Abstract
The clustered regularly interspaced short palindromic repeats and their associated proteins (CRISPR/Cas) constitute a recently identified prokaryotic defense mechanism against invading nucleic acids. Activity of the CRISPR/Cas system comprises of three steps: (i) insertion of alien DNA sequences into the CRISPR array to prevent future attacks, in a process called ‘adaptation’, (ii) expression of the relevant proteins, as well as expression and processing of the array, followed by (iii) RNA-mediated interference with the alien nucleic acid. Here we describe a robust assay in Escherichia coli to explore the hitherto least-studied process, adaptation. We identify essential genes and DNA elements in the leader sequence and in the array which are essential for the adaptation step. We also provide mechanistic insights on the insertion of the repeat-spacer unit by showing that the first repeat serves as the template for the newly inserted repeat. Taken together, our results elucidate fundamental steps in the adaptation process of the CRISPR/Cas system.
INTRODUCTION
The clustered regularly interspaced short palindromic repeats (CRISPR) and their associated proteins comprise a significant prokaryotic defense system against viruses and horizontally transferred nucleic acids (1–4). This defense system consists of a CRISPR array that is usually preceded by a leader sequence and located near a cluster of CRISPR-associated (cas) genes (5–7). RNA transcribed from the CRISPR array is processed by Cas proteins and directs interfering proteins to target nucleic acids matching the sequences between the repeats. These sequences, called spacers, often originate from plasmids and phages, and thus the system adaptively targets these invaders.
The adaptation process of the CRISPR system, i.e. acquisition of new spacers into the genome, is still poorly understood. Barrangou et al. were the first to report spacer acquisition into the CRISPR array of Streptococcus thermophilus (2). They showed that bacteria surviving a phage challenge expanded their CRISPR array with spacers identical to small DNA regions from the challenging phage, termed protospacers. Spacer acquisition seemed polarized toward the leader end of the array. Their study did not identify a bias of sampled protospacers from a specific strand nor a preference for a specific region in the phage DNA. Knock out of csn2 [previously annotated cas7 (8)] dramatically reduced spacer acquisition, providing an indirect evidence that the product of csn2 is essential for adaptation of the CRISPR array in S. thermophilus. Later, van der Ploeg characterized in vivo spacer acquisition in Streptococcus mutans. He observed acquisition of new spacers in ∼25% of phage-resistant mutants. The acquired spacers, in this case too, corresponded to randomly distributed protospacers with regard to strand or position in the phage genome (9). These studies did not address the roles of the repeats, leader and the core Cas proteins in the acquisition process.
The core Cas proteins, Cas1 and Cas2, were hypothesized to play a major role in the acquisition process. This hypothesis is based on the fact that these two proteins have no role in the interference stage, yet they are conserved in most CRISPR loci (2,3,10). Involvement of Cas1 and Cas2 in the acquisition step is supported by the fact that both proteins show endonuclease activities. Cas1 of Pseudomonas aeruginosa and of Escherichia coli was shown to function as a metal-dependent DNA endonuclease (11,12), and Cas2 of Sulfolobus solfataricus and other strains was shown to be an ssRNA-specific endonuclease (13). Nevertheless, direct evidence for Cas1 and Cas2 involvement in the adaptation process has not yet been provided.
It was suggested from DNA sequence analyses, and later shown experimentally, that short, 2–5 bp sequences found near the protospacer, called protospacer adjacent motifs (PAMs), are crucial for the interference step (14,15). Requirement of PAMs for the interference stage suggests that acquisition of new spacers requires DNA sequences having PAMs. Indeed, it was demonstrated that spacers conferring phage resistance were identical in sequence to protospacers with PAMs (2,14). Moreover, some phage mutants escaping CRISPR/Cas-interference harbored mutations in the PAMs, indicating that PAMs play a role in both interference and adaptation steps (14).
Insights on the adaptation process were also obtained from in silico analyses. These analyses determined PAM sequences for six different CRISPR types based on sequence conservations adjacent to protospacers (15,16) [most recent classification system (8) in parentheses]: 1, 2 (I-E), 3 (I-C), 4 (I-F), 7 and 10 (II). For example, the study identified that CRISPR-2 type (I-E), to which E. coli arrays I and II belong, contained a PAM of the sequence 5′-AWG. Sequence analyses of CRISPR arrays also indicated that the leader sequence may orient the acquisition of new spacers, yet no direct experimental evidence for these analyses was provided.
Partially due to lack of a robust experimental system to study adaptation, several primary questions have not yet been experimentally addressed: How are spacers incorporated into the genome? Which proteins are essential for this process? Are the leader or repeat sequences important for this process? What elements in the leader sequence are required? We describe a robust assay to study the adaptation process in E. coli and provide insights on the essential proteins, DNA elements and insertion mechanism of repeat-spacer units.
MATERIALS AND METHODS
Reagents, strains and plasmids
LB medium (10 g/l tryptone, 5 g/l yeast extract and 5 g/l NaCl) was from Acumedia, agar was from Difco, and antibiotics, isopropyl-β-d-thiogalactopyranoside (IPTG) and l-arabinose were from Sigma-Aldrich. Restriction enzymes were from New England Biolabs. Rapid ligation kit was from Roche. The bacterial strains, plasmids and oligonucleotides used in this study are listed in Supplementary Table S1.
Spacer acquisition assay
E. coli BL21-AI or IYB5101 harboring pCas1+2 plasmid were aerated at 37°C in LB medium containing 50 µg/ml streptomycin with or without 0.2% l-arabinose + 0.1 mM IPTG for 10 to 16 h; the culture was diluted 1:300, grown for an additional 10–16 h, and the procedure repeated for a total of three times. A sample of the culture was used as template in a PCR amplifying CRISPR array I using primers RE10R/MG7F or 260 F/IY13R for non-manipulated BL21-AI and IYB5101, respectively (Supplementary Table S1). For experiments presented in Figures 3, 4B, and 5, primers WIS75188/RE10R were used to detect array expansion, in addition to RE10R/MG7F, and for experiments presented in Figure 4A, primers WIS75188/MG7F were used in addition to RE10R/MG7F.
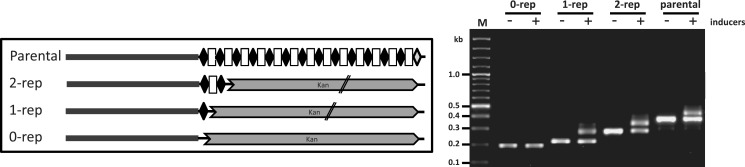
Figure 3.
Minimal repeat sequence essential for spacer acquisition. Schematics of the constructs in CRISPR array I of genetically engineered E. coli BL21-AI. Repeats are marked as diamonds, spacers as rectangles. Kanamycin-resistance cassette used to construct the different strains is marked as gray chevron. Parental: non-manipulated E. coli BL21-AI; 2-rep: one spacer between two repeats, other repeats removed. 1-rep: only one repeat, no spacer, other repeats removed. 0-rep: complete array replaced with a kanamycin-resistance cassette. Parallel lines on the kan cassette indicate that only a partial gene is depicted. Gel shows PCR products amplified from the indicated cultures of E. coli BL21-AI harboring plasmid pCas1+2 and grown in the presence or absence of inducers (0.2% l-arabinose and 0.1 mM IPTG) for 10–16 h for three cycles. Gel is representative of two experiments yielding similar results.
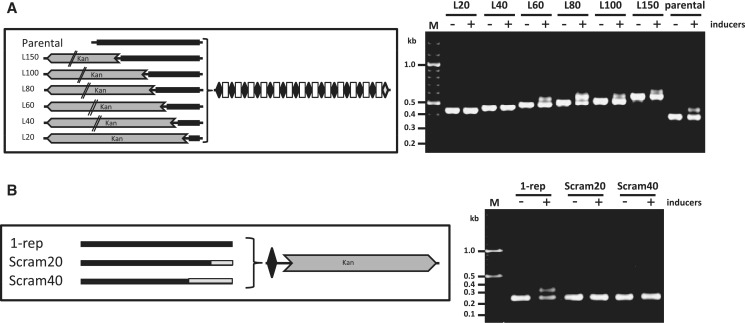
Figure 4.
Regions in the leader sequence essential for spacer acquisition. (A) Schematics of the constructs in CRISPR array I of genetically engineered E. coli BL21-AI. Repeats are marked as diamonds, and spacers as rectangles. Parental: non-manipulated array; LN: N – number of nt upstream of first repeat left intact. Parallel lines on the kan cassette indicate that only a partial gene is depicted. Gel shows PCR products amplified from the indicated cultures of E. coli BL21-AI harboring plasmid pCas1+2 and grown in the presence or absence of inducers (0.2% l-arabinose and 0.1 mM IPTG) for 10–16 h for three cycles. (B) Schematics of constructs as above. Scram40: 40 nt upstream of first repeat scrambled, other nucleotides not manipulated; Scram20: 20 nt upstream of first repeat scrambled, other nucleotides not manipulated. Gel shows PCR products amplified from the indicated cultures of E. coli BL21-AI harboring plasmid pCas1+2 and grown in the presence or absence of inducers (0.2% l-arabinose and 0.1 mM IPTG) for 10–16 h for three cycles. Gels are representative of two experiments yielding similar results.
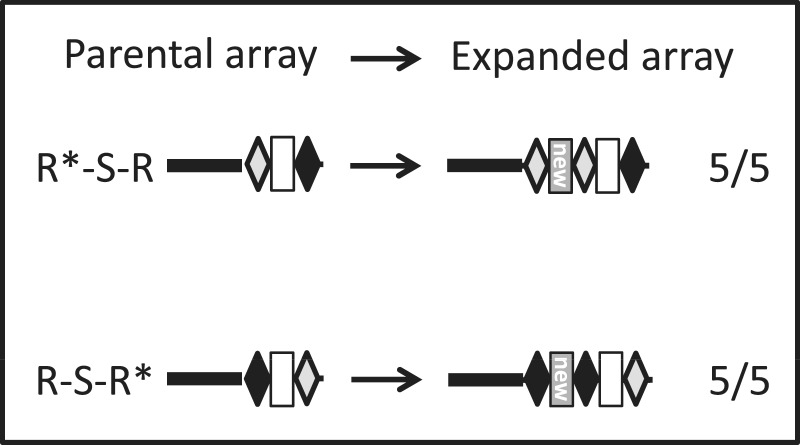
Figure 5.
Determination of the DNA template for newly inserted repeats. Schematic of the parental and expanded arrays in CRISPR I of E. coli BL21-AI. Repeats are marked as black or gray diamonds, depending on the nucleotide ‘label’, parental spacer as a blank rectangle whereas newly inserted spacer as gray rectangle marked ‘new’. R*-S-R: first repeat from BL21-AI consensus, second from K-12 consensus; R-S-R*: first repeat from K-12 consensus, second repeat from BL21-AI consensus. Indicated cultures of E. coli BL21-AI harboring plasmid pCas1+2 and grown in the presence or absence of inducers (0.2% l-arabinose and 0.1 mM IPTG) for 10–16 h for three cycles were streaked to isolate single colonies. Five isolated colonies from each culture showing insertion of a spacer in a PCR were DNA-sequenced. Five out of five colonies of each strain harbored labeled repeats, as indicated.
Strain and plasmid construction
Construction of strains and plasmids is detailed in the Supplementary Methods.
RESULTS AND DISCUSSSION
Establishment of an acquisition assay
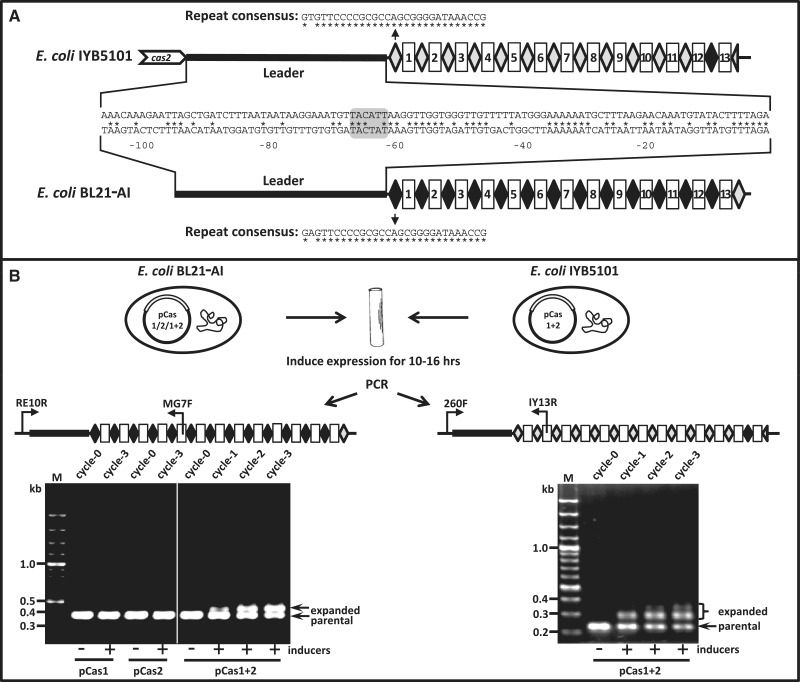
To study the adaptation process, we developed an assay for detection of the insertion of new spacers into CRISPR array I of E. coli. We used strains derived from E. coli B (BL21-AI) or K-12 (IYB5101), lacking or encoding the endogenous cas genes, respectively (Figure 1A). Both strains encode T7 RNA polymerase under an l-arabinose-induced promoter. We introduced plasmids pCas1, pCas2 or pCas1+2, encoding K-12-derived Cas1, Cas2 or both, respectively, under a T7-lac promoter, into these strains. Cas1 and Cas2 were tested since they are found in almost all CRISPR/Cas systems, and it has been hypothesized that since these proteins are not required in the expression and interference step, they might play a role in the adaptation step (2,3,10). Induced expression of E. coli Cas1 and Cas2 resulted in acquisition of spacers, as determined by PCR amplification of the repeat-spacer units adjacent to the leader terminus in CRISPR array I of both strains (Figure 1B). The size of each repeat-spacer unit is 61 bp, and accordingly PCR amplification from array I of cultures induced for Cas1 and Cas2 expression showed a band representing an increase of approximately this size (Figure 1B). Acquisition was detected at significant levels during the 10–16 h course of the assay by comparing the intensities of the parental-size band versus the higher MW band (expanded with a newly inserted spacer). Continuous growth of bacteria overexpressing Cas1 and Cas2 resulted in even higher MW bands, indicating that even more than a single spacer could be added into the CRISPR array under conditions of Cas1 and Cas2 overexpression (Figure 1B). The assay could detect acquisition events occurring in <1% of the total bacterial suspension, as determined by a titration experiment in which a known number of cells having expanded array were serially diluted with cells having the parental array and subjected to PCR analysis (Supplementary Figure S1). Strains harboring plasmids encoding Cas1 or Cas2 alone (pCas1 and pCas2, respectively) did not show observable expansion of their array (Figure 1B). Moreover, no expansion of the array was detected when pCas1+2 was mutated to encode Cas1D221A, Cas1 with a residue reported in P. aeruginosa and in E. coli to abolish the DNase activity of the protein without loss of protein stability (11,12) (Supplementary Figure S2A). Strains harboring pCas1, or pCas1D221A+2 or pCas2 plasmids showed expression levels of Cas1, Cas1D221A or Cas2 similar to or higher than their expression level detected in the strain harboring pCas1+2, in which adaptation does occur, indicating that lack of adaptation in these strains was not due to lower expression levels of these proteins (Supplementary Figure S2B). These results indicate that Cas1 and Cas2 are essential for the adaptation process and that the DNase activity of Cas1 is essential for the acquisition activity. The fact that adaptation occurs in BL21-AI, a strain lacking casABCDE genes (3,17), and in K-12 strain in which these genes are silenced by H-NS (18–20), indicates that these genes are dispensable for the adaptation step. They also indicate that despite a 1-nt difference at position 2 between the consensus repeats of CRISPR array I of IYB5101 and BL21-AI (Figure 1A), Cas1 and Cas2 can efficiently process both repeats. Interestingly, IYB5101 harbors an additional CRISPR array, array II, with repeats and leader sequences identical to those found in BL21-AI. In line with the observation that Cas1 and Cas2 processes the BL21-AI array efficiently, we observed significant expansion of IYB5101 array II, as expected. On the other hand, CRISPR array II in BL21-AI, which does not have conserved leader sequence upstream the repeats, did not show acquisition of new spacers (Supplementary Figure S3).
Figure 1.
Detection of spacer acquisition in BL21-AI and IYB5101. (A) Schematics of CRISPR I arrays and leader sequences from E. coli BL21-AI and IYB5101 (based on NCBI reference sequences of NC_012947.1 positions 1002800–1003800 and NC_000913.2 positions 2875600–2876800, respectively). Repeats are marked as gray diamonds for IYB5101 consensus and black diamonds for BL21-AI consensus. Last repeat in IYB5101 is marked as half a diamond to indicate that only half a repeat is present. Spacers are marked as numbered rectangles. The leader and repeat sequences of each strain are specified. Asterisk below or above a base indicates its conservation between the two strains. Gray highlighting indicates the (-10)-TATA box (18). (B) Schematics and results of the spacer-acquisition assay. Cultures of E. coli BL21-AI or IYB5101 harboring the indicated plasmids were grown in the presence of inducers (0.2% l-arabinose and 0.1 mM IPTG) for the indicated cycles. Each cycle represents dilution of 1:300 of a previously 10–16 h induced culture grown for additional 10–16 h in medium containing inducers. PCR products using the primers indicated as flagged arrows, from samples taken from cultures at the indicated cycle, were electrophoresed on an agarose gel and imaged. Parental and expanded bands are indicated. Gels are representative of two experiments yielding similar results.
Analysis of acquired spacers
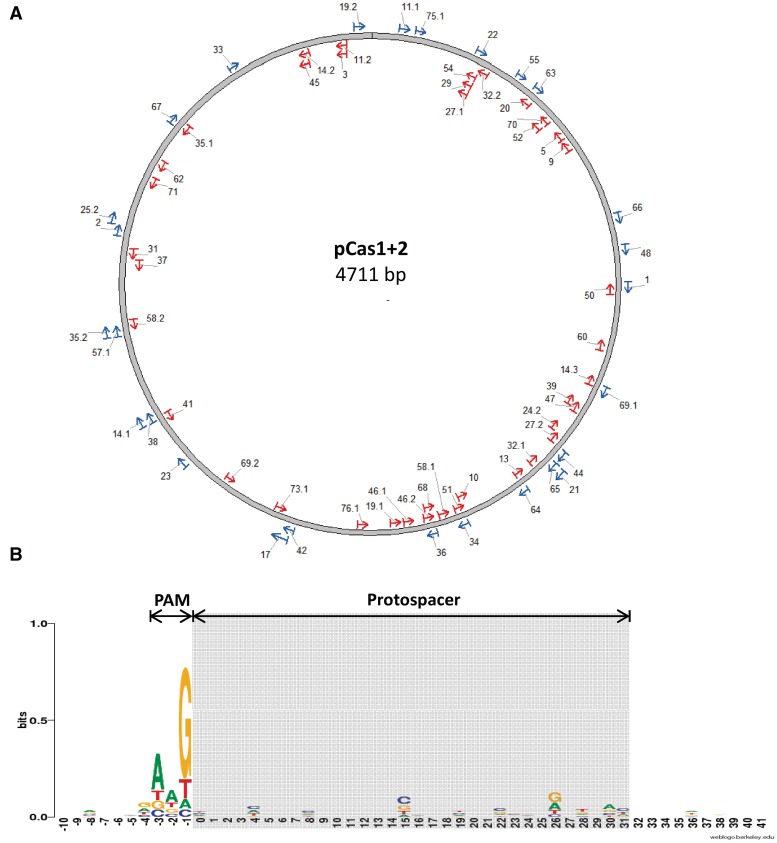
Detection of higher MW bands in PCR amplifying the CRISPR array suggested that new spacers inserted in the array. However, other possibilities exist for this observation. For example, rearrangement of spacers within the array could show these expansion patterns. To prove that the expanded array contained newly acquired spacers and to gain more insights on their nature, we sequenced DNA from the examined strains having expanded arrays. We used two approaches to isolate DNA for sequencing. The first was to ligate PCR-amplified DNA from bacterial cultures that underwent adaptation into plasmid vectors, and then transform and sequence the DNA inserts. The other was to streak the cultures on agar plates for isolation of individual clones and then to sequence DNA of individual clones showing expansion of the CRISPR array (see Supplementary Methods). Both approaches yielded DNA sequences of a total of 94 new spacers. The source of the new spacers was from pCas1+2 and also from genomic DNA, an expected observation, since these were the only DNA sources in the culture. Figure 2A shows the protospacer location and orientation on the plasmid DNA, whereas Supplementary Table S3 provides a detailed list of all sequenced spacers. For an unknown reason, the sequences originating from the plasmid DNA were highly overrepresented in the new spacers. The expected ratio of plasmid-derived spacers versus genome-derived spacers is ∼1:100, as the plasmid has on average 10 copies per cell (21) and its length is 4711 bp (yielding a total length of ∼4.5 × 104 bp) compared with a genome length of ∼4.5 × 106 bp. Nevertheless, the observed fraction of plasmid-derived spacers is 42/57 for BL2-AI and 24/37 for IYB5101, ∼200 folds more than expected. This result suggests that an active mechanism selectively acquires spacers from extrachromosomal DNA or that spacer acquisition from the genome kills the bacteria and thus reduces the observable occurrences of genomic spacers. The newly acquired genomic spacers cannot kill the bacteria using the CRISPR/Cas system in both the BL21-AI and in IYB5101 because in the former there are no cas genes, whereas in the latter, the presence of hns in the genome silences the activity of the system (18–20). In all instances in which a single spacer inserted, it was in the first position next to the leader. In a few cases, we observed expansion of up to three spacers, and these were in all cases located at the first, second and third positions adjacent to the leader, as observed in other systems (2). The length of most spacers was 32–33 bp, consistent with the observable length of spacers in the CRISPR array, except one instance of a spacer of 49 bp (clone 17, Supplementary Table S3). The observed PAM was AWG, in accordance with the reported motif (16) as analyzed by Weblogo (22) (Figure 2B). Nevertheless, the first two bases of the PAM, AW, were significantly less conserved than the third base, G. Analysis of motifs in the protospacer and in the 10 nt flanking it showed no significant conversation in other positions. We could not identify a bias for acquisition of spacers from any strand of DNA (Figure 2A, Supplementary Table S3), indicating that strand selection is random under the tested conditions. The number of clones acquiring new spacers following pCas1+2 overexpression for 10 h were 121/207 and 105/278 for E. coli BL21-AI and IYB5101, respectively. The high acquisition observed corroborates the intensity of the expanded band compared to the parental band in the PCR analysis in Figure 1B. This robust assay allowed us to study several aspects of the acquisition mechanism, as described below.
Figure 2.
Position and orientation of protospacers on plasmid DNA and analysis of protospacer adjacent motifs (PAMs). (A) Protospacers matching the sequenced spacers are marked on the pCas1+2 plasmid map according to their position and orientation (blue arrows pointing clockwise and red arrows pointing counterclockwise). Protospacers are numbered according to the clone from which the spacers were sequenced, as listed in Table S3. Spacer position in a sequenced array is separated from the clone number by a period, where applicable. (B) WebLogo (22) was used to analyze PAMs from sequenced spacers that were 32 bp long. The first nt of the protospacer is at position 0. Protospacer region is highlighted in gray. Weblogo of the protospacer as well as 10 nt upstream and downstream of the protospacer are shown. Relative letter size indicates the base frequency in that position.
Elements in the repeats essential for acquisition
We wanted to define the minimal number of repeats that is essential for acquisition. Therefore, we deleted most of the repeats, leaving 0, 1 or 2 of them in the array (Figure 3). The constructed strains were tested for their capacity to acquire new spacers using the assay described above. Results showed that the process of spacer acquisition required at least one repeat (Figure 3). Moreover, the efficiency of adaptation into an array having one or two repeats was similar to that of the parental array. These results indicated that a specific DNA sequence in the repeat is essential for adaptation (presumably a motif recognized by the acquisition machinery), but that repetition by itself is not required. Strikingly, as shown in the gel, the size of the inserted repeat-spacer unit into the single repeat array (1-rep) was ∼61 bp (size of inserted repeat-spacer unit), despite the absence of a spacer in this array. This indicates that the mechanism by which a spacer length is determined does not rely on previous spacer-repeat units in the array, suggesting that an inherent mechanism in the protein machinery dictates the size of the spacer. The inverse process of spacer acquisition—spacer deletion—probably occurs through recombination or slippage of the DNA polymerase during replication and requires at least two repeats (10,23). If deletion of spacers occurs through recombination of repeats, then theoretically, a deletion event could result in only a single repeat being left in the array. The fact that adaptation does not require more than one repeat explains how the CRISPR array may still expand, even if all of the CRISPR spacers are deleted by such an event, and this observation thus has physiological significance.
Elements in the leader sequence essential for acquisition
The leader sequence has been shown to promote transcription of the CRISPR array (18) and has been postulated to direct the orientation of the newly acquired spacers (2). We wanted to test whether the leader sequence is indeed essential to the adaptation process and to determine the essential DNA elements within the leader required for acquisition of new spacers. The leader sequences of E. coli BL21-AI and K-12 have extensive similarities in the ∼90 nt upstream of the first repeat, and thus we hypothesized that the essential region for spacer acquisition is found in these sequences (Figure 1A). We therefore systematically deleted DNA segments of the leader sequence at short intervals within these 90 nt, and a larger interval of 50 nt upstream of the first 100 nt. Initially, segments of 20, 40, 60, 80, 100 and 150 nt upstream of the first repeat were left intact in the genome of E. coli BL21-AI, by inserting a kanamycin-resistance cassette exactly upstream of these locations (Figure 4A). We then assayed for acquisition capability using our developed assay. The results of the acquisition assays carried out for BL21-AI-derived-strains indicated that within the tested intervals, 60 bp is the minimal length required for acquisition: leader sequences of 40 and 20 bp showed no acquisition at all (Figure 4A). It is interesting to note that deletion of the (-10)-TATA box [position (-61)–(-66) upstream of the first repeat (Figure 1A, (18)], required for transcription of the array, did not reduce the acquisition efficiency, suggesting that transcription may not be essential for the adaptation process.
To find out if elements within the 60-bp segment upstream of the first repeat are essential, we replaced 40 and 20 bp of the 3′-end of the leader sequence adjacent to the repeat, with the original DNA sequence, scrambled. This produced a leader with similar length as the parent but with a different sequence (Figure 4B). For technical reasons, we constructed these in a CRISPR array containing a single repeat, which was shown to be as functional as a complete array (Figure 3). In this case, no acquisition was observed when replacement of even 20 bp was tested. This indicated that at least some elements in the 20-bp segment upstream of the first repeat are essential for acquisition and that the mere presence of similar nucleotides of similar length is not sufficient for acquisition. This result was expected considering the high-sequence conservation (∼65%) between the leader sequences of BL21-AI and K-12 in this region (Figure 1A).
Mode of insertion of the repeat sequence
To further elucidate the insertion mode of the repeat, we took advantage of the fact that two variants of the functional repeats exist, one starting with the sequence 5′-GAG (e.g. repeat 1 and most repeats in E. coli BL21-AI), and one starting with 5′-GTG (e.g. repeat 14 of E. coli BL21-AI and most repeats in E. coli K-12) (Figure 1A). These variants were used as genetic labels of a two-repeat array (Figure 5). Labeling the repeats enabled us to determine which one serves as a template for replicating the newly inserted repeat, and whether the sequence of the template or its position in the array influences the outcome. Another insight that might be deduced from these experiments is whether the new repeat is synthesized de-novo or perhaps synthesized from a genetic source other than the array. If the repeat is always replicated from a single position in the array, then the label of the new repeat should change when positions are switched. Following labeling of the two repeats in the two possible positions, we sequenced five randomly selected colonies of each strain showing insertion of one repeat-spacer unit in the PCR amplification. Sequencing of all 10 colonies showed that the first and second repeats adjacent to the leader in the expanded array always carry the same label, regardless of the label of the third repeat in the array (Figure 5). This indicates that the new repeat is always replicated from repeat #1 (starting from the leader end) and not generated “de-novo” or from another genetic reservoir (e.g. repeat #2 or repeats from CRISPR array II).
Overall, our assay provides a robust tool for studying the adaptation process; using this tool, we define the minimal requirements for the process. We provide first direct evidence for the following: Cas1 and Cas2 are both essential for efficient adaptation of the CRISPR array, the leader has a direct role in spacer acquisition, and a single repeat is sufficient for spacer acquisition. In addition, we demonstrate that the inserted repeat is always replicated from the first repeat in the array proximal to the leader. We believe that these insights will significantly facilitate research on the adaptation process in E. coli, and consequently in other prokaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Tables 1–3, Supplementary Figures 1–3, DNA sequence of plasmid pCas1+2, and Supplementary References [18,23–26].
FUNDING
Funding for open access charge: The Israel Science Foundation (611/10); the Binational Science Foundation (2009218); and a Marie Curie International Reintegration Grant (PIRG-GA-2009-256340).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nir Osherov for critical reading of the manuscript and Camille Vainstein for professional language editing.
REFERENCES
- 1.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 6.Sorek R, Kunin V, Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Ploeg JR. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology. 2009;155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- 10.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 11.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 16.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 17.Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156:1351–1361. doi: 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- 18.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc. Natl Acad. Sci. USA. 2011;108:20136–20141. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 21.Kool AJ, Nijkamp HJ. Isolation and characterization of a copy mutant of the bacteriocinogenic plasmid Clo DF13. J. Bacteriol. 1974;120:569–578. doi: 10.1128/jb.120.2.569-578.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.