were determined using the standard digestions of isolated genomes with DNase, RNase and MBN as described in the Materials and Methods and in the previous reports for HRPV-1 and HHPV-1 (6,20). The results summarized in Table 2 show that the pleomorphic viruses contain either single-stranded or double-stranded DNA genomes. The dsDNA genomes of HRPV-3 and HGPV-1, however, were digested into distinct fragments by MBN, indicating possible single-stranded regions in the genomes (see below). All genomes are circular molecules varying in length between 8000 and 11 000 nt (Table 2). The circularity of the HRPV-1 and HHPV-1 genomes was demonstrated earlier (6,20). HRPV-2 and HRPV-6 genomes were shown to be circular by exonuclease I digestion (data not shown). Isoforms of HRPV-3 and HGPV-1 genomes were visualized in 2D agarose gel (44) (Supplementary Materials and Methods, Supplementary Figure S1). Analysis revealed the existence of two major isoforms, one of which was attributed to relaxed circular, whereas the other one to linear form. Based on the results, we suggest that HRPV-3 and HGPV-1 genomes are also circular. The GC percentages of the viral genome sequences are shown in Table 2. Putative double-strand origins of replication (dso) are reported in the Supplementary results and the Supplementary Table S3. Analyses of the promoter and Shine-Dalgarno (SD) sequences are described in the Supplementary results. At the nucleotide level, the genomes show similarity reaching up to 74% along relatively short stretches (∼400 nt). HRPV-2 and HRPV-6 genomes are an exception, as they seem to share high identity (up to 97%) over DNA regions up to 4000 nt (Figure 1).
Table 2.
Properties of the characterized genomes
The haloarchaeal pleomorphic viruses can be divided into three subgroups
The genomes were analysed and annotated as described in the Materials and Methods. The properties of the putative and identified proteins are shown in Supplementary Tables S4–S7. The start site for the linearized nucleotide sequence is either the start site of the predicted ORF encoding the putative replication initiation protein or in the cases of HRPV-3 and HGPV-1 the first predicted ORF transcribed in the same direction as the genes coding for the major structural proteins (Figure 1).
Genomic comparison (Figure 1) showed that HRPV-1, HHPV-1 and haloarchaeal pleomorphic virus genomes reported in this study share a cluster of five conserved genes, starting with genes encoding the VP3-like proteins and ending with genes coding for the VP8-like proteins. These viruses, although related, can be divided into two subgroups according to their genome organization. The first subgroup contains viruses HRPV-1, HRPV-2, HRPV-6 and HHPV-1. The genomes of HRPV-2 and HRPV-6 conform to the gene order previously reported for HRPV-1 and HHPV-1. In these genomes, the first predicted open reading frame (ORF1) encodes a putative RCR Rep (24) and is located one or two ORFs upstream of the conserved cluster.
HRPV-3 and HGPV-1 form the other subgroup of haloarchaeal pleomorphic viruses. The most notable difference of HRPV-3 and HGPV-1 genomes compared to the first subgroup is that they do not contain the homologue of the replication initiation protein. These genomes are clearly divided into two regions that are transcribed in opposite directions. In addition to the conserved cluster, these two viruses share two ORFs, not found in the genomes of the first subgroup of viruses (Figure 1).
The earlier reported His2 virus (5) has four homologues belonging to the conserved cluster of pleomorphic viral genes. However, it cannot be included in either one of the subgroups and clearly forms a third subgroup, in which it is currently the only member. In addition to the conserved cluster of genes, His2 major structural protein VP27 is a putative homologue of HGPV-1 major structural protein VP3 (21) (Figure 1). These two proteins are not homologues of VP3-like proteins but were suggested to serve similar function (21).
Related genetic elements in haloarchaeal genomes
In addition to the already reported Hfx. lucentense plasmid pHK2 and proviral regions in Haloferax volcanii DS2 (region 2) (20) and Halomicrobium mukohataei DSM 12286 (region 2) (23), there are several putative proviral elements found in the sequenced genomes of different haloarchaeal species (Table 3, Supplementary Figure S2, Supplementary Results) that are related to the viruses described in this study. Based on the colinearity, putative proviral regions can also be assigned to above-mentioned viral subgroups. Hmc. mukohataei region 2, Hfx. volcanii region 2 and Hfx. lucentense plasmid pHK2 can be assigned to the first subgroup, and the rest fall into the second subgroup (Table 3).
Table 3.
The coordinates and the content of putative proviral regions related to haloarchaeal pleomorphic viruses
| Sub group | Archaeal strain (GenBank accession no) and proviral region | Positions of the proviral region (nt) | Genes included in proviral region |
|---|---|---|---|
| I | Haloferax volcanii DS2 (NC_013967) region 2 | 1 307 561-1 295 035 | HVO_1434-HVO_1422 |
| Halomicrobium mukohataei DSM 12286 (CP001688) region 2 | 912 487-926 019 | Hmuk_0952-Hmuk_0965 | |
| II | Haloarcula hispanica ATCC 33960 (CP002923)a | 2 720 469-2 730 360 | HAH_2828-HAH_2836 |
| Haloarcula marismortui ATCC43049 (AY596297) region 1 | 2 031 007-2 046 290 | rrnAC2284-rrnAC2297 | |
| Haloarcula marismortui ATCC43049 (AY596297) region 2a | 2 134 852-2 146 679 | rrnAC2395-rrnAC2404 | |
| Haloferax volcanii DS2 (NC_013967) region 1a | 231 538-252 098 | HVO_0258-HVO_0280A | |
| Halomicrobium mukohataei DSM 12286 (CP001688) region 1 | 435 432-448 356 | Hmuk_0455-Hmuk_0467 | |
| Halopiger xanaduensis SH-6 (CP002839) region 1 | 631 787-647 728 | Halxa_1313-Halxa1331 | |
| Halopiger xanaduensis SH-6 (CP002839) region 2 | 3 612 420-3 632 698 | Halxa_4253-Halxa_4277 | |
| Halorhabdus utahensis DSM 12940 (NC_013158) | 771 758-788 395 | Huta_0789-Huta_0809 | |
| Natrialba magadii ATCC 43099 (CP001932) | 284 491-266 777 | Nmag_0295-Nmag_0272 |
aThese putative proviral regions are referred to as proviral remnants.
Replication initiation proteins
The genomes of the first subgroup of viruses contain the ORF1 encoding the putative RCR Reps (24). Despite almost 100% identity between Reps of HRPV-2 and HRPV-6, our data confirm that replication initiation proteins are more diverged than other parts of the haloarchaeal pleomorphic virus genomes (20). As described in the Supplementary Results, the multiple sequence alignment suggested that the putative Rep proteins of the viruses and their homologues from the proviral elements could be divided into two clusters.
The conserved cluster of putative and identified genes
The first protein encoded by a conserved cluster of genes is VP3-like protein (Figure 1). It is a major structural protein of haloarchaeal pleomorphic viruses, predicted to contain several transmembrane regions. VP3-like proteins of the first subgroup of viruses and putative proviruses share extensive identity at the amino acid level (∼50% and above, Supplementary Figure S3). In contrast, VP3-like proteins of the second subgroup show only ∼20% identity to each other and to homologues of the first subgroup members.
The next protein encoded in the conserved cluster of genes is VP4-like major structural protein (Figure 1), forming the spike structure on the virion surface. This protein is processed during maturation. In order to predict the translation start site, the empirically determined N-termini (Table 1) together with Signal P and Tat find programs were used. Three of the manually annotated VP4-like protein precursors were predicted to have a twin-arginine signal sequence suggesting export of the protein in a folded state. Most of the VP4-like protein precursors share only ∼20% identity (Supplementary Table S8). Even though VP4-like polypeptides are much more diverse than VP3-like proteins, the secondary structures are predicted to be similar. All of them are also predicted to contain a coiled-coil region in the C-terminus of the protein just preceding the transmembrane domain that serves as a membrane anchor.
The following members of the conserved cluster are ORF6-like and ORF7-like putative genes (Figure 1), encoding products of unknown function. Both of the putative proteins are predicted to have transmembrane domains. Putative ORF6-like gene products show rather low conservation among haloarchaeal viruses and proviral regions (Supplementary Table S9). Between the majority of these proteins amino acid sequence identity varies in the range of 20–30%. It seems that the tendency of higher protein conservation among the same viral subgroup members observed for VP3-like protein does not apply to ORF6-like gene products. On the contrary, ORF7-like putative gene products do show higher conservation among the members of the same viral subgroup with most of pairwise amino acid identities ranging between 30% and 40% (Supplementary Figure S4).
The last predicted ORF that belongs to the conserved set of viral genes encodes VP8-like protein, a putative P-loop NTPase. Similarly to ORF7-like gene products, VP8-like proteins are more conserved among the members of the same viral subgroup with amino acid sequence identities varying from 40% to 50% (Supplementary Figure S5).
His2, the only representative of the third subgroup of haloarchaeal pleomorphic viruses, also contains a related conserved cluster (Figure 1). However, its identity to other pleomorphic viruses is low.
Comparatively high conservation of VP3-like, VP8-like proteins and ORF7-like gene products among the viruses and proviral regions within the first two subgroups allowed the reconstruction of the phylogenetic history of these proteins (Supplementary Figures S3–S5). Analysis revealed that the phylogenetic history of VP3-like, VP8-like proteins and ORF7-like gene products (Supplementary Figures S3–S5) is essentially the same. HGPV-1 and proviral remnant in Hfx. volcanii region 1 were left out from this analysis, because their proteins share rather low identity with the proteins of the other second subgroup members.
HRPV-3 ORF9 and HGPV-1 ORF14
All the haloarchaeal pleomorphic viruses and proviral regions belonging to the second subgroup share a predicted ORF that is encoded by the negative strand (Figure 1). These proteins are often predicted to contain winged helix-turn-helix (wHTH) DNA binding domain in their C-termini (Conserved Domain Database search E-values: 0.06 for HGPV-1 ORF14 product and 1.27×10−3 for HRPV-3 ORF9 product). Putative HRPV-3 ORF9 and HGPV-1 ORF14 products share 50% identity (Figure 1). In addition, HRPV-3 ORF9 and HGPV-1 ORF14 products also show ∼43% identity to the products of ORFs Hqrw_6002 and Hqrw_7002 in Haloquadratum walsbyi plasmids PL6A and PL6B, respectively (45).
HRPV-3 and HGPV-1 genomes are discontinuous dsDNA molecules
The unusual nature of HRPV-3 and HGPV-1 genomes was initially noticed during the routine analysis of the genomes using different nucleases. Digestion of the genomes with MBN gave a set of distinct fragments suggesting that the genomes consist mostly of dsDNA with some regions of ssDNA (Figure 2A). In the case of discontinuities in HRPV-3 and HGPV-1 genomes, free 3′OH ends should be able to serve as substrates for the TdT. Indeed, TdT was able to incorporate DIG-11-dUTP into the genomes of HRPV-3 and HGPV-1 as detected using anti-DIG-antibody. The label was incorporated roughly throughout the genome (data not shown).
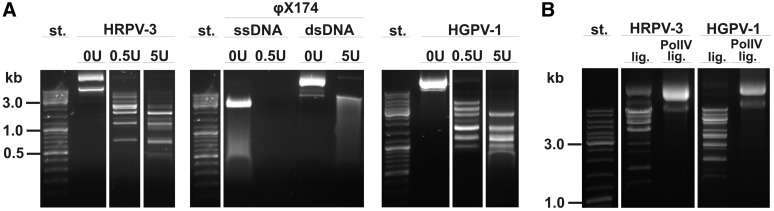
Figure 2.
Mung bean nuclease (MBN) analyses of HRPV-3 and HGPV-1 genomes. (A) Digestion of HRPV-3 and HGPV-1 genomes using concentrations of either 0.5 or 5 units (U) of MBN per 1 µg of genomic DNA. Virion φX174 DNA (ssDNA) and its replicative form RFII (dsDNA) were used as controls. (B) MBN digestions (0.5 U of MBN/1 µg of genomic DNA); the genomes were treated with either T4 DNA ligase (lig.) or Sulfolobus DNA polymerase IV and T4 DNA ligase (PolIV lig.). GeneRulerTM DNA ladder mix was used as a standard (‘st.’).
In order to study the nature of the discontinuities more closely, a wide range of MBN concentrations was used, starting from concentrations digesting only ssDNA (0.025 U/1 µg of DNA), to concentrations that also act on nicked dsDNA (0.5 U/1 µg of DNA) and on dsDNA molecules (5 U/1 µg of DNA). The single-stranded φX174 virion DNA and the nicked dsDNA replicative form RFII were used as controls to show specificities of different MBN concentrations. Clear fragmented patterns of the HRPV-3 and HGPV-1 genomes were obtained when 0.5 U and 5 U of MBN were used (Figure 2A), whereas smaller MBN concentration (0.025 U/1 µg of DNA) did not have any effect on the genomes (data not shown). The results suggest that the viral genomes contain nicks or short stretches of ssDNA. In order to resolve this, HRPV-3 and HGPV-1 genome were treated with either T4 DNA ligase alone or with Sulfolobus DNA polymerase IV and then T4 ligase followed by MBN digestion (Figure 2B). Ligase alone was not able to repair the genomes since MBN was still able to digest the genomes into separate fragments. However, the genomic DNA treated with Sulfolobus DNA polymerase IV and ligase was resistant to MBN concentrations digesting ssDNA. Hence, the data suggest that the putative discontinuities in the dsDNA genomes are short regions of ssDNA.
Discontinuities in HRPV-3 genome are preceded by conserved DNA motif
In order to localize MBN-resistant fragments in HRPV-3 and HGPV-1 genomes, the fragments were cloned into pJET1.2/blunt vectors and the ends of the cloned fragments were sequenced (data not shown). Some of the mapped MBN-resistant fragments were overlapping and some parts in both genomes were not covered. Thus, the results were inconclusive. Alignment of the sequences around the ends of the cloned fragments of HRPV-3 genome, however, indicated the presence of the DNA motif GCCCA (Figure 3A). In HGPV-1 genome the motif is less clear, but examination of sequences located at varying distances from the ends of the cloned fragments revealed a putative motif, which is currently under study (Sencilo and Roine, unpublished).
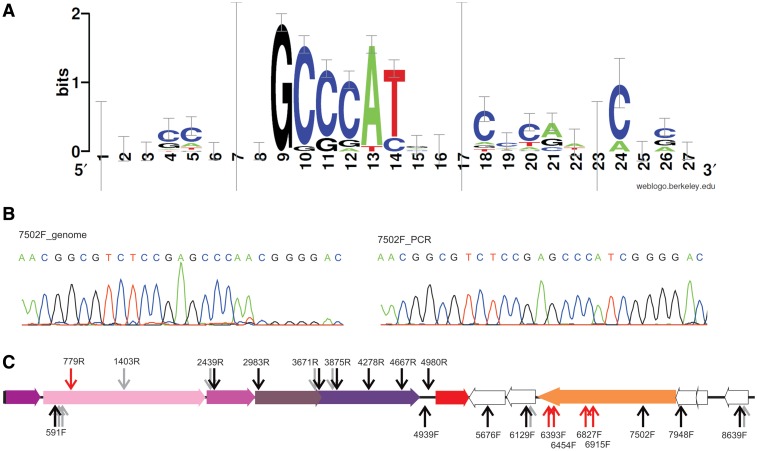
Figure 3.
Mapping of the discontinuous sites in the HRPV-3 genome. (A) Conserved nucleotides of the aligned sequences showing the identified motif found in the MBN-resistant fragment ends of HRPV-3. (B) Sanger sequencing of the genomic site with predicted discontinuity. A drop in sequencing signal was observed after the motif GCCCA when HRPV-3 genome was used as a template (7502 F_genome). Sequencing of corresponding HRPV-3 fragment amplified by PCR (7502 F_PCR) showed no decrease in the signal. (C) HRPV-3 genome map with arrows marking predicted sites of discontinuities (GCCCA motif). The names of primers used to sequence the regions with putative discontinuities are noted above or below the arrow. Sites which were not possible to sequence because of the proximity of the other interruption are marked with grey arrow. Black arrows indicate the predicted sites of discontinuity where a sudden drop of sequencing signal was observed as exemplified by the above-placed sequencing chromatograph (7502F_genome). Red arrows indicate positions where no sequencing signal drop was detected despite the predicted motif for discontinuity.
HRPV-3 genome contains 27 sites with the motif GCCCA (Figure 3C). In a few cases, there are two and in one case there are three motifs in close proximity to each other. In order to verify that the sites with the motif or its reverse complement contain discontinuous regions, we first tested two sites by performing Sanger sequencing of the HRPV-3 genomic DNA with the primers reading through the motif along both DNA strands. In both sites, the sequencing signal decreased dramatically after the GCCCA motif, suggesting a discontinuous region in the opposite strand. When the same genomic regions were sequenced from generated PCR template, the drop in sequencing signal was not observed. This suggests that the drop of signal is caused by an inherent discontinuous property of the HRPV-3 genome (Figure 3B). In order to test the rest of the sites, we designed primers ∼100 nt upstream of the GCCCA motif when possible (Table S3). In 13 cases out of 19, a decrease in sequencing signal after the GCCCA motif was observed (Figure 3C). In most of the cases, we could also observe a 3′dA overhang produced by Taq polymerase during Sanger sequencing after the motif, indicating a discontinuity in the template strand (Figure 3B).
The distribution of the mapped discontinuities clearly exhibits polarity, which coincides with the switches of transcription from one strand to another (Figure 3C). All the discontinuities are located in the coding strand with one exception: a triplet of consecutive GCCCA motifs starting at the position 591 is located in the region of the strand which codes for VP4-like protein, meaning that the discontinuities are in the template strand.
HPLC analysis of nucleoside composition
The nucleoside composition of the viral genomes of HRPV-3 and HGPV-1 was analysed after digestion to nucleosides and subsequent separation by chromatography. Detection of eluting nucleosides was done with a diode array detector and a mass spectrometer (HPLC-DAD-MS). In addition to the four major nucleosides, two peaks appeared in the UV-chromatogram at 6.3 min and 9.9 min (Figure 4C), the molecular masses of which correspond to methylated derivatives of cytidine and adenosine, respectively. Using the multiple fragmentation reaction monitoring (MRM) mode of the Triple Quadrupol, the fragmentation patterns of all nucleoside peaks were analysed. The mass transitions of the two methylated nucleosides (Figure 4A and B) showed that the methylgroups are located on the base and not on the sugar moiety. Retention time and fragmentation of the peak at 6.3 min were identical to methylated nucleoside 5-methylcytidine (mC). The peak of the base-methylated adenosine at 9.9 min, also present in both viral DNA digests, corresponds most probably to N6-methyladenosine (mA) which is already well described (46). Figure 4C shows the chromatograms taken at 254 nm of the viral digests used for calculation of relative nucleoside amounts. Both viral digests contain similar amounts of mC (0.58% and 0.66%) and slightly different amounts of mA (0.20% and 1.35%).
DISCUSSION
This study extends the genomic description of the new group of haloarchaeal pleomorphic viruses by adding the characterization of HRPV-2, HRPV-3, HRPV-6 and HGPV-1 genomes. Comparative genomic analysis of these new genomes together with the previously published HRPV-1 (6), HHPV-1 (20) and His2 (5) shows that, in general, this group of viruses has a collinear cluster of conserved genes. Analysis of the genomic content shows that although similar viruses can be found from wide geographical areas (3), the closely related ones were found from geographically close regions.
Annotation of the genomes in this study relies not only on the predictions but also on the experimental data obtained by protein chemistry (N-terminal sequences of proteins). Since the N-termini of the mature VP4-like proteins are known, it was possible to conclude that the automated prediction of translation initiation sites for these proteins often resulted in unusually long signal peptides. Thus, alternative start sites that retained the usual length of the signal peptides were chosen. Experimental data of the viral proteins also give us useful tools for assessing the original annotation of related proviral regions in haloarchaeal genomes. For example, the VP3 homologue of Hfx. volcanii DS2 (HVO_1432) has been annotated as a fibronectin type III domain (Fn3)-containing protein (47). Due to the reasons outlined in the Supplementary Discussion, we suspect that the annotation of the HVO_1432 as a Fn3 domain containing protein is erroneous.
All the haloarchaeal pleomorphic viruses and related proviral elements contain a genomic region encoding the two major structural proteins and three putative gene products, possibly involved in viral assembly. We define this region as a conserved gene cluster. Among the encoded proteins, VP3-like, VP8-like and ORF7-like gene products show the highest degree of conservation, and preliminary phylogenetic analyses suggest similar evolutionary histories for these proteins. The set of genome sequences described here, however, is still too small and diverse to enable a proper phylogenetic analysis of genes encoded by the conserved cluster and their products. Good examples for this are VP4-like proteins. As discussed before (20,23), the VP4-like proteins do not usually show high identity because they are likely to serve as receptor recognition proteins. Exceptions to this are the VP4-like proteins of HRPV-2, HRPV-3 and HRPV-6 (∼50–60% identity, Supplementary Table S8). This case most probably reflects the correlation between the relatedness of the VP4-like protein and the hosts of these viruses (Halorubrum). HRPV-1 also infects a host strain that has been designated as Halorubrum sp., but the VP4 protein similarity to those of HRPV-2, HRPV-3 and HRPV-6 is much lower (∼20%). Since the HRPV-1 VP4 is the only protein among the VP4-like proteins that is shown to be glycosylated and glycans are often involved in specific recognition events, glycosylation may be the factor responsible for the observed difference. Host may not be the only selection pressure on the VP4-like proteins, however, because the VP4-like proteins of HHPV-1 and His2 infecting Har. hispanica are rather diverged.
The pleomorphic viruses seem to be simple in terms of both the virion architecture (21,22) and the genome composition. The replicative form of the circular pleomorphic viral genomes is similar to the haloarchaeal plasmids. Thus, analogies to membrane vesicles containing plasmid DNA have been drawn (1,48). The pleomorphic viruses retard the host cell growth (6,20,21), which is also usual for many high-copy-number plasmids. Although the distinction between plasmids and replicating virus genomes can be difficult (49) and hybrids of plasmids and viruses have been reported (50), the pleomorphic haloarchaeal viruses described in this report fall into the category of viruses on the basis of at least two criteria. First, the replicon is packaged into a particle (51), and second, the replicon packaged encodes the structural proteins and the proteins required for the assembly of these particles.
Although the modes of replication for the two viral subgroups identified in this study have not yet been elucidated, it is likely that they are different. The first subgroup members encode putative replication initiation protein (Rep) of rolling circle replication (RCR), suggesting that these viruses replicate their genomes via RCR mechanism. Viruses belonging to the second subgroup do not encode Rep. The third subgroup member, His2, is predicted to use protein-primed replication for its linear genome (5,52). Thus, this is yet another excellent example of the related viruses having adopted different genome types and possibly also replication strategies (20,51).
For the first two subgroups of pleomorphic viruses (Figure 1), there are many related putative proviruses or proviral remnants (6,20,23) (Table 3). Proviruses contain an integrase, and in most cases, we can identify a tRNA on the other end of the proviral region. The putative proviral regions usually have longer variable part of the genomes than the pleomorphic viruses do. It seems that the described pleomorphic viruses have systematically lost the integrase encoding genes and many putative genes from the regions with variable gene content. The only exception is HRPV-2 which still seems to have many putative genes that may not be necessary for the pleomorphic viral lifecycle sensu stricto.
Although the genomes of HRPV-3 and HGPV-1 are dsDNA molecules they contain localized single-strand interruptions (Figures 2 and 3). In HRPV-3 genome, they are preceded by a specific motif GCCCA. Localized single-strand interruptions have been identified before in the tailed ‘phiKMV-like viruses’ of Pseudomonas aeruginosa (53,54) and T5 (55–57). In those viruses, the discontinuities were shown to be nicks that were located in one strand only, and in phage φkF77, they were shown to be located in intergenic regions (54). The function of the nicks for earlier reported viruses is not known, and the candidates for gene products generating these nicks have been hypothesized to be of viral origin (54). The results obtained in this study show that the interruptions in HRPV-3 and HGPV-1 genomes are not nicks, but longer stretches of single-stranded DNA. Also, we do not have a candidate gene product for causing these interruptions in the HGPV-1 or HRPV-3 genome, and we speculate that in this case the putative enzyme is host encoded. Both HRPV-3 and HGPV-1 show methylation patterns of cytidine (mC) and adenosine (mA) which are well-known in DNA research and have not been found to influence the genomic integrity. Hence, there is no evidence that modified nucleosides are related to the discontinuous nature of the HRPV3 and HGPV1 virus genomes. Further studies are needed to elucidate the mechanism causing these short stretches of ssDNA and most importantly to determine their biological function.
This study describes comparative genomic analyses of six pleomorphic viruses containing a conserved cluster of two identified and three predicted genes involved in the virion structure and assembly. His2 also contains homologues of the same conserved cluster of genes. Although His2 virus is currently grouped with spindle-shaped virus His1 to the genus Salterprovirus, these two viruses share only one homologue, type B DNA polymerase (5). This suggests that His2 virus is more closely related to haloarchaeal pleomorphic viruses, than to spindle-shaped virus His1. A parallel study reports the analysis of virion morphology of these newly identified haloarchaeal pleomorphic viruses and earlier reported His2 (5) by quantitative biochemical dissociation and electron cryo-microscopy (cryo-EM) (21). It shows that the architectural principles of His2 are the same as those of the other haloarchaeal pleomorphic viruses. Taken together, our results are consistent with the above-mentioned findings (21) and allow us to further classify the haloarchaeal pleomorphic viruses into subgroups according to genome organization. On the basis of these results, we can now propose a new group of viruses, the group of seven pleomorphic haloarchaeal viruses, having similar principles of virion architecture, but different types of genomes.
ACCESSION NUMBERS
HRPV-2, HRPV-3, HRPV-6 and HGPV-1 viral genomes were submitted to GenBank under the accession numbers JN882264, JN882265, JN882266 and JN882267.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–9, Supplementary Figures 1–5, Supplementary Materials and Methods, Supplementary Results, Supplementary Discussion and Supplementary References [3,20–22,44,47,58–63].
FUNDING
The University of Helsinki three year grant 2010–2012 (to E.R.) and Academy of Finland Centre of Excellence Program in Virus Research grant 11296841 (2006–2011). Funding for open access charge: University of Helsinki Three Year Grant to Elina Roine
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Hanna M. Oksanen for coordination of the virus production. Gunilla Rönnholm and Professor Nisse Kalkkinen are acknowledged for the determination of the N-terminal sequences of the structural proteins. We thank Maija K. Pietilä, Nina S. Atanasova, Violeta Manole, Lassi Liljeroos, Professor Sarah J. Butcher, Dr. Hanna M. Oksanen and Professor Dennis H. Bamford who have conducted a parallel study, for providing the additional N-terminal sequences of the structural proteins. Professor Sarah J. Butcher and Professor Dennis H. Bamford are also thanked for the useful comments on the manuscript. Sari Korhonen and Eija Stenius are thanked for the excellent technical assistance. Kirsi Lipponen, Eeva-Marja Turkki, Päivi Laamanen and Hanna Turunen are acknowledged for excellent technical assistance in DNA sequencing work. A. S. is a fellow of Viikki Graduate School in Molecular Biosciences. We also thank University of Helsinki for the support to EU ESFRI Instruct Associate Centre for Virus Production and Purification used in this study.
REFERENCES
- 1.Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol. Rev. 2011;35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 2.Mochizuki T, Sako Y, Prangishvili D. Provirus induction in Hyperthermophilic Archaea: Characterization of Aeropyrum pernix Spindle-Shaped Virus 1 and Aeropyrum pernix Ovoid Virus 1. J. Bacteriol. 2011;193:5412–5419. doi: 10.1128/JB.05101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 2011;14:426–440. doi: 10.1111/j.1462-2920.2011.02603.x. [DOI] [PubMed] [Google Scholar]
- 4.Krupovič M, Forterre P, Bamford DH. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 2010;397:144–160. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 5.Bath C, Cukalac T, Porter K, Dyall-Smith ML. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 2009;72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- 7.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 8.Redder P, Peng X, Brügger K, Shah SA, Roesch F, Greve B, She Q, Schleper C, Forterre P, Garrett RA, et al. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ. Microbiol. 2009;11:2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 9.Vestergaard G, Aramayo R, Basta T, Häring M, Peng X, Brügger K, Chen L, Rachel R, Boisset N, Garrett RA, et al. Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J. Virol. 2008;82:371–381. doi: 10.1128/JVI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J. Virol. 2004;78:1954–1961. doi: 10.1128/JVI.78.4.1954-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl Acad. Sci. USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang SL, Nuttall S, Dyall-Smith M. Haloviruses HF1 and HF2: evidence for a recent and large recombination event. J. Bacteriol. 2004;186:2810–2817. doi: 10.1128/JB.186.9.2810-2817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forterre P, Prangishvili D. The origin of viruses. Res. Microbiol. 2009;160:466–472. doi: 10.1016/j.resmic.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 15.Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- 16.Gardner AF, Prangishvili D, Jack WE. Characterization of Sulfolobus islandicus rod-shaped virus 2 gp19, a single-strand specific endonuclease. Extremophiles. 2011;15:619–624. doi: 10.1007/s00792-011-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goulet A, Pina M, Redder P, Prangishvili D, Vera L, Lichière J, Leulliot N, van Tilbeurgh H, Ortiz-Lombardia M, Campanacci V, et al. ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease. J. Virol. 2010;84:5025–5031. doi: 10.1128/JVI.01664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prangishvili D, Quax TE. Exceptional virion release mechanism: one more surprise from archaeal viruses. Curr. Opin. Microbiol. 2011;14:315–320. doi: 10.1016/j.mib.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Kukkaro P, Bamford DH. Virus-host interactions in environments with a wide range of ionic strengths. Environ. Microbiol. 2009;1:71–77. doi: 10.1111/j.1758-2229.2008.00007.x. [DOI] [PubMed] [Google Scholar]
- 20.Roine E, Kukkaro P, Paulin L, Laurinavičius S, Domanska A, Somerharju P, Bamford DH. New, closely related haloarchaeal viral elements with different nucleic Acid types. J. Virol. 2010;84:3682–3689. doi: 10.1128/JVI.01879-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietilä MK, Atanasova NS, Manole V, Liljeroos L, Butcher SJ, Oksanen HM, Bamford DH. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic archaea. J. Virol. 2012;86:5067–5079. doi: 10.1128/JVI.06915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietilä MK, Laurinavičius S, Sund J, Roine E, Bamford DH. The single-stranded DNA genome of novel archaeal virus Halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J. Virol. 2010;84:788–798. doi: 10.1128/JVI.01347-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roine E, Oksanen HM. Viruses from the hypersaline environments: current research an future trends. In: Ventosa A, Oren A, Ma Y, editors. Halophiles and Hypersaline Environments. Heidelberg: Springer; 2011. pp. 153–172. [Google Scholar]
- 24.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes ML, Pfeifer F, Dyall-Smith ML. Analysis of the halobacterial plasmid pHK2 minimal replicon. Gene. 1995;153:117–121. doi: 10.1016/0378-1119(94)00761-g. [DOI] [PubMed] [Google Scholar]
- 26.Porter K, Dyall-Smith ML. Transfection of haloarchaea by the DNAs of spindle and round haloviruses and the use of transposon mutagenesis to identify non-essential regions. Mol. Microbiol. 2008;70:1236–1245. doi: 10.1111/j.1365-2958.2008.06478.x. [DOI] [PubMed] [Google Scholar]
- 27.Nuttall SD, Dyall-Smith ML. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology. 1993;197:678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 30.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 2002;45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 32.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 34.Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 35.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 36.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 41.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 42.Dunn DB, Hall RH. Purines, pyrimidines, nucleosides, and nucleotides: physical constants and spectral properties, and natural occurrence of the modified nucleosides. In: Fasman GD, editor. Handbook of Biochemistry and Molecular Biology. Cleveland: CRC Press; 1975. pp. 65–215. [Google Scholar]
- 43.Bamford DH, Ravantti JJ, Rönnholm G, Laurinavičius S, Kukkaro P, Dyall-Smith M, Somerharju P, Kalkkinen N, Bamford JK. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 2005;79:9097–9107. doi: 10.1128/JVI.79.14.9097-9107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 45.Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One. 2011;6:e20968. doi: 10.1371/journal.pone.0020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soler N, Gaudin M, Marguet E, Forterre P. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem. Soc. Trans. 2011;39:36–44. doi: 10.1042/BST0390036. [DOI] [PubMed] [Google Scholar]
- 49.Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnold HP, She Q, Phan H, Stedman K, Prangishvili D, Holz I, Kristjansson JK, Garrett R, Zillig W. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 1999;34:217–226. doi: 10.1046/j.1365-2958.1999.01573.x. [DOI] [PubMed] [Google Scholar]
- 51.Krupovič M, Bamford DH. Does the evolution of viral polymerases reflect the origin and evolution of viruses? Nat Rev Microbiol. 2009;7:250. doi: 10.1038/nrmicro2030-c1. [DOI] [PubMed] [Google Scholar]
- 52.Porter K, Russ BE, Yang J, Dyall-Smith ML. The transcription programme of the protein-primed halovirus SH1. Microbiology. 2008;154:3599–3608. doi: 10.1099/mic.0.2008/019422-0. [DOI] [PubMed] [Google Scholar]
- 53.Kulakov LA, Ksenzenko VN, Kochetkov VV, Mazepa VN, Boronin AM. DNA homology and adsorption specificity of Pseudomonas aeruginosa virulent bacteriophages. Mol. Gen. Genet. 1985;200:123–127. doi: 10.1007/BF00383324. [DOI] [PubMed] [Google Scholar]
- 54.Kulakov LA, Ksenzenko VN, Shlyapnikov MG, Kochetkov VV, Del Casale A, Allen CC, Larkin MJ, Ceyssens PJ, Lavigne R. Genomes of “phiKMV-like viruses” of Pseudomonas aeruginosa contain localized single-strand interruptions. Virology. 2009;391:1–4. doi: 10.1016/j.virol.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Abelson J, Thomas CA. The anatomy of the T5 bacteriophage DNA molecule. J. Mol. Biol. 1966;18:262–291. [Google Scholar]
- 56.Shaw AR, Lang D, McCorquodala DJ. Terminally redundant deletion mutants of bacteriophage BF23. J. Virol. 1979;29:220–231. doi: 10.1128/jvi.29.1.220-231.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Jiang Y, Vincent M, Sun Y, Yu H, Bao Q, Kong H, Hu S. Complete genome sequence of bacteriophage T5. Virology. 2005;332:45–65. doi: 10.1016/j.virol.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 58.Zhou L, Zhou M, Sun C, Han J, Lu Q, Zhou J, Xiang H. Precise determination, cross-recognition, and functional analysis of the double-strand origins of the rolling-circle replication plasmids in haloarchaea. J. Bacteriol. 2008;190:5710–5719. doi: 10.1128/JB.00596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brenneis M, Hering O, Lange C, Soppa J. Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 2007;3:e229. doi: 10.1371/journal.pgen.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sartorius-Neef S, Pfeifer F. In vivo studies on putative Shine-Dalgarno sequences of the halophilic archaeon Halobacterium salinarum. Mol. Microbiol. 2004;51:579–588. doi: 10.1046/j.1365-2958.2003.03858.x. [DOI] [PubMed] [Google Scholar]
- 61.Tolstrup N, Sensen CW, Garrett RA, Clausen IG. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles. 2000;4:175–179. doi: 10.1007/s007920070032. [DOI] [PubMed] [Google Scholar]
- 62.Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 63.Bork P, Doolittle RF. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl Acad. Sci. USA. 1992;89:8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.