Abstract
Modification of ribosomal RNA is ubiquitous among living organisms. Its functional role is well established for only a limited number of modified nucleotides. There are examples of rRNA modification involvement in the gene expression regulation in the cell. There is a need for large data set analysis in the search for potential functional partners for rRNA modification. In this study, we extracted phylogenetic profile, genome neighbourhood, co-expression and phenotype profile and co-purification data regarding Escherichia coli rRNA modification enzymes from public databases. Results were visualized as graphs using Cytoscape and analysed. Majority linked genes/proteins belong to translation apparatus. Among co-purification partners of rRNA modification enzymes are several candidates for experimental validation. Phylogenetic profiling revealed links of pseudouridine synthetases with RF2, RsmH with translation factors IF2, RF1 and LepA and RlmM with RdgC. Genome neighbourhood connections revealed several putative functionally linked genes, e.g. rlmH with genes coding for cell wall biosynthetic proteins and others. Comparative analysis of expression profiles (Gene Expression Omnibus) revealed two main associations, a group of genes expressed during fast growth and association of rrmJ with heat shock genes. This study might be used as a roadmap for further experimental verification of predicted functional interactions.
INTRODUCTION
Modification of ribosomal RNA is ubiquitous among all living organisms. The proportion of modified residues roughly correlates with the complexity of an organism (1). Many efforts were invested in the determination of specific genes coding for rRNA modification enzymes and the mapping of modified residues in rRNA (2–4). A common opinion is that rRNA modification is needed for fine-tuning ribosome structure and function. All modifications of rRNA residues studied so far in Escherichia coli are dispensable. It is likely that modified nucleotides could be involved in the functioning of specific regulatory mechanisms of gene expression. In particular, modification of ribosomal RNA could be used for functional diversification of ribosomes. Hence, the existence of different ribosome species and the appearance of ‘altered’ ribosomes is well documented for eukaryotes (5). Plasmodium species express different rRNA genes when infecting either vertebrate or mosquito hosts (6). Ribosomal rpS6 phosphorylation alters translation efficiency (7). In yeast, ribosomal proteins encoded in the paralogue genes were shown to be specialized for mRNA-specific translation (8).
When speaking about possible ribosome specialization by rRNA modification, we should distinguish two possibilities. Firstly, ‘constitutive’ rRNA modification, independent of regulation by itself, could be necessary for the functioning of a specific regulatory mechanism. In this case, rRNA modification would be necessary for alteration of translation regulation but does not regulate it in a strict sense. This type of function could be revealed by a comparison of gene expression in the wild-type strain and the rRNA modification-deficient strain. Several laboratories, including our own, are involved in systematic analysis of gene expression differences caused by specific rRNA modifications. Second, ‘conditional’ rRNA modification dependent on some other factor would in turn influence translation of some mRNAs. This would be regulation by rRNA modification in a strict sense. In this case, not only the influence of rRNA modification on gene expression, but also some conditions influencing the extent of particular rRNA modification should be demonstrated.
In this study, we used mining several large data sets in search for genes/proteins functionally related to rRNA modification. As queries for this analysis, we chose known genes coding for rRNA modification enzymes of the most studied bacteria, E. coli (Table 1). The results of this study could be used to guide hypothesis-driven studies on the specific functional role of rRNA modification. An equally probable result of this study might be to suggest a secondary function of rRNA modification enzymes.
Table 1.
Known E. coli rRNA modification enzymes and modifications they made
| Enzyme | Nucleotide | Reference |
|---|---|---|
| Pseudouridine synthases | ||
| RsuA (YejD) | 16S rRNA 516Ψ | (25) |
| RluA (YabO) | 23S rRNA 746Ψ | (26) |
| RluB (YciL) | 23S rRNA 2605Ψ | (22) |
| RluC (YceC) | 23S rRNA 955Ψ, 2504Ψ, 2580Ψ | (17,27) |
| RluD (YfiI) | 23S rRNA 1911Ψ, 1915Ψ, 1917Ψ | (17,18) |
| RluE (YmfC) | 23S rRNA 2457Ψ | (22) |
| RluF (YjbC) | 23S rRNA 2604Ψ | (22) |
| Methyltransferases | ||
| RsmG (GidB) | 16S rRNA m7G527 | (28) |
| RsmD (YhhF) | 16S rRNA m2G966 | (3) |
| RsmB (YhdB) | 16S rRNA m5C967 | (29,30) |
| RsmC (YjjT) | 16S rRNA m2G1207 | (31) |
| RsmI (YraL) | 16S rRNA Cm1402 | (32) |
| RsmH (MraW) | 16S rRNA m4C1402 | (32) |
| RsmF (YebU) | 16S rRNA m5C1407 | (33) |
| RsmE (YggJ) | 16S rRNA m3U1498 | (34) |
| RsmA (KsgA) | 16S rRNA m62A1518, m62A1519 | (35,36) |
| RlmAI (RrmA,YebH) | 23S rRNA m1G745 | (37) |
| RlmC (YbjF, RumB) | 23S rRNA m5U747 | (38) |
| RlmF (YbiN) | 23S rRNA m6A1618 | (2) |
| RlmG (YgjO) | 23S rRNA m2G1835 | (39) |
| RlmH (YbeA) | 23S rRNA m3Ψ1915 | (40,41) |
| RlmD (YgcA, RumA) | 23S rRNA m5U1939 | (38,42) |
| RlmI (YccW) | 23S rRNA m5C1962 | (43) |
| RlmB (YjfH) | 23S rRNA Gm2251 | (44) |
| RlmL (YcbY) | 23S rRNA m2G2445 | (45) |
| RlmM (YgdE) | 23S rRNA 2498Cm | (4) |
| RlmN (YfgB) | 23S rRNA m2A2503 | (46) |
| RlmE (FtsJ, RrmJ) | 23S rRNA Um2552 | (47,48) |
MATERIALS AND METHODS
Database mining
Analysis of phylogenetic profiling was done using mutual information criteria using the STRING database search system (9), utilizing the probability assessment algorithm provided in the database engine. Significance criteria used was a probability of the functional linkage being more than or equal to 0.4. Data of gene co-occurrence in the bacterial genomes and all other data on functional association of genes were visualized by Cytoscape (10). Genome neighbourhood was analysed using the STRING database using significance criteria ≥0.7. Operone structure of rRNA modification genes was checked manually on the basis of published results of a deep sequencing analysis of the complete E. coli transcriptome (11). Co-expression profiling was analysed by the microbesonline database (12) on the basis of gene expression omnibus (GEO) database (13) entries related to E. coli. Only genes whose co-expression correlation coefficients with rRNA modification genes were ≥0.6 were selected. Protein co-purification data were extracted from the microbesonline database (12) based on the original data (14,15). Phenotype profile similarity data were extracted from http://ecoliwiki.net/tools/chemgen/ based on the original work (16). The phenotype landscape of rRNA modification gene knockout strains was extracted from an original published data set (16) and visualized by Microsoft Excel.
RESULTS AND DISCUSSION
Co-occurrence with rRNA modification genes in bacterial genomes
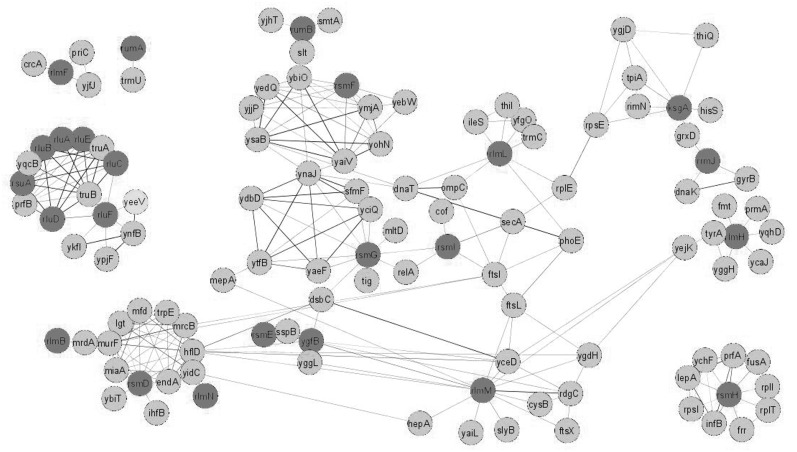
If enzymes are used in the same metabolic pathway, their genes’ presence across genomes is strictly correlated. A mutual information criteria were used as implemented into the STRING database (9) search engine. It is a more flexible criterion that accommodates not only simultaneous presence or absence of a pair of genes in a number of genomes but also similar degrees of conservation between pairs in a wide range of species. A similar degree of conservation reflects a co-evolution of genes. The confidence cut-off was set at 0.4 probability of functional relation. Mutual information analysis of genes responsible for rRNA modification revealed several independent clusters and clusters connected by a few interactions (Figure 1 and Supplementary File S1). In addition, nodes are observable, which connect several otherwise independent clusters. All rRNA pseudouridine synthases, several tRNA pseudouridine synthases and a translation termination protein RF2, formed a cluster. Clusterization of all pseudouridine synthases with the gene coding for RF2 raise the question of their functional connection. This issue was recently addressed for the RluD pseudouridine synthetase responsible for formation of three pseudouridine residues in the helix 69 of 23S rRNA (Table 1) (17,18). It was experimentally shown that the requirement of the rluD gene presence’s correlates with RF2 activity. Knockout of rluD caused a severe growth defect in E. coli K12 cells (19) due to increased UGA and, to a lesser extent, UAA stop codon read-through (20). This phenotype could be suppressed by a mutation in prfB gene coding for the RF2 translation termination factor. Moreover, in an E. coli strain B and Salmonella enterica, which both possess the prfB gene coding for an RF2 factor of higher activity, rluD inactivation has a much milder phenotype (21). In addition, the presence of rluF gene coding for 23S rRNA pseudouridine 2604 synthetase (22) correlates with the presence of several phage-related toxin genes, such as ykfI, ypjF, yeeV and the gene ynfB. These toxins are related to each other (23) and interfere with the bacterial cytoskeleton proteins FtsZ and MreB (24).
Figure 1.
Genes whose presence in bacterial genomes coordinated with the presence of rRNA modification genes. Shown are genes whose presence in bacterial genomes correlated with the presence of rRNA modification genes (dark grey) at the confidence level 0.4 and higher. Confidence measure was based on mutual information. Edge width corresponds to the confidence of interaction. Figure was created with the help of Cytoscape.
Mutual information analysis of nearly universal among bacteria methyltransferase RsmH responsible for 16S rRNA C1402 N4-modification (32) revealed a link with several translation-related proteins of high conservation, such as ribosomal proteins S9, L9 and L20 and translation factors EF-G, IF2, RRF, LepA, RF1 and ribosome-related GTPase YchF. Another cluster is formed by RsmD, RlmB and RlmN methyltransferases connected with a set of genes involved in a number of unrelated processes, namely cell wall biosynthesis, transcription, DNA repair and amino acid biosynthesis.
The RsmG gene responsible for N7-methylation of the16S rRNA nucleotide G527 (28,49) is linked with rsmI, which is responsible for ribose methylation of the 16S rRNA nucleotide C1402 (28). The RelA gene links both modification enzyme genes by an additional connection. Putative involvement of rsmG and rsmI methyltransferases in relA-dependent stringent response (50) could be suggested. RumB and RsmF methyltransferase genes form a poorly defined cluster with a number of genes with an unknown function. RlmL, responsible for N2-methylation of the 23S rRNA nucleotide G2445 (45), connects with the isoleucyl-tRNA synthetase gene, the ribosomal protein L5 gene and genes involved in 4-thiouridine biosynthesis. The KsgA methyltransferase gene forms a cluster with the rimN, gene coding for the ribosome maturation protein histidyl-tRNA synthetase gene, and several genes unrelated to translation. RrmJ forms a cluster with glutaredoxin 4 and the dnaK chaperone. The RlmH gene responsible for m3Ψ1915 formation in the 23S rRNA (40,41) forms a cluster with formyltransferase, tRNA (m7G46) methyltransferase, ribosomal protein L11-specific methyltransferase (prmA) and several genes unrelated to translation.
Neighbourhood in bacterial genomes
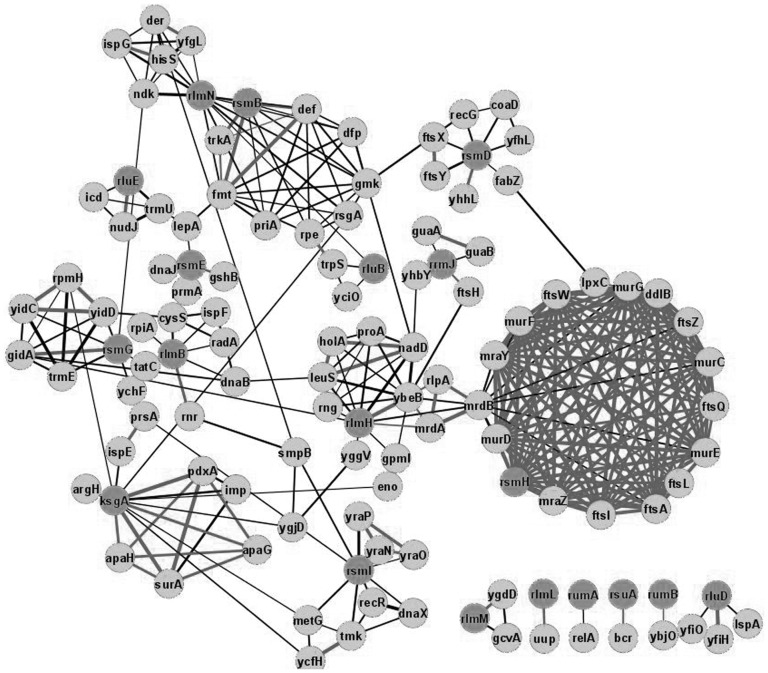
Knowing the operon structure of rRNA modification enzyme genes can be informative for formulating an ‘educated guess’ of regulatory processes involving rRNA modification (Figure 2 and Supplementary File S2). In Figure 2, a genomic neighbourhood analysis of rRNA modification genes among bacterial genomes is presented. The operon structure of rRNA modification genes in E. coli (11) is marked by bold edges connecting the genes on the same operon. Among rRNA modification genes, only few are coded as stand-alone cistrons. Several rRNA modification enzymes are co-expressed with tRNA modification enzymes. RsmG, responsible for 16S rRNA m7G527 formation (28), is located in the operon with mnmG, the gene coding for the tRNA modification enzyme, necessary for carboxymethylaminomethyl (cmnm) modification of U34 residue of certain tRNAs (51). This arrangement is conserved according to the large-scale genomes analysis. Although there is no direct contact between m7G527 and U34 of A-site-bound tRNA (52), modified nucleotides might ‘communicate’ via G530 undergoing syn-anti flip upon decoding (52).
Figure 2.
Neighbours of rRNA modification genes in bacterial genomes. Shown are genes and neighbours of rRNA modification genes (dark grey) in bacterial genomes. Confidence level cut-off was 0.7 and more. Edge width corresponds to the confidence of interaction. Bold edges correspond to genes located on the same operone as evidenced by E. coli transcriptome deep sequencing. Figure was created with the help of Cytoscape.
RluE, the gene coding for pseudouridine synthase, required for U to Ψ conversion at 23S rRNA position 2457 (22), is located adjacent to mnmA, a gene responsible for 2-thiouridine formation at tRNA nucleotide 34 (53). However, analysis of E. coli transcriptome does not reveal existence of the joint operon (11). The s2U residue could be subsequently converted to s2U residue, modified at position 5, depending on tRNA identity. It is difficult to imagine any communication between position 34 of tRNA and Ψ2457 of the 23S rRNA, except for communication through the tRNA body during the process of accommodation (54), which might be relevant to translation fidelity (55). A gene located in the same operon with rluE is nudJ, coding for a member of Nudix hydrolases family, responsible for catabolism of damaged and modified nucleoside triphosphates (56). Similarly, rsuA coding for the 16S rRNA Ψ516 synthetase (25) could be found on the same operon with a number of genes coding for pseudouridine metabolism.
HepA, located in the same transcript with rluA, is a protein involved in transcription termination (57), whereas RluD forms an operon with yfiH as evidenced from complete transcriptome sequencing (11). RsmB methyltransferase is coded in an operon (11) after protein deformylase and formyltransferase, involved in initiator fMet residue cleavage (58) and synthesis (59). After these two genes a strong Rho-independent terminator is encoded, followed by the rsmB gene that is not preceded by any Shine–Dalgarno-like sequence (60). RsmB works early in assembly of the small ribosomal subunit to form nucleotide m5C967 of the 16S rRNA (61). It is interesting how the RsmB enzyme could act in time for the modification of 16S rRNA if its expression has so many obstacles leading apparently to low enzyme concentration. RsmB has a NusB-like RNA-binding domain that could potentially serve as an assembly chaperone. A functional link between the initiator fMet-tRNAfMet tRNA and the 16S rRNA modified nucleotide m5C967 is quite logical. This modified residue is in direct contact with the initiator fMet-tRNAfMet tRNA during the process of translation initiation (62).
A surprisingly high proportion of rRNA modification genes are co-expressed with various genes coding for transmembrane proteins, proteins involved in cell wall synthesis, transmembrane transport, etc. The YhhL gene that partially overlaps with the 16S rRNA G966 N2-methyltransferase gene rsmD (3,60) and YbjO, located in the same operon (11) with the 23S rRNA U747 C5-methyltransferase gene rumB (38), are considered to be putative membrane proteins (63). RsmH is located in a large operon coding for many cell wall biosynthetic genes (11,63). The operon structure of rsmH is conserved and shows high significance by large-scale genome analysis (9). This arrangement suggests an experimental verification of a mutual influence of the 16S rRNA N4-C1402 methylation and cell wall biosynthesis. RlmH, which is involved in methylation of 23S rRNA nucleotide Ψ1915 (40,41), is located in an operon with several cell wall biosynthesis genes (11,63), but its nearest operon neighbour ybeB codes for a protein physically associated with 50S ribosomal subunits (64,65). The YbeB protein resembles plant Iojap protein associated with chloroplast ribosomes (66); however, its function remains unknown and is certainly worth experimental investigation.
KsgA, one of the most universal rRNA modification genes common in all living organisms, is responsible for formation of modified nucleotides m26A1518 and m26A1519 in the 16S rRNA (35,36). According to a recent finding, KsgA is not simply a modification enzyme but rather a switch protein sequestering incompletely assembled 30S subunits from being involved in translation (67–69). KsgA is encoded in an operon with a periplasmic peptidyl-prolyl isomerase (surA) (63) and apaH, coding for AppppA hydrolase (70). Diadenosine tetraphosphate (AppppA) is an ubiquitous alarmone molecule produced by lysyl-tRNA synthetase (71). Little is known about its function in bacteria, but its possible involvement in stress response was suggested in scientific literature (72). Several other stress–-response genes are located on the same operons with rRNA modification genes. The GshB, enzyme necessary for the last step in glutathione biosynthesis (73), is located in an operon with rsmE (11), whereas the RelA protein involved in ppGpp alarmone biosynthesis in response to amino acid starvation (28,74) is located next to the rumA gene. The RrmJ gene is located in the same operon with ftsH, a membrane-bound protease (47,48). Both genes are induced upon heat shock (75). Involvement of RrmJ, responsible for formation of Um2552 of the 23S rRNA (47,48), is certainly possible in a heat shock response. What is mysterious, however, is the fact that Um2552 is methylated constitutively, independent of heat shock (47).
The observed genomic neighbourhood of the 23S rRNA nucleotide G2251 ribose methyltransferase gene rlmB (44) with rnr gene, coding for RNase R involved in rRNA maturation and RNA decay under cold shock conditions and in a stationary phase, seems logical (76). Both proteins are involved in rRNA maturation. RNase R co-purifies with several ribosome maturation proteins, such as RlmN, RluB, RluC, RlmI and RlmL (14,15).
The Uup gene is linked with the rRNA methyltransferase gene rlmL, responsible for formation of the m2G2445 modified nucleotide in 23S rRNA (45). Uup codes for a soluble ATP-binding cassette (ABC) protein (77). Such proteins, exemplified with ABCE1 (78), could be involved in regulation of translation. Deletion of uup makes cells susceptible to contact-dependent inhibition (CDI) by the parent strain (77). Although the mechanism of such inhibition is unknown and involvement of rRNA modification in this process is questionable, it opens a possibility for further studies. It might be that this mechanism involves inactivation of rRNA either by a small molecule or enzyme and rRNA modification is relevant to this process.
Similar patterns of expression
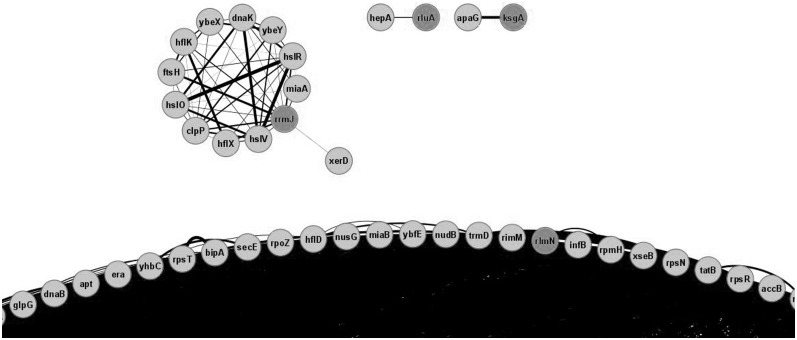
Expression pattern similarity data were extracted from the GEO database (13) with the help of www.microbesonline.org website (12). Co-expression in a number of growth conditions might reflect similar mechanisms of gene expression regulation, which, in turn speaks in favour of co-involvement in the same functional pathway. Using a confidence cut-off value of 0.6, one can observe that expression profiles for rRNA modification genes are separated into several uneven clusters (Figure 3, Supplementary Table S1 and Supplementary File S3). Two stand-alone binary clusters were visible reflecting co-expression based on the operon structure: ksgA with apaG and rluA with hepA. A larger cluster contains the rrmJ methyltransferase gene and several heat shock-related chaperone and protease genes. This link is expected one since rrmJ is located in the heat shock-induced operon and its expression increases with the temperature (75). All other rRNA modification genes form a huge cluster of a total 235 gene associated with exponential growth. This clusterization seems absolutely logical because exponential growth requires massive ribosome assembly and thus ribosome modification factors.
Figure 3.
Genes sharing same transcription profiles with rRNA modification genes in E. coli. Shown is part of the graph connecting rRNA modification genes (dark grey) with genes that have similar expression profiles. Confidence level cut-off was 0.6 and more. Visible are clusters formed by rrmJ and heat shock and protease genes, binary clusters based on an operone structure of rluA and ksgA genes and part of a huge cluster formed by genes relevant for the exponential phase growth. Edge width corresponds to the confidence of interaction. Figure was created with the help of Cytoscape.
At the co-expression profile confidence level cut-off of 0.7, several ribosome modification genes fall off the main cluster. RsmH forms a separate cluster with its operon neighbours mraZ and ftsL (11,63), and similarly rsmE clusters with gshB (11,63). Ribosomal RNA modification genes that remained associated with other genes induced in an exponential growth phase are rluC, rlmH, rlmI, rsmB, rsmG, rlmN and rlmL.
Phenotypes of knockout strains
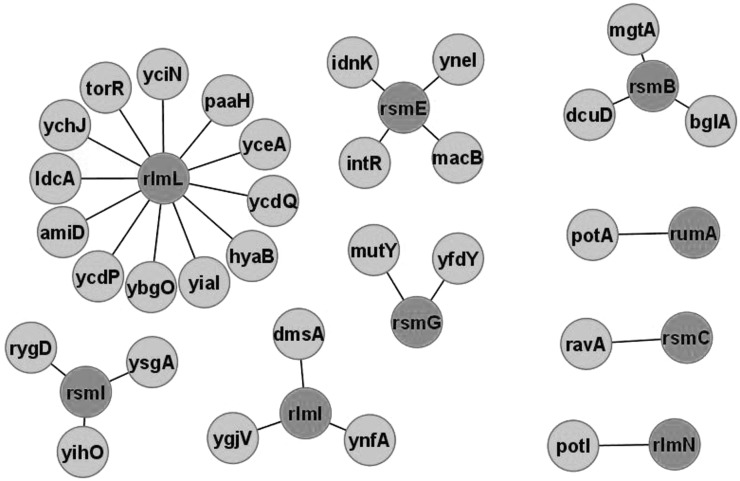
The availability of a comprehensive E. coli gene knockout collection (Keio) (79) made it possible to check gene influence on cell fitness under various stress conditions in high-throughput experiments (16). Phenotype profiles of knockout strains could be compared and this new measure could be used to find a functional relation between the genes. Analysis of gene knockout phenotype similarity revealed several genes whose knockout phenotypes resembles that for rRNA modification gene knockouts (Figure 4 and Supplementary File S4). Analysis of genes associated with rRNA modification genes according to phenotype profiling does not reveal any correlation with other data sets. Attention can be given to genes potA, potI and mgtA involved in transport of polyamines and magnesium ions (46). Both magnesium ions and polyamines associate with the ribosome and are required for the folding if rRNA (80). Genetic interaction between polyamine biosynthesis and rRNA modification was previously reported for KsgA rRNA methyltransferase and SpeD S-adenosylmethionine decarboxylase essential for spermidine biosynthesis (81). Double ksgA/speD knockout had a synergistic effect on kasugamicin resistance (81). Since KsgA is a switch protein involved in 30S subunit assembly, its functional link with the synthesis of essential rRNA counterions is logical. It should be noted, however, that phenotype profiling analysis does not link ksgA and speD (16), but other rRNA modification and polyamine-related genes (Figure 4).
Figure 4.
Genes sharing same phenotype profiles with rRNA modification genes in E. coli. Shown are the genes whose knockout phenotypes in a model set of conditions resemble those for the knockouts of rRNA modification genes (dark grey). Confidence level cut-off was 0.6 and more. Figure was created with the help of Cytoscape.
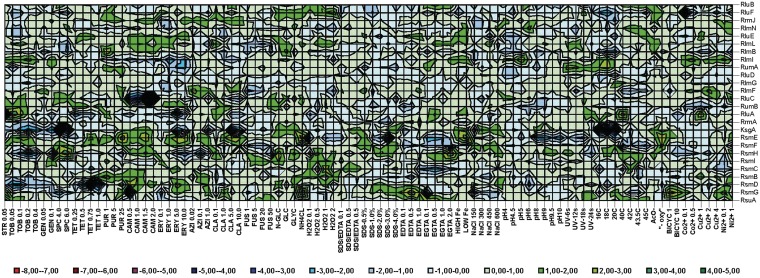
Despite the low efficiency of phenotype profile comparison for mining of functional relations between rRNA modification genes and other genes, phenotype profiles themselves give some insight into the functional role of rRNA modification. Figure 5 and Supplementary File S5 presents a phenotype landscape of rRNA modification gene knockouts in a subset of conditions relevant for translation. Several results were expected and match previously published experimental data, namely increased susceptibility of the rluC knockout strain to peptidyltransferase-associated antibiotics (82), streptomycin resistance of the rsmG knockout strain (28) and extreme cold sensitivity of the ksgA knockout strain (68).
Figure 5.
Phenotype landscape of rRNA modification gene knockout strains of E. coli. Shown are logarithms of E. coli knockout strains fitness at a number of conditions relevant to translation. Genes inactivated in a particular strain are indicated on the right. Conditions used are indicated below the landscape. Colour corresponds to the fitness of particular strain at particular condition. The colour scheme key is presented below the landscape. Figure was created with Microsoft Excel.
Modification of rRNA in some cases increases susceptibility and in some other cases resistance to antibiotics. Not only the rsmG knockout strain, but also the rsmC, and to some extent, the rsmF knockout strain, were found to be resistant to streptomycin (16). Knockouts of rsmD, rsmH and ksgA make cells hypersensitive to tobramycin, whereas knockouts of rluA and rlmI lead to moderate tobramycin resistance (16). Extreme and moderate spectinomycin sensitivity were found for the ksgA and rsmG knockout strains, whereas rsmH and to some extent rluC and rluB knockouts lead to spectinomycin resistance (16). High tetracycline sensitivity could be ascribed to the rsmD knockout strain, which is not surprising, because the tetracycline binding site is juxtaposed with a methylated nucleotide m2G966 of 16S rRNA that is formed by RsmD methyltransferase (83,84). Several strains defective in rRNA modification genes have altered sensitivity towards erythromycin. Strains devoid of rsmH, rluA, rumA and rlmI are hypersensitive, whereas knockouts of rsmE and rluE are moderately resistant to erythromycin. Susceptibility patterns for azitromycin and claritromycin resemble, but not perfectly match those for erythromycin (16).
Curiously, the rsmG knockout strain gains advantage in a number of conditions, such as the presence of EDTA, bicyclomycin and absence of a nitrogen source except ammonium chloride. Only in the presence of spectinomycin and a low concentration of chloramphenicol does the rsmG knockout has a disadvantage in comparison with the parental strain. It seems that the reason behind selective pressure to retain rsmG in bacterial genomes is not yet known.
Proteins co-purified with rRNA modification enzymes
Several massive co-purification studies to investigate E. coli interactome were done in recent years (14,15). Data regarding rRNA modification enzymes were extracted from databases using the www.microbesonline.org search engine (12). Most likely, interactions of the rRNA modification enzymes are formed in the context of the small and large ribosomal subunit assembly intermediates. Modification enzymes acting on rRNA utilize a complete range of assembly intermediates starting with deproteinized rRNA (39,45) and ending with associated 70S ribosomes (40,41). Apart from serving solely for rRNA modification, these enzymes might serve as switch proteins for assembly checkpoints [see ref. (85) for discussion]. The best documented case of rRNA methyltransferase involvement in assembly is KsgA, serving to prevent immature 30S subunits to start translation (67–69).
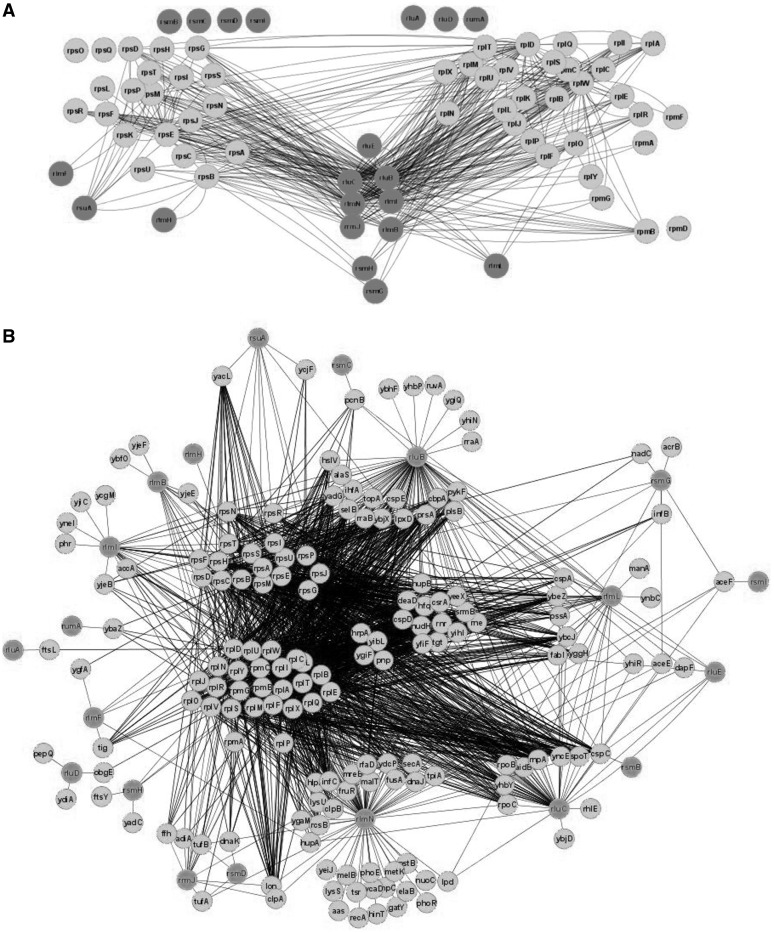
Firstly, interactions between rRNA modification enzymes and ribosomal proteins were analysed (Figure 6A and Supplementary File S6). Ribosomal proteins were sorted on the basis of small and large ribosomal subunit assembly maps (86,87). It was interesting to see if one can determine the protein composition of an assembly intermediate that is utilized by certain rRNA modification enzymes. The result was somewhat puzzling. Proteins that are exclusively co-purified with small subunit proteins are RsuA, RlmF and RlmH. RsuA is involved in the 30S subunit maturation (25), RlmH interacts with 70S ribosomes (41), whereas RlmF acts on the assembly intermediate of the large subunit (2). RlmL, 23S rRNA G2445 N2- methyltransferase (45) is exclusively associated with the 50S protein L1 protein, which binds independently with other proteins and away from the RlmL modification site (88). A number of 16S rRNA modification enzymes (RsmG and RsmH) and 23S rRNA modification enzymes (RluB, RluC, RlmB, RlmN, RlmI, RrmJ) co-purify with ribosomal protein components of both subunits (14,15). RluB, RluC and RluE, pseudouridine synthetases that modify residues in the peptidyltransferase centre form single complex.
Figure 6.
Proteins co-purified with rRNA modification enzymes in E. coli. Shown are rRNA modification proteins, ribosomal proteins and proteins co-purified with rRNA modification proteins. (A) Co-purification of rRNA modification enzymes with ribosomal proteins. Ribosomal proteins are located roughly as they are located in small (86) and large (87) subunit assembly map. Proteins involved in rRNA modification are dark grey and connected to its purification partners. (B) Co-purification of rRNA modification enzymes with both ribosomal and non-ribosomal proteins. Ribosomal proteins of small and large ribosomal subunits are grouped in the centre—to left part of figure. Proteins involved in rRNA modification are located on the periphery of the figure and dark grey. Proteins co-purified with rRNA modification enzymes are located in accordance with their co-purification partners. Those proteins that co-purifies with both rRNA modification enzymes and ribosomal proteins are located halfway between them, whereas proteins co-purified exclusively with rRNA modification enzymes but not with ribosomal proteins are located further on periphery, close to its interaction partners. Figure was created with the help of Cytoscape.
Proteins that co-purify with ribosomal proteins and rRNA modification enzymes (Figure 6B, Supplementary Table S2 and Supplementary File S7) are likely candidates for assembly chaperones. Most probably, co-purification results reflect not binary protein–protein interactions but a separation of a complete assembly intermediate. This category is populated by ribonuclease Rnr (76), helicases DeaD (89) and SrmB (90), chaperones DnaJ and DnaK (91,92), GTPase ObgE (93), GTPase activating protein YihI (94) and the RNA-binding proteins YhbY (95) and YibL (64) of yet unknown function. Somewhat surprisingly, factors involved in interaction with mature ribosomes, such as IF2, IF3, EF-Tu, SelB and EF-G could also be found among interaction partners. In addition, aminoacyl-tRNA synthetases and proteins involved in protein export could be found as interacting with rRNA modification enzymes and ribosomal proteins. It is possible that rRNA modification enzymes could still bind to completely assembled and functionally active ribosomes. Hypothetically, this could indicate possible involvement of rRNA modification enzymes not only in the assembly of the ribosome but also in post-assembly regulation of ribosomal function. Several DNA binding proteins could be found in the list. Most prominently, the HU protein was shown to interact with ribosomal proteins and as much as four rRNA modification enzymes. Nearly the entire set of cold shock protein family members co-purifies with rRNA modification enzymes and ribosomal proteins. Proteases and chaperones are also present in the set of co-purified proteins as well as a number of proteins with RNA degradation/processing and other RNA-related activities. NudH pyrophosphohydrolase initiates mRNA decay by removal of the triphosphate ‘cap’ from mRNA 5′-end (96). Co-purification of NudH (RppH) with ribosomal proteins and three rRNA modification enzymes indicates that it could act on rRNA as well as on mRNA. Similar conclusion could be made for HrpA helicase (97). A set of uncharacterized proteins were co-purified with high confidence with ribosomal proteins and several rRNA modification enzymes, putative methyltransferase YfiF, the RNA binding protein YbcJ, ATPase YjeE that was later shown to regulate tRNA N6-threonylcarbamoylation at adenosine residue (98), the predicted adenylate cyclase YgiF, conserved proteins YeeX and the putative DNA-binding protein YncE (99). These proteins are potential ribosome biogenesis factors.
From the set of proteins that co-purified solely with rRNA modification enzymes, one should also mention a complete set of subunits of the pyruvate dehydrogenase complex. This result might be considered artificial if not for the observation that three–five different rRNA modification enzymes independently co-purified with the pyruvate dehydrogenase complex. Jiang et al. (64) noticed large quantities of pyruvate dehydrogenase complex in their preparations of 50S subunit precursors, but they ascribed it to a co-migration of 50S subunits and multimeric pyruvate dehydrogenase on sucrose density centrifugation. Affinity purification (14,15) should not allow co-purification solely on the basis of physical dimensions. This interaction certainly deserves more detailed experimental investigation. After enolase was shown to be an integral part of the RNA degradosome (100), pyruvate dehydrogenase association with rRNA modification enzymes does not seem so strange.
Proteins and genes whose functional relations to rRNA modification enzymes were shown by several methods
We performed an analysis of data obtained from large biological datasets in a search for genes and proteins functionally related to rRNA modification enzymes. Several genes (proteins) could be identified as potential functional partners of rRNA modification enzymes by more than one method. The intersection of genes co-expressed, and proteins co-purified with rRNA modification enzymes, is not informative, since ribosomal proteins and translation components/ribosome assembly factors belong to the cluster of genes expressed in the logarithmic growth phase. Many of the corresponding proteins co-purify with rRNA modification enzymes.
More informative could be a comparison of proteins co-purified with rRNA modification enzymes and proteins whose genes are located close to the correspondent genes in the genomes. A number of putative functionally linked partners of rRNA modification enzymes could be identified in such a way: FtsY, FtsL, Rnr, DnaJ, YhbY and PrsA. FtsY is a signal recognition particle receptor, the protein involved in co-translational protein export (101). Ribonuclease R (Rnr) is involved in rRNA maturation (76), whereas chaperone DnaJ is involved in ribosome assembly (91,92). The role of protein YhbY in ribosome assembly is not clear yet, but its involvement in this process was demonstrated (64). Two proteins that were not yet shown to be involved in translation machinery function or assembly could be extracted from the list—the membrane-bound cell division protein FtsL and the phosphoribosylpyrophosphate synthase PrsA. Specific experiments are required to determine whether ribosome assembly/function is a secondary activity of these proteins or if it is an artefact of the performed experiments
CONCLUSIONS
Analysis of genes/proteins associated with rRNA modification enzymes by phylogenetic profiling, genome neighbourhood, co-expression profiling, similarity in the phenotype profile and co-purification revealed a substantial number of putative partners. Many of them are undoubtedly false positives, i.e. proteins or genes accidentally linked to rRNA modification. The majority of linked genes/proteins belongs to the translation apparatus, but does not have any specific relations to the process of rRNA modification. However, in the list of putative functional interactions that are listed in this work, there are undoubtedly proteins functionally related to the modification of rRNA. Among the co-purification partners of rRNA modification enzymes, there are many proteins known to participate in rRNA maturation and ribosome assembly. Several proteins whose involvement in assembly or maturation of the ribosome was not demonstrated are still good candidates for experimental validation. In this category one would list NudH, HrpA, YfiF, YbcJ, YjeE, YgiF, YeeX and YncE. Association of the HU protein and the pyruvate dehydrogenase complex with assembled ribosomes might reflect some unknown function of these well-characterized proteins.
Phylogenetic profiling (co-occurrence) of rRNA modification enzymes revealed several interesting links: pseudouridine synthetases with RF2, RsmH with translation factors IF2, RF1 and LepA and RlmM with DNA-binding protein RdgC. The latter is most puzzling and deserves experimental examination.
Genome neighbourhood analysis revealed a number of genes predicted to be functionally linked to rRNA modification. Several genes of this subset belong to other rRNA maturation processes, like ybeB and rnr. Plenty of genome neighbourhood connections deserve experimental verification of their functional linkage. Specifically, the conserved involvement of rlmH gene into an operone coding for cell wall biosynthetic proteins and ksgA involvement into a complex operone with a periplasmic peptidyl-prolyl isomerase (surA), AppppA hydrolase (apaH), the enzyme involved in vitamin B6 synthesis (pdxA) and a protein of unknown function (apaG) can bear further research. Several binary associations are also suggested to study experimentally, such as rsmD–yhhL, rrmJ–ftsH, rluE–nudJ, rluD–yfiH, rlmL–uup and rlmM–ygdD.
Comparative analysis of expression profiles through GEO revealed two main associations, the group of genes expressed during the fast growth, including rluC, rlmH, rlmI, rsmB, rsmG, rlmN and rlmL and the association of rrmJ with heat shock genes. The first association seems logical, since ribosome maturation should be especially effective during the logarithmic growth phase. The second association deserves special attention: are there any functional links between 23S rRNA U2552 ribose methylation and heat shock response?
The application of database mining in the search for potential functional links of rRNA modification enzymes allowed us to reveal many potential functional interaction partners. The results of this study might be used to plan further experimental verification of predicted functional interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1 and S2, Supplementary Cytoscape Files S1–S6.
FUNDING
Russian Foundation for Basic Research (grant numbers 10-04-01345-a, 11-04-01314-a, 11-04-01018-a and 11-04-12060-ofi); Russian Ministry of Science (grant 16.512.11.2108), Federal Agency for Science and Innovations (grant 02.740.11.0706); Moscow University Development Program (grant PNR 5.13). Funding for open access charge: Lomonosov Moscow State University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are very thankful to M. Gelfand for his advice on database selection and to Alexander Lebedeff for his efforts on improving the language of the article.
REFERENCES
- 1.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Sergiev PV, Serebryakova MV, Bogdanov AA, Dontsova OA. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 2008;375:291–300. doi: 10.1016/j.jmb.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 3.Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J. Biol. Chem. 2007;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purta E, O’Connor M, Bujnicki JM, Douthwaite S. YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 2009;72:1147–1158. doi: 10.1111/j.1365-2958.2009.06709.x. [DOI] [PubMed] [Google Scholar]
- 5.Ramagopal S. Are eukaryotic ribosomes heterogeneous? Affirmations on the horizon. Biochem. Cell Biol. 1992;70:269–272. doi: 10.1139/o92-042. [DOI] [PubMed] [Google Scholar]
- 6.Li J, McConkey GA, Rogers MJ, Waters AP, McCutchan TR. Plasmodium: the developmentally regulated ribosome. Exp. Parasitol. 1994;78:437–441. doi: 10.1006/expr.1994.1051. [DOI] [PubMed] [Google Scholar]
- 7.Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. Int. Rev. Cell Mol. Biol. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 8.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 15.Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene PJ, Santi DV. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry. 1998;37:15951–15957. doi: 10.1021/bi981002n. [DOI] [PubMed] [Google Scholar]
- 18.Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutgsell NS, Deutscher MP, Ofengand J. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA. 2005;11:1141–1152. doi: 10.1261/rna.2550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ejby M, Sorensen MA, Pedersen S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl Acad. Sci. USA. 2007;104:19410–19415. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor M, Gregory ST. Inactivation of the RluD pseudouridine synthase has minimal effects on growth and ribosome function in wild-type Escherichia coli and Salmonella enterica. J. Bacteriol. 2011;193:154–162. doi: 10.1128/JB.00970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Campo M, Kaya Y, Ofengand J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA. 2001;7:1603–1615. [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JM, Shaw KJ. A novel family of Escherichia coli toxin–antitoxin gene pairs. J. Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 2011;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrzesinski J, Bakin A, Nurse K, Lane BG, Ofengand J. Purification, cloning, and properties of the 16S RNA pseudouridine 516 synthase from Escherichia coli. Biochemistry. 1995;34:8904–8913. doi: 10.1021/bi00027a043. [DOI] [PubMed] [Google Scholar]
- 26.Wrzesinski J, Nurse K, Bakin A, Lane BG, Ofengand J. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for psi 746 in 23S RNA is also specific for psi 32 in tRNA(phe) RNA. 1995;1:437–448. [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad J, Sun D, Englund N, Ofengand J. The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23 S ribosomal RNA. J Biol Chem. 1998;273:18562–18566. doi: 10.1074/jbc.273.29.18562. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- 29.Tscherne JS, Nurse K, Popienick P, Michel H, Sochacki M, Ofengand J. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:1884–1892. doi: 10.1021/bi981880l. [DOI] [PubMed] [Google Scholar]
- 30.Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:4053–4057. doi: 10.1021/bi982364y. [DOI] [PubMed] [Google Scholar]
- 31.Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J Biol Chem. 1999;274:924–929. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 32.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J Mol Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poldermans B, Roza L, Van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16 S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30 S interactions. J. Biol. Chem. 1979;254:9094–9100. [PubMed] [Google Scholar]
- 36.Helser TL, Davies JE, Dahlberg JE. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 37.Gustafsson C, Persson BC. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S. Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003;31:4738–4746. doi: 10.1093/nar/gkg657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sergiev PV, Lesnyak DV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m2G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23 S rRNA. J. Mol. Biol. 2006;364:26–31. doi: 10.1016/j.jmb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Douthwaite S. YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA. 2008;14:2234–2244. doi: 10.1261/rna.1198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ero R, Peil L, Liiv A, Remme J. Identification of pseudouridine methyltransferase in Escherichia coli. RNA. 2008;14:2223–2233. doi: 10.1261/rna.1186608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwalla S, Kealey JT, Santi DV, Stroud RM. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J Biol Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- 43.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 44.Lovgren JM, Wikstrom PM. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 2001;183:6957–6960. doi: 10.1128/JB.183.23.6957-6960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesnyak DV, Sergiev PV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m2G methyltransferases: I. The ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J. Mol. Biol. 2006;364:20–25. doi: 10.1016/j.jmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Toh SM, Xiong L, Bae T, Mankin AS. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 2000;275:16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
- 48.Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 49.Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2) J. Bacteriol. 2007;189:3876–3883. doi: 10.1128/JB.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Block R, Haseltine AW. Purification and properties of stringent factor. J. Biol. Chem. 1975;250:1212–1217. [PubMed] [Google Scholar]
- 51.Yim L, Moukadiri I, Bjork GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 53.Kambampati R, Lauhon CT. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry. 2003;42:1109–1117. doi: 10.1021/bi026536+. [DOI] [PubMed] [Google Scholar]
- 54.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl Acad. Sci. USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Connor M, Dahlberg AE. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 56.McLennan AG. The Nudix hydrolase superfamily. Cell Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sukhodolets MV, Jin DJ. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 1998;273:7018–7023. doi: 10.1074/jbc.273.12.7018. [DOI] [PubMed] [Google Scholar]
- 58.Bingel-Erlenmeyer R, Kohler R, Kramer G, Sandikci A, Antolic S, Maier T, Schaffitzel C, Wiedmann B, Bukau B, Ban N. A peptide deformylase–ribosome complex reveals mechanism of nascent chain processing. Nature. 2008;452:108–111. doi: 10.1038/nature06683. [DOI] [PubMed] [Google Scholar]
- 59.Kahn D, Fromant M, Fayat G, Dessen P, Blanquet S. Methionyl-transfer-RNA transformylase from Escherichia coli: purification and characterisation. Eur. J. Biochem. 1980;105:489–497. doi: 10.1111/j.1432-1033.1980.tb04524.x. [DOI] [PubMed] [Google Scholar]
- 60.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 61.Weitzmann C, Tumminia SJ, Boublik M, Ofengand J. A paradigm for local conformational control of function in the ribosome: binding of ribosomal protein S19 to Escherichia coli 16S rRNA in the presence of S7 is required for methylation of m2G966 and blocks methylation of m5C967 by their respective methyltransferases. Nucleic Acids Res. 1991;19:7089–7095. doi: 10.1093/nar/19.25.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 63.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J. Bacteriol. 2007;189:3434–3444. doi: 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walbot V, Coe EH. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl Acad. Sci. USA. 1979;76:2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Z, O’Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 68.Connolly K, Rife JP, Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mangat CS, Brown ED. Ribosome biogenesis; the KsgA protein throws a methyl-mediated switch in ribosome assembly. Mol. Microbiol. 2008;70:1051–1053. doi: 10.1111/j.1365-2958.2008.06484.x. [DOI] [PubMed] [Google Scholar]
- 70.Blanchin-Roland S, Blanquet S, Schmitter JM, Fayat G. The gene for Escherichia coli diadenosine tetraphosphatase is located immediately clockwise to folA and forms an operon with ksgA. Mol. Gen. Genet. 1986;205:515–522. doi: 10.1007/BF00338091. [DOI] [PubMed] [Google Scholar]
- 71.Zamecnik PC, Stephenson ML, Janeway CM, Randerath K. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 1966;24:91–97. doi: 10.1016/0006-291x(66)90415-3. [DOI] [PubMed] [Google Scholar]
- 72.Varshavsky A. Diadenosine 5′, 5′-P1, P4-tetraphosphate: a pleiotropically acting alarmone? Cell. 1983;34:711–712. doi: 10.1016/0092-8674(83)90526-3. [DOI] [PubMed] [Google Scholar]
- 73.Apontoweil P, Berends W. Glutathione biosynthesis in Escherichia coli K 12. Properties of the enzymes and regulation. Biochim. Biophys. Acta. 1975;399:1–9. doi: 10.1016/0304-4165(75)90205-6. [DOI] [PubMed] [Google Scholar]
- 74.Atherly AG. Ribonucleic acid regulation in amino acid-limited cultures of Escherichia coli grown in a chemostat. J. Bacteriol. 1974;120:1322–1330. doi: 10.1128/jb.120.3.1322-1330.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R: comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 77.Murat D, Goncalves L, Dassa E. Deletion of the Escherichia coli uup gene encoding a protein of the ATP binding cassette superfamily affects bacterial competitiveness. Res. Microbiol. 2008;159:671–677. doi: 10.1016/j.resmic.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartetzko A, Nierhaus KH. Mg2+/NH4+/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol. 1988;164:650–658. doi: 10.1016/s0076-6879(88)64075-4. [DOI] [PubMed] [Google Scholar]
- 81.Ochi K, Kim JY, Tanaka Y, Wang G, Masuda K, Nanamiya H, Okamoto S, Tokuyama S, Adachi Y, Kawamura F. Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance. Antimicrob. Agents Chemother. 2009;53:193–201. doi: 10.1128/AAC.00873-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toh SM, Mankin AS. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. J. Mol. Biol. 2008;380:593–597. doi: 10.1016/j.jmb.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 84.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sergiev P, Golovina A, Prokhorova I, Sergeeva O, Osterman I, Nesterchuk M, Burakovsky D, Bogdanov A, Dontsova O. (2011) Modifications of Ribosomal RNA: From Enzymes to Function. Wien: Springer; [Google Scholar]
- 86.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30 S ribosomal particles from RNA and proteins. Proc. Natl Acad. Sci. USA. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl Acad. Sci. USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nevskaya N, Tishchenko S, Volchkov S, Kljashtorny V, Nikonova E, Nikonov O, Nikulin A, Kohrer C, Piendl W, Zimmermann R, et al. New insights into the interaction of ribosomal protein L1 with RNA. J. Mol. Biol. 2006;355:747–759. doi: 10.1016/j.jmb.2005.10.084. [DOI] [PubMed] [Google Scholar]
- 89.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50 S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009;37:6540–6549. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rene O, Alix JH. Late steps of ribosome assembly in E. coli are sensitive to a severe heat stress but are assisted by the HSP70 chaperone machine. Nucleic Acids Res. 2011;39:1855–1867. doi: 10.1093/nar/gkq1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Al Refaii A, Alix JH. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 2009;71:748–762. doi: 10.1111/j.1365-2958.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- 93.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 94.Hwang J, Inouye M. A bacterial GAP-like protein, YihI, regulating the GTPase of Der, an essential GTP-binding protein in Escherichia coli. J. Mol. Biol. 2010;399:759–772. doi: 10.1016/j.jmb.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 95.Ostheimer GJ, Barkan A, Matthews BW. Crystal structure of E. coli YhbY: a representative of a novel class of RNA binding proteins. Structure. 2002;10:1593–1601. doi: 10.1016/s0969-2126(02)00886-9. [DOI] [PubMed] [Google Scholar]
- 96.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 97.Koo JT, Choe J, Moseley SL. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- 98.Hashimoto C, Sakaguchi K, Taniguchi Y, Honda H, Oshima T, Ogasawara N, Kato J. Effects on transcription of mutations in ygjD, yeaZ, and yjeE genes, which are involved in a universal tRNA modification in Escherichia coli. J. Bacteriol. 2011;193:6075–6079. doi: 10.1128/JB.05733-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kagawa W, Sagawa T, Niki H, Kurumizaka H. Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE. Acta Crystallogr. D Biol. Crystallogr. 2011;67:1045–1053. doi: 10.1107/S0907444911045033. [DOI] [PubMed] [Google Scholar]
- 100.Chandran V, Luisi BF. Recognition of enolase in the Escherichia coli RNA degradosome. J. Mol. Biol. 2006;358:8–15. doi: 10.1016/j.jmb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 101.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.