Abstract
Synthetic biology applications call for efficient methods to generate large gene cassettes that encode complex gene circuits in order to avoid simultaneous delivery of multiple plasmids encoding individual genes. Multiple methods have been proposed to achieve this goal. Here, we describe a novel protocol that allows one-step cloning of up to four gene-size DNA fragments, followed by a second assembly of these concatenated sequences into large circular DNA. The protocols described here comprise a simple, cheap and fast solution for routine construction of cassettes with up to 10 gene-size components.
INTRODUCTION
Construction of large gene cassettes that encode entire gene circuits is in acute demand in synthetic biology. Traditional cloning techniques make this process extremely laborious due to the step-wise nature of these protocols and the increasing dearth of unique restriction sites as the constructs become larger. As a result, the last two decades have seen the search for more efficient methods, with the first major advance being the invention of Ligase-independent cloning (LIC) (1–4). That method circumvented the use of restriction sites by generating long, unique single-stranded overhangs using the 3′→5′ exonuclease activity of T4 DNA polymerase in combination with flanking dsDNA termini lacking one of the four nucleotides (‘chew-back’) (Figure 1). The original report demonstrated the ligation of an insert into a vector, i.e. the complexity of the process did not go beyond traditional restriction–ligation cloning. To the best of our knowledge, the first attempt to combine three fragments with long ssDNA overhangs was described by Donahue et al. (5). However, generating the overhangs was enabled by including ribose residues in the PCR primers, which would not allow a hierarchical assembly of larger constructs without recurrent PCR steps since the ribose residues are lost after bacterial amplification. Performing PCRs after each assembly step is unfavourable when the assembly intermediates reach a certain size and it may introduce additional mutations. A different strategy was shown in a report by Geu-Flores et al. (6), where overhangs were generated by uracil excision-based cloning. Four fragments were assembled in a single step; however, the requirement to include dU residues in the primers results in the same PCR dependency, which is even further restricted to a special DNA polymerase.
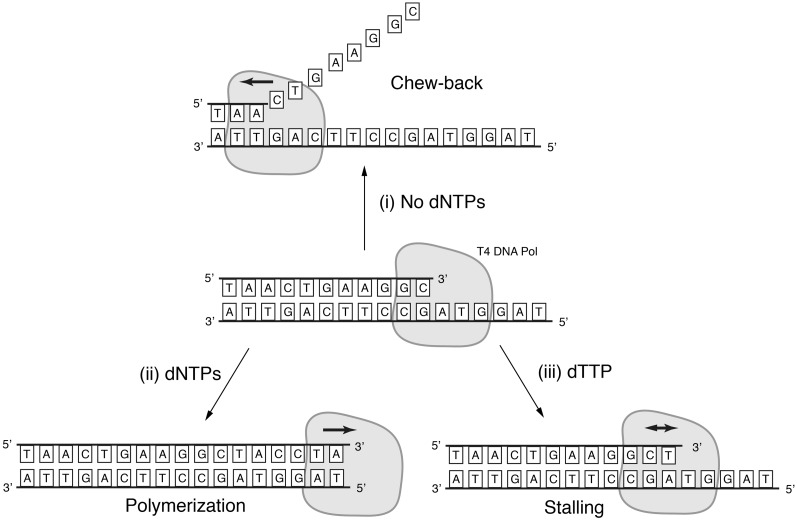
Figure 1.
T4 DNA polymerase is a proof-reading polymerase. In the absence of deoxyribonucleotide triphosphates, it digests DNA strands in the 3′→5′ direction (i). In the presence of all four deoxyribonucleotide triphosphates and a partially single-stranded template, it extends the recessed 3′-end of DNA strand (ii). Both enzymatic activities compete with each other when only some of the deoxyribonucleotide triphosphates are present. In the example shown in the figure, the presence of dTTP causes the polymerase to stall (iii).
A number of breakthroughs in high-throughput DNA assembly were reported in the context of whole-genome synthesis by Gibson et al. Two alternative methods were put forward. The first is an extension of LIC, with a major difference being the non-specific chew-back of overlapping DNA termini, and the reliance on DNA repair machinery of the bacterial host to deal with imperfect annealing of the resulting overhangs. Overlaps of at least 40 bp were shown to allow assembly of up to four fragments in a single cloning reaction (7,8). Another feature of the method is the hierarchical assembly, where the cloning vectors that contain the assembled fragments contain NotI restriction sites that are used to excise the combined sequences for the next cloning level. While the method is highly efficient in producing very long DNA from synthetic starting materials of a few kilobases, it is unclear whether the protocols could efficiently assemble shorter building blocks of fewer than 1000 base pairs due to the risk of complete DNA degradation by non-specific chew-back. Besides, generating overhangs of 40 bp in a PCR reaction requires relatively expensive primers of at least 60 nt in length. Moreover, including additional functional sequences in the primers, a common practice in recombinant DNA work, can easily push the total primer length to 100 nt.
The second method recently shown by the same group demonstrates concurrent assembly of up to 25 DNA fragments in yeast using recombination of overlapping DNA termini (9). While being a tour-de-force of high-throughput assembly, a few features of the process might pose problems in gene circuit assembly. First, the overlaps are at least 80 bp long and thus may not be readily introduced via PCR primers. Second, a sequence that appears more than once in different building blocks (such as a common promoter) could lead to undesirable recombination and a compromised final product.
The method we describe here uses short overlaps of ∼20 bp and specific chew-back to accomplish hierarchical assembly of about 10 gene-size DNA fragments in a two-step process.
MATERIALS AND METHODS
Primer phosphorylation
Primers were phosphorylated in 20 μl reactions containing 5 μM primer, 1× PNK buffer (NEB), 1 mM ATP and 8 U PNK for 1 h at 37°C. The enzyme was heat-inactivated for 20 min at 65°C.
PCR amplification
PCR amplification was performed according to the manufacturer’s protocol with either Pfu Ultra II Fusion HS (Inverter circuit, reprogramming circuits 1 and 2); KOD extreme DNA polymerase (for the CAG promoter containing amplicons in reprogramming circuits) or with Phusion DNA polymerase (reprogramming circuit 3). The heat-inactivated phosphorylation reactions were used as the source of primers without further purification. Where DpnI digest was performed, 50 µl of PCR reaction were mixed with 5.5 µl of Fermentas FastDigest Green buffer and 2 µl DpnI (Fermentas FastDigest). The reactions were incubated at 37°C for 1 h.
Pfu Ultra II Fusion HS (Stratagene)
A 100 μl reaction mix contained: 1 × Pfu Ultra II buffer, 250 μM dNTPs each, 5 ng plasmid template, 4 μl each primer phosphorylation mixture, unpurified, and 2 μl Pfu Ultra II Fusion HS polymerase. The temperature cycling protocol was:
2 min at 95°C,
20 s at 95°C,
20 s at 68°C,
15 s/kb at 72°C,
3 min at 72°C and
hold at 4°C.
Steps (2)–(4) were repeated 30 times.
Phusion HF polymerase (Finnzymes)
A 50 μl reaction contained: 1 × Phusion HF buffer, 200 μM each dNTP, 5 μl each primer phosphorylation mixture, unpurified, 20 ng plasmid template DNA and 0.02 U/μl Phusion HF polymerase. The temperature cycling protocol was:
30 s at 98°C,
10 s at 98°C,
20 s at 62°C
20 s/kb at 72°C
5 min at 72°C and
hold at 4°C.
Steps (2)–(4) were repeated 30 times.
KOD extreme polymerase (Merck)
A 50 μl reaction contained: 1× KOD extreme buffer, 400 μM each dNTP, 3 μl each primer phosphorylation mixture, unpurified, 5 ng plasmid template DNA and 0.02 U/μl KOD extreme hotstart polymerase. The ‘step down’ temperature cycling protocol was used with KOD polymerase:
2 min at 95°C,
1 s at 98°C,
1 min/kb at 74°C,
1 s at 98°C,
1 min/kb at 72°C,
1 s at 98°C,
1 min/kb at 70°C,
1 s at 98°C,
1 min/kb at 68°C and
hold at 4°C.
Steps (2) and (3) were repeated 5 times;
Steps (4) and (5) were repeated 5 times;
Steps (6) and (7) were repeated 5 times; and
Steps (8) and (9) were repeated 15 times.
AarI digestion
One microgram of plasmid DNA was digested by AarI in a 20 μl reaction containing 1× AarI buffer (Fermentas) supplemented with 1× oligonucleotide supplied by the manufacturer and 1 μl of AarI (Fermentas). After incubation at 37°C, the reactions were purified by agarose gel electrophoresis.
PacI digestion
PacI enzyme (NEB) was used according to the manufacturer’s instructions.
Gel purification of fragments
PCR products or restriction reactions were run on 1% agarose gels containing EtBr at 80 V for 45 min. Bands of correct size were excised and purified with a Qiagen QIAquick gel extraction kit according to the manufacturer’s protocol.
Concentration estimation
Fragment concentrations were measured using a NanoDrop 1000 (Thermo Scientific).
Oligonucleotides in assembly reactions
Two microlitres of the sense oligo phosphorylation reaction were mixed with 2 μl of the antisense oligo phosphorylation reaction and 246 μl water. Two microlitres of this mixture were used in a regular assembly reaction to yield a final concentration of 2 nM in 40 μl.
Chew-back reactions
All DNA fragments of a single assembly that were to be chewed in the presence of the same extension nucleotide were included at 2 nM concentration in a 20 μl reaction containing 1× NEB2 buffer (NEB), 0.1 mg/ml BSA (NEB), 1 U T4 DNA polymerase (NEB) and 1 mM extension nucleotide triphosphate (Invitrogen). Reactions were incubated at 27°C for 5 min and put on ice until the PCR block reached the inactivation temperature of 75°C. Reactions were put back to the block at 75°C. Immediately, corresponding chew-back reactions were pooled for each assembly in a 1:1 ratio. The reactions were kept at 75°C for 20 min and then slowly cooled to 55°C at a ramp rate of 1°C/min. After incubation for 20 min at 55°C, the samples were slowly cooled to room temperature by 0.4°C/min. Note that incubation below room temperature is not advised.
Bacterial transformation
Fifty microlitre aliquots of chemically competent Escherichia coli DH10b were thawed on ice, mixed with 4 μl of assembly mixes and incubated immediately at 42°C for 1 min. After putting on ice for 2 min, cells were allowed to recover for 1 h at 37°C while shaking in 500 μl LB medium without antibiotics. After that, tubes were shortly spun down and the pellet was resuspended in 40 μl of medium in order to be plated on LB agar plates containing the appropriate antibiotic.
Liquid culture, miniprep of plasmid DNA
Colonies were picked with a clean pipette tip to inoculate 3 ml liquid culture in LB medium with the appropriate antibiotic. Roughly, after 16 h of incubation, bacteria were pelleted and purified using a plasmid Miniprep kit (Qiagen) according to the manufacturer’s manual.
Test restrictions
Test restrictions were carried out in 10 μl volumes using NEB or Fermentas enzymes and the buffer suggested by the manufacturer for 20 min at 37°C. Samples were run on 1% agarose gels containing EtBr to visualize DNA bands under UV light.
RESULTS
The basic features of the chew-back technique
The ability of proof-reading DNA polymerases like T4 DNA polymerase to chew-back the 3′ ends of double-stranded DNA molecules makes it possible to generate well-defined ssDNA overhangs of arbitrary length at the termini of dsDNA molecules. Since the enzyme’s exonucleolytic activity constantly competes with its polymerase activity, the 3′→5′ strand digestion of dsDNA molecules stops as soon as the just-removed base is also present in the reaction as a deoxyribonucleotide triphosphate. Thus, one can engineer dsDNA termini such that they are only composed of three or fewer types of DNA bases, and flank those restricted sequences with the omitted bases. If mononucleotides complementary to the flanking bases are added to a T4 polymerase reaction mix, the 3′→5′ exonuclease is stopped exactly when the flanking bases are reached (Figures 1 and 2). The same mononucleotide can counteract the exonuclease activity in both the top and the bottom strands of a dsDNA molecule if the sequences are properly designed.
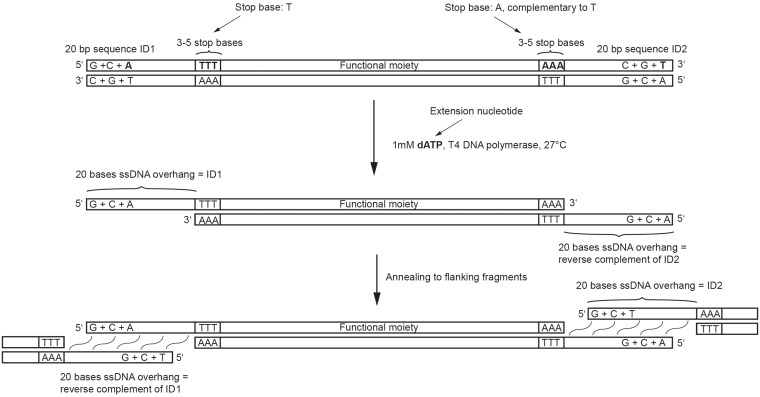
Figure 2.
A sequence of interest contains at both ends a stretch of sequence with restricted nucleotide composition (ID sequence) that comprises only three out of four nucleotide types. Between this sequence and the region to be cloned are three consecutive nucleotides of the type missing from the ID sequence. T4 DNA polymerase treatment generates two 20-mer single-stranded overhangs, allowing annealing to two other DNA fragments with complementary overhangs.
As an example, the 5′ nuceleotide-restricted sequence portion of the dsDNA top strand may contain dG, dC and dA followed by one or more dT bases. In this case, the extension nucleotide dATP should be added to the chew-back reaction. Accordingly, the 5′→3′ sequence of the bottom strand at the opposite dsDNA end must likewise contain only dG, dC and dA followed by dT. Once the first dT is exposed in the template strands (top strand at the 5′-end of the dsDNA substrate, and bottom strand at the 3′-end of the dsDNA substrate), dA is incorporated into the digested strand by the polymerase activity of the enzyme, thus effectively stopping the digestion process. To summarize, the dsDNA molecule should have the following structure, where the bases in grey will be removed during chew-back:
[G/C/A]-[T]-[NNNN]-[A]-[G/C/T]
[C/G/T]-[A]-[NNNN]-[T]-[C/G/A]
When designing cloning experiments using computer software, only the top strand of dsDNA molecules is normally used; to avoid confusion due to chew-back of both the top and the bottom strands, we use the following terminology throughout this report: the sequences with restricted nucleotide composition are called ‘ID sequences’; an ID sequence is always defined using the top strand of the dsDNA to which it is attached so that if the same ID sequence is placed at the 3′-end of molecule A and at the 5′-end of molecule B, the resulting complementary overhangs will anneal and concatenate A with B after the digested fragments are mixed. A ‘stop base’ of an ID sequence is a nucleotide that is not used in this ID, always determined using the top strand of a dsDNA. In the example above, the stop nucleotide for the ID sequence at the 5′-end of the dsDNA molecule is dT; it is dA for the ID at the 3′-end. A mononucleotide used to counteract the chew-back reaction is called an ‘extension nucleotide’. When an ID sequence is placed at the 5′-end of the top strand, the extension nucleotide is complementary to the stop base. When an ID sequence is placed at the 3′-end of the top strand, the extension nucleotide is identical to the stop base. Since the same extension nucleotide must control the chew-back at both dsDNA termini in a reaction, the stop bases at both ends must be complementary to each other (Figure 2).
Assembly reaction design
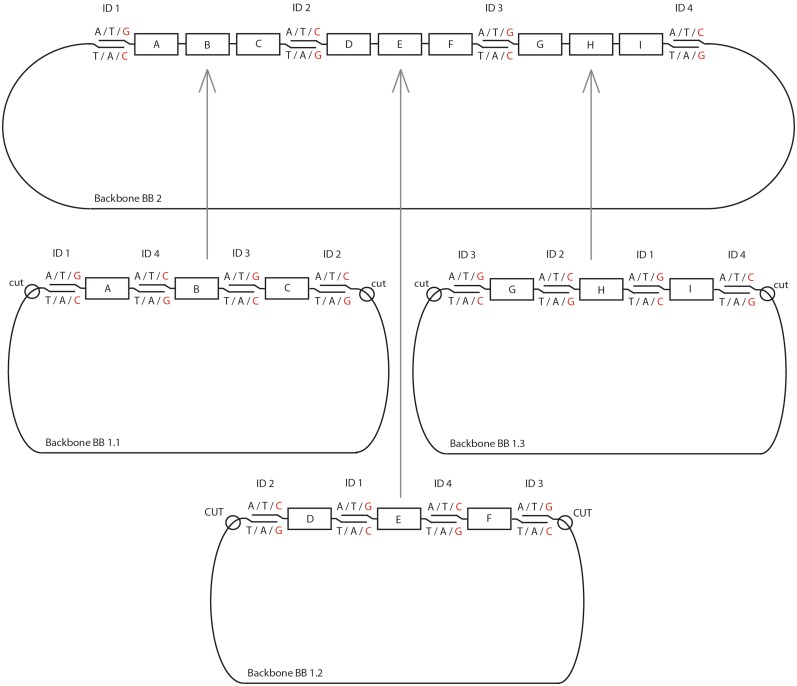
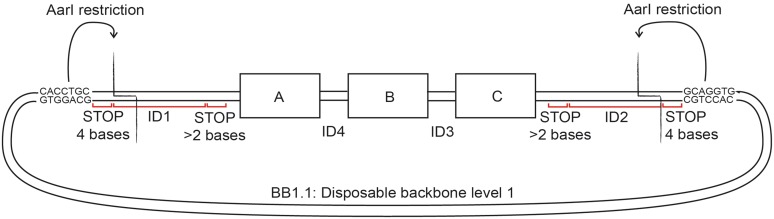
In our hands, we could assemble up to four gene-size fragments in a single reaction to create a circular plasmid that can be propagated in bacteria. The limit on the fragment number of this method remains an open question since assemblies with more DNA fragments have not been tested. For larger assemblies, the process is performed in a hierarchical fashion as shown in Figure 3. Composite constructs A–C, D–F and G–I are assembled from individual fragments A, B,… I and cloned into vector backbones BB1.1, BB1.2 and BB1.3. For the second level of assembly, these composite constructs are excised from their backbones by enzymatic digestion and are assembled together with the new backbone BB.2 in another four-part assembly step.
Figure 3.
Schematics of the assembly process. A 10-part assembly is created hierarchically, first by cloning concatenated three-fragment constructs in appropriate backbones flanked by restriction sites. Subsequently, the partial constructs are released from the backbones and assembled together in a separate process, which reuses the terminal ID sequences, to render the desired cassette.
The ID sequence design is dictated by the order in which the fragments are to be assembled in the full-size cassette. In the example of composite construct A–C in Figure 3, fragment A has to anneal to fragments B and to BB1.1, fragment B has to anneal to fragment A and C, and so forth. As discussed above, ID sequences on both termini of each dsDNA are chewed-back simultaneously so that the same extension nucleotide must be used for both. For example, the single-stranded overhangs generated on fragment A may contain dA, dT and dG, lack dC, and require dG as an extension nucleotide. Therefore, the fragments that anneal to both sides of A − BB1.1 and B − must contain nucleotides dA, dT and dC in their overhangs, lack dG, and use dC as the extension nucleotide. Likewise, fragment B should anneal to A and C; as we have just determined that B must use dC as an extension nucleotide, fragment C must use dG as an extension nucleotide. In summary, this requires that the extension nucleotides used to generate ssDNA overhangs alternate between adjacent fragments, either between dC and dG or between dA and dT, which in turn dictates the ID base compositions as well as the requirement that only even numbers of DNA fragments be joined in a single assembly step.
A separate assembly of sub-modules of the final construct allows reusing all ID sequences. This circumvents the need for an additional PCR amplification step prior to the second level of assembly. The rules that dictate use and reuse of the ID sequences are explained using a specific example in Figure 3. Four different ID sequences are required for the second-level assembly, with two IDs used to flank each first-level construct. In our example, A–C is flanked by ID1 and ID2, D–F is flanked by ID2 and ID3, G–I is flanked by ID3 and ID4 and lastly BB2 is flanked by ID4 and ID1. Accordingly, in the first-level assembly of A, B, C and BB1.1, A must be flanked at the 5′-end by ID1, C must be flanked at the 3′-end by ID2 and BB1.1 must be flanked by ID2 and ID1 at its 5′- and 3′-ends, respectively. The remaining junctions are those between A and B, and B and C, and the IDs 3 and 4 can be used to form these junctions in an order determined by the alternating extension nucleotides. The above analysis leads to in silico specification of the first-level fragments with ID junctions and stop sequences.
If the functional moiety of the fragment excluding the IDs is novel, the entire sequence can be ordered from a synthetic genes supplier. In most cases, however, the sequence may already exist either in nature or in previously cloned constructs, thus making PCR the method of choice for fragment generation. PCR primers used to amplify the fragments and to introduce the ID sequences are designed as follows: For every functional moiety, the usual forward and reverse primers are designed with a predicted melting temperature close to 60°C. The forward primer sequences are extended at their 5′-ends by adding the appropriate ID sequence followed by three consecutive stop bases comprising the base omitted from the ID sequence (Figure 4). If the primer binding site starts with the same nucleotide as the stop sequence, the stop sequence can be shortened so that the total number of stop bases is at least 3. The reverse primers are extended at their 5′-end by adding the reverse complement of the chosen ID sequence, followed by at least three bases complementary to the stop base of this ID. Sets of four ID sequences that we have successfully used are given in Tables 1 and 2 (see below for more details).
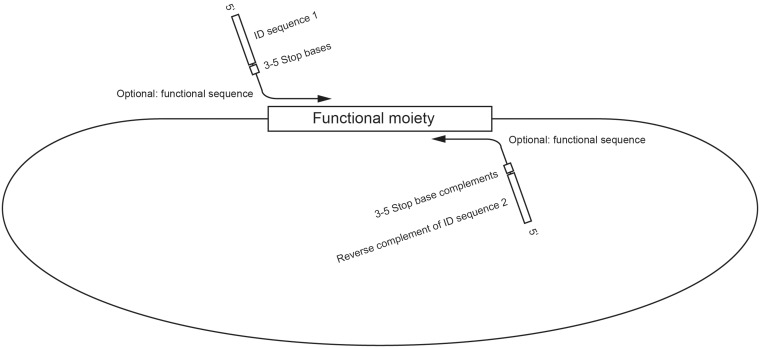
Figure 4.
Anatomy of PCR primers. Functional fragments (i.e. genes) as well as backbone sequences are amplified via PCR using primers composed of an ID sequence (or its complement in a reverse primer) followed by a stretch of stop bases (or their complement in a reverse primer), by an optional functional sequence such as AarI binding site, and by a template-specific sequence.
Table 1.
ID sequences for the inverter circuit
| ID number | ID sequence |
|---|---|
| ID1 | TTGTCTCTTGCTGGTGTTCG |
| ID2 | AACACCGGAACAAGAAAGGC |
| ID3 | GGTTCTTTTTCGTTGGGCGT |
| ID4 | GAGAGGCAGCAAGCAACGAA |
Table 2.
ID sequences for the reprogramming circuits
| ID number | ID sequence |
|---|---|
| ID1′ | CCACTCTCCATCAACACCTA |
| ID2′ | GGTGTTAAGGTGGAGGGAAT |
| ID3′ | AACCTCTCCCTACCAAATAC |
| ID4′ | AGAGAATGATGGATGGTAGG |
Theoretically, any plasmid template containing a bacterial origin of replication and an antibiotic resistance gene can be used as the backbone. The primers for amplifying the backbone elements are designed in the same way as those for the functional moieties but with exactly four stop bases followed by the reverse complement of the recognition site of the SII-type restriction enzyme AarI, GCAGGTG, 3′ of the stop bases (Figure 5). This restriction site is used in the second assembly level; we note that the site might be present in the amplified functional moieties or the backbone, which requires to either remove the sequence by site-directed mutagenesis or to use a different enzyme like PacI as described later.
Figure 5.
AarI-based regeneration of linear DNA fragments for the next-level assembly. AarI cuts four bases downstream of its CACCTGC recognition sequence on the sense strand so that the necessary stop bases for the first level of assembly can be fit in between. AarI digestion removes the stop bases and the ID can be reused in the next level of assembly.
Assembly reaction set-up
To perform first-level assemblies, all primers are phosphorylated using T4 polynucleotide kinase (PNK) at a primer concentration of 5 µM. PNK has to be heat-inactivated after the reaction to prevent it from dephosphorylating the primers in a PCR reaction that follows. The primer pairs are used for PCR with a proofreading polymerase such as Phusion (Finnzyme). If possible, high annealing temperature is used, usually ranging from 62–65°C. Very low amounts of template plasmid should be used to prevent template contamination in PCR products, which could otherwise give rise to colonies after the transformation of an assembly reaction. A convenient way to eliminate remaining template plasmid after PCR is digestion with a methylation-sensitive enzyme, such as DpnI. Difficult GC-rich amplicons can be efficiently amplified by KOD-extreme polymerase. The PCR products are purified using 1% agarose gel electrophoresis and a silica column purification kit. DNA concentrations are estimated by absorption measurements at 260 nm.
A chew-back reaction is set up using 2–4 nM final concentration of the PCR-amplified DNA fragments in 20 µl. For every first-level assembly, two chew-back reactions are set up such that all PCR products that have the same extension nucleotide are processed together. For example, one reaction will contain the products that use dATP as an extension nucleotide, while the second reaction will contain the ones that use dTTP. The reactions are incubated for 5 min at 27°C and then heated to 75°C to inactivate the polymerase. Subsequently, chew-back reactions corresponding to the same assembly are mixed and the temperature is slowly lowered from 75°C to room temperature at a ramp rate of −0.4°C/min. After reaching room temperature, the reaction mixtures are transformed into chemically competent E. coli using standard protocols and plated on LB-agar plates containing an appropriate antibiotic for selection, typically resulting in 100 colonies (Supplementary Figure S1). Colonies are expanded in liquid culture and are checked by test restrictions or by sequencing, as mutations could be introduced during PCR. By using a high fidelity DNA polymerase the proportion of mutant clones is minimized (see below).
A correct clone of every assembly reaction is digested with AarI in the presence of an auxiliary oligonucleotide that aids AarI restriction. When the disposable backbone fragment is similar in size to the excised composite fragment, an additional digestion can be used to cut the backbone and reduce the fragment size to enable efficient gel purification. The composite first-level assembly fragments are gel-purified. Second-level assembly reactions of purified composite fragments are performed as described for first-level assemblies, again using a PCR-amplified second-level backbone (i.e. BB2). The appropriate extension nucleotides for chew-backs of digested composite fragments can be determined by the ID sequences used in the bottom-level assemblies, since those IDs are reused in the second level. After transformation, colonies can be screened for correct assemblies by expanding them in liquid culture and performing multiple restriction enzyme digestions or by functional screening. Sequencing of the regions created by enzymatic digestion is usually not necessary while PCR-amplified parts like the backbone can be sequence-verified if needed.
Specific experiments
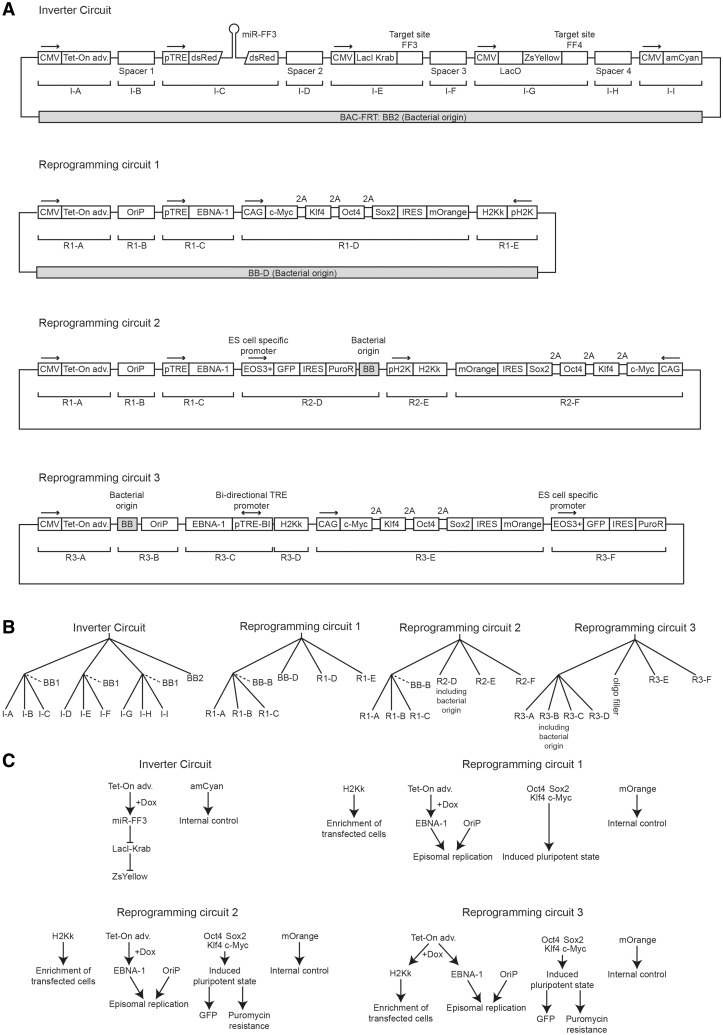
Inverter circuit
In the following section, we describe a number of constructs that we assembled with the chew-back method. Their schematics are given in Figure 6A and a detailed description of their constituent parts is given in Supplementary Table S1. The inverter circuit was built using two levels of hierarchical assembly. The ID sequences used in the first level were reused by utilizing the SII-type restriction enzyme AarI which recognizes a relatively long non-palindromic sequence of seven bases and cuts four bases outside its recognition sequence (Figure 5). This makes it an ideal enzyme to release composite fragments from first-level assemblies, regenerating the terminal ID sequences for reuse in the next level of assembly. The backbone for the second level of assembly was PCR amplified from a bacterial artificial chromosome (BAC) vector that had previously been modified by inserting an FRT site for stable genomic integration into Flp-In cells. The nine parts of interest were intended to compose a synthetic five-gene circuit whose functional characterization will be described elsewhere. The remaining four fragments were spacers of roughly 1 kb, intended to minimize promoter crosstalk. These spacers were PCR-amplified from the mouse genome.
Figure 6.
Schematics of the assembled circuits and the assembly protocols. (A) Annotated diagrams of the fully assembled DNA cassettes with different building blocks indicated by abbreviations. Bacterial origins of replication are indicated. (B) Assembly trees for different DNA cassettes. Backbones discarded at a later stage are indicated with a dotted line. (C) Network diagrams of the assembled circuits depict the intended roles of the different gene products.
To design a set of four ID sequences, we first generated long random DNA sequences using a random generator (http://www.faculty.ucr.edu/~mmaduro/random.htm). Four 20-mers with a GC content of 47–53% were chosen arbitrarily; in two of the sequences named ID1 and ID3 all A bases were replaced with T bases; conversely, in the remaining two sequences, named ID2 and ID4, all T bases were replaced with A bases. In addition, the lack of secondary structure was confirmed by simulation with DNAman software (http://en.bio-soft.net/format/DNAMAN.html). The resulting IDs are given in Table 1. The prospective junctions between the parts of the second-level assembly were sequentially assigned the ID sequences ID1, ID2, ID3 and ID4. We then labelled the prospective junctions in the first-level assembly reactions by reusing the same four ID sequences as described above. Primers augmented with ID sequence precursors or their reverse complements plus 5 bp stop sequences or their complements were designed as described above (Supplementary Table S2). Primers used to amplify the plasmid backbones were augmented with AarI sites and the stop sequence was reduced to four bases in length. To verify the integrity of the design, we constructed in silico plasmid maps for every step of the assembly process.
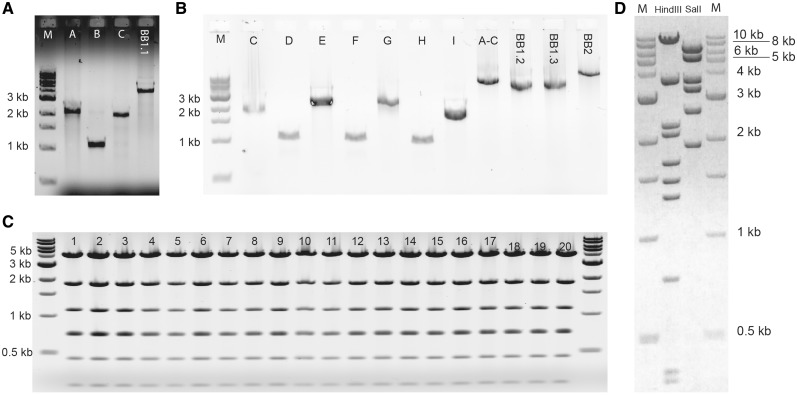
Desalted primers were phosphorylated directly (see ‘Primer phosphorylation’ section). PCR reactions were performed according to ‘PCR amplification’ section with Pfu Ultra II HS DNA polymerase using templates and primers in Supplementary Table S2 (Figure 7A and B). The PCR products were then gel-purified (see ‘Gel purification of fragments’ section). Typical DNA yields were 5–40 ng/µl in a total elution volume of 20 µl. The amplicons were combined in chew-back reactions as follows: fragments I–A (Inverter-A) and I–C with extension nucleotide dTTP (reaction 1), I–B and BB1.1 with dATP (reaction 2), I–D and I–F with dATP (reaction 3), I–E and BB1.2 with dTTP (reaction 4), I–G and I–I with dTTP (reaction 5) and I–H and BB1.3 with dATP (reaction 6). Subsequently, reactions 1 and 2 (assembly 1), 3 and 4 (assembly 2) and 5 and 6 (assembly 3) were annealed (see ‘Chew-back reactions’ section). Transformation into chemically competent E. coli strain XL-1 blue was done as described in ‘Bacterial transformation’ section. We typically observed 100–200 colonies in first-level assemblies when using concentrations of ∼4 nM of purified DNA fragments. DNA was isolated from expanded clones (see ‘Liquid culture, miniprep of plasmid DNA’ section) and digested for verification (see ‘Test restrictions’ section, Figure 7C), resulting in a correct band pattern. Next, we performed AarI digestions of correct clones obtained in first-level assemblies 2 and 3 (see ‘AarI digestion’ section) and gel-purified the composite fragments of expected size. Since assembly 1.1 did not contain AarI restriction sites in the primers used to amplify the backbone, we re-amplified the composite fragment I-[A–C] using Pfu Ultra II Fusion HS DNA polymerase (see ‘PCR amplification’ section) and primers A_fwd and C_rev; the product was gel-purified as well (Figure 7B). For the second-level assembly, the PCR-amplified I-[A-C] composite fragment was mixed with the AarI-digested I-[G-I] composite fragment for a chew-back reaction in the presence of dTTP as the extension nucleotide; in parallel, the AarI-digested composite fragment I-[D–F] and the PCR-amplified backbone BB2 were combined in a chew-back reaction with dATP. After chew-back, annealing of the mixed reactions, transformation and clonal expansion were performed as described above. However, the number of colonies was only in the range of 10 for this much larger construct. A correct clone was identified by restriction analysis (Figure 7D), completing the construction of this DNA cassette.
Figure 7.
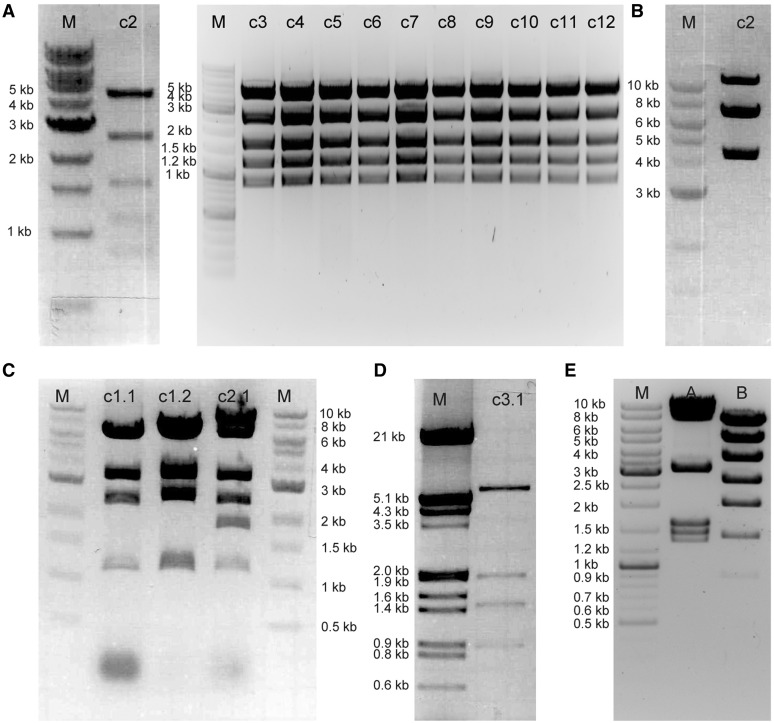
Agarose gels of the inverter circuit assembly. (A) PCR products for the first-level assembly no. 1. Expected bands are 2.2 kb (fragment A), 1.1 kb (fragment B), 2.0 kb (fragment C) and 4 kb (backbone BB1.1). M, DNA size marker. (B) PCR product used in the first-level assemblies nos 2 and 3, and the second-level assembly. Previously gel-purified Fragment C was included to assure the efficiency of the DNA extraction. Expected bands are 2.0 kb (fragment C), 1.0 kb (fragment D), 2.4 kb (fragment E), 1.0 kb (fragment F), 2.2 kb (fragment G), 1.0 kb (fragment H), 1.6 kb (fragment I), 5.0 kb (composite construct A–C), 4 kb (fragment BB1.2), 4 kb (fragment BB1.3) and 9.5 kb (fragment BB2). For fragment and assembly numbering refer to Figure 6. (C) EcoRI test restrictions from 20 independent random clones of first-level assembly no. 1. Expected band sizes are 4.3, 1.9, 1.1, 0.74, 0.72, 0.46 and 0.25 kb. The resolution of the gel does not allow differentiating the 0.74 kb from the 0.72 kb band. (D) Digestion tests for the second-level assembly clone. Expected bands are 10.0, 3.8, 2.3, 2.1, 1.7, 1.5, 1.3, 0.75, 0.47 and 0.36 kb with Hind III; and 7.0, 5.6, 3.7, 3.3, 2.6 and 1.9 kb with SalI.
Reprogramming circuits
We constructed three large DNA cassettes using two-level assemblies with the long-term goal to induce reprogramming of human primary fibroblasts to pluripotency (10,11) without the need for a viral vector (Figure 6A, reprogramming circuits). Difficult PCR amplicons of 9 kb and a high GC content, which contain the CAG promoter, could be amplified using the KOD extreme polymerase. Remarkably, the three final constructs of more than 25 kb each were assembled and maintained in E. coli using a standard pUC backbone rather than a BAC, thus allowing more efficient DNA preparation yet not resulting in clonal instability as one might have expected.
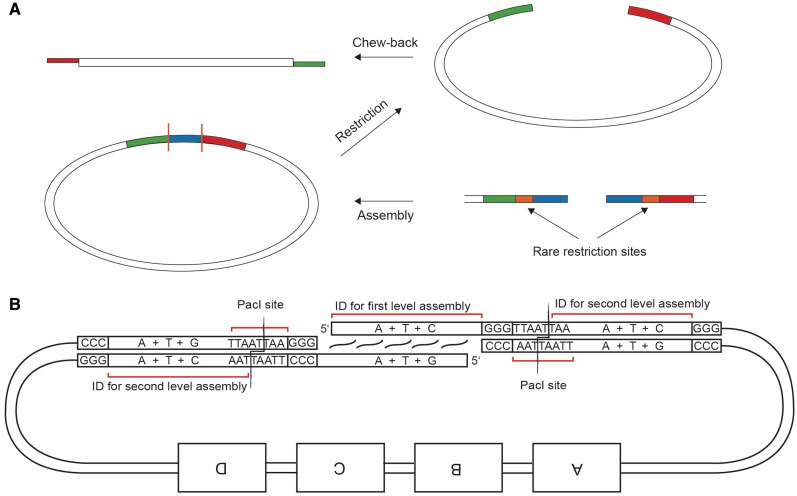
One of the assemblies employed a modification to the protocol. Instead of discarding the backbone portion of the first-level assembly and using a new backbone cassette in the second level, we preserved the backbone from the first level by introducing two inert ID precursors next to an active ID sequence at the 5′-end of the PCR product in the left-most position and the 3′-end of the PCR product in the right-most position (usually, the backbone) within the assembly, respectively. Inert and active ID sequences are separated by a stop sequence and the recognition site of a restriction enzyme PacI (Figure 8). Thus, after the first-level chew-back assembly, the resulting junction is flanked by two PacI sites and can be completely removed by PacI digestion, thereby exposing the previously inert ID precursors.
Figure 8.
A multiple-level assembly strategy that allows reusing backbone sequences. (A) Overview of the assembly strategy. The first-level assembly introduces three consecutive ID sequences with interspersed enzymatic cleavage sites. While the blue ID is used for the first assembly, the red and green IDs are used for the second level of assembly after enzymatic excision of the blue ID. (B) Detailed view of an exemplary ID design: PacI sites between the three IDs partially overlap with the outer two IDs, thereby determining the three types of bases used in their design. By PacI digestion of the first-level assembly, the middle ID is discarded in order to set free the outer ones while retaining the plasmid backbone portions for the next level of assembly. Note that the 3′ overhangs generated by PacI digestion are removed during chew-back.
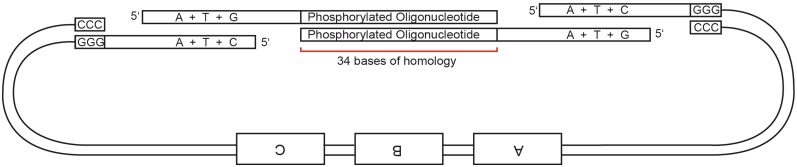
As a further extension of the protocol, we enabled the assembly of an odd number of DNA fragments by adding an auxiliary synthetic oligonucleotide spacer. Two pre-phosphorylated oligos are annealed to each other forming a 34-bp helix and two single stranded 5′ overhangs which are compatible with the adjacent ID sequences (Figure 9). The spacer oligos do not have to contain stop bases since they do not participate in the chew-back reaction, but are added in an equimolar concentration to the digested fragments for annealing.
Figure 9.
Schematics of odd-part assembly using a synthetic oligonucleotide spacer.
The genes Oct-4, c-myc, Klf-4 and Sox-2 were included in the constructs in order to reprogram transfected primary fibroblasts to a state of induced pluripotency as previously shown (10,11). At the same time, the embryonic stem cell-specific EOS promoter driving a puromycin resistance gene (12) was included to allow antibiotic selection of successfully reprogrammed cells. The H2k gene is a murine MHC surface antigen that allows dramatically enriching for transfection-positive cells by magnetic cell separation using antibody-coupled nanospheres (Miltenyi GmbH). Furthermore, the EBV OriP region as well as the doxycycline-inducible expression cassette of the EBNA1 gene were intended to allow controllable, cell-cycle synchronous episomal replication of the plasmids for long-term gene expression (13). The chosen OriP sequence was an improved version that had been shown to bear a higher affinity for the EBNA1 protein thus possibly increasing replication efficiency (14).
Reprogramming circuit 1
Four ID sequences were designed as described before but with the difference that the stop bases were dC and dG instead of dA and dT. The IDs are shown in Table 2. Primers were designed and phosphorylated as described (Supplementary Table S3). The first-level PCR fragments were generated using Pfu Ultra II Fusion HS DNA polymerase, and were gel-purified as described. The first-level assembly was performed by chew-back of fragments R1-A and R1-C with dCTP as the extension nucleotide, and of R1-B and BB-B using dGTP. The reactions were then mixed, annealed and transformed as described. Test restrictions and AarI restrictions were performed as described in ‘Test restrictions’ and ‘AarI digestion’ section; the digestion patterns were consistent with expectation (Figure 10A and B). A composite fragment released by AarI restriction of a first-level assembly clone was gel-purified. For the second-level assembly, PCR-amplified fragment R1-E and AarI restriction fragment R1-[A-C] were chewed-back using dCTP as the extension nucleotide, while PCR-amplified R1-D and BB-D were digested with dGTP as the extension nucleotide. The reactions were combined, annealed, transformed and subsequently treated as described (see ‘Materials and Methods’ section). BamHI test restrictions of two clones revealed the correct band pattern (Figure 10C).
Figure 10.
Structure confirmation of the reprogramming circuit assembly reactions. (A) 11 clones of the assembly R1-A+R1-B+R1-C+BB-B digested with BamHI + MluI. Expected bands are 4.9, 2.6, 1.6, 1.2 and 0.86 kb. All clones are correct. For assembly numbering refer to Figure 6 (B) Assembly R1-A+R1-B+R1-C+BB-B digested with AarI. Expected bands are 6.7 and 4.2 kb. Note that the band at 11 kb represents the linearized plasmid due to incomplete digestion (C) Lanes c1.1, c1.2: reprogramming circuit 1 digested with BamHI. Expected bands are 6.4, 3.3, 2.5 and 1.2 kb Lane c2.1: reprogramming circuit 2 digested with BamHI. Expected bands are 8.4, 6.4, 3.3, 2.5, 1.8, and 1.2 kb (D) A representative clone of the assembly R3-A+R3-B+R3-C+R3-D digested with XhoI and MluI. Expected bands are 5.9, 1.9, 1.4 and 0.9 kb (E) Reprogramming circuit 3 digested with NotI and MluI (lane A) and XhoI and NheI (lane B). Expected bands are 8.4, 6.9, 3.0, 1.54, 1.4 and 1.28 kb with NotI, MluI; and 6.8, 4.8, 3.5, 2.6, 1.9, 1.34, 0.86, 0.28 and 0.17 kb with XhoI and NheI.
Reprogramming circuit 2
The first-level composite fragment as well as all ID sequences were reused from reprogramming circuit 1. Templates and primers for the additional gene fragments used are given in Supplementary Table S4. The primers were phosphorylated (see ‘Primer phosphorylation’ section) and the fragments of interest were amplified using Pfu Ultra II Fusion HS DNA polymerase (see ‘Pfu Ultra II Fusion HS (Stratagene)’ section). Gel purification was performed as described (see ‘Gel purification of fragments’ section). Subsequently, the AarI-digested first-level fragment and the amplicon R2-E were chewed-back in the presence of the extension nucleotide dCTP; amplicons R2-D and R2-F were chewed-back in the presence of dGTP (see ‘Chew-back reactions’ section). The subsequent treatment was identical to the one described above (see ‘Materials and Methods’ section); a digestion pattern of one of the clones is shown in Figure 10C.
Reprogramming circuit 3
The ID sequences were reused from reprogramming circuit 1. Templates and primers for the gene fragments are given in Supplementary Table S5. The primers were phosphorylated and the products were amplified using Phusion DNA polymerase (see ‘Phusion HF polymerase (Finnzymes)’ section). Gel purification was performed as described (see ‘Gel purification of fragments’ section). Subsequently, the amplicons R3-A and R3-C were chewed-back in the presence of dCTP as the extension nucleotide; in parallel, amplicons R3-B and R3-D were chewed-back in the presence of dGTP (see ‘Chew-back reactions’ section). Annealing of the mixture and subsequent treatment was identical to the one described above (see ‘Materials and Methods’ section); the digestion pattern of one resulting clone is shown in Figure 10D.
PacI digestion was performed according to ‘PacI digestion’ section. The composite first-level restriction fragment was gel-purified as described. The digested product as well as the amplicon R3-F were chewed-back with dCTP as the extension nucleotide; PCR product R3-E was chewed-back using dGTP. These reactions were mixed and annealed together with the synthetic oligonucleotide spacer (see ‘Oligonucleotides in assembly reactions’ section); digestion patterns of one of the clones are shown in Figure 10E.
Efficiency measurement and quality control
The first-level assembly for reprogramming circuit 1 was repeated with varying input DNA concentrations in quadruplicates in order to assess the efficiency of the cloning technique. The results given in Supplementary Figure S1 show that ∼200 colonies can be expected if the fragments are included at a final concentration of 4 nM in the chew-back reaction (for the specific assembly, this corresponds to roughly 20 ng of each of the constituent DNA fragments in the transformation mixture). The same figure shows that the apparent cloning efficiency in CFU/microgram units is about 3000, although it depends non-linearly on the absolute DNA amount.
From the same assembly reactions, 10 clones were sequenced using 16 Sanger sequencing reactions per clone. In over 100 kb of sequence, only two point mutations were found (Supplementary Table S6). One of the mutations fell within the stop sequence of a fragment and thus seems to originate from imperfect primer synthesis. The second mutation is probably a result of imperfect PCR amplification. Not a single mutation was detected in the 40 separate sequenced ID junctions.
DISCUSSION
The method described in this study allows rapid, easy and flexible assemblies of PCR amplicons or synthetic DNA inserts of diverse lengths ranging from 30 bp to 10 kbp. In contrast to multiple-fragment yeast recombination, our method is easily performed in standard E. coli strains and requires PCR amplification primers that are only 45 bases long for creating overlapping 20-mer ID sequences. In addition, the technique is not hindered by internal homologies of the fragments, which poses an obstacle for in vivo and in vitro recombination-based cloning methods.
Our protocol exhibits nearly undetectable background colony formation likely due to the specific annealing of 20-nt long overhangs and to the clearance of PCR template DNA by DpnI digestion. This renders the technique well suited for high-throughput applications and robotic automation. We use the SII-type restriction enzyme AarI for hierarchical assemblies, whose 7-bp long recognition sequence is expected only once in every 16 kb of random DNA sequence. When AarI is not available for cloning, we demonstrated the feasibility of an alternative approach, utilizing the SI-type restriction enzyme PacI, which has an 8 bp recognition site.
Our method exhibits an efficiency sufficient to assemble >25 kb plasmids, while still using standard chemical transformation techniques. The maximum size of plasmids that can be assembled using our protocol was not determined. To further increase the number of colonies obtained when assembling larger constructs, it may be advantageous to increase the length of the overhangs to 30 or 40 nt.
One disadvantage of the present method is the fact that it introduces >20 bases of sequence between the DNA parts of interest. While this is not an issue when constructing artificial gene circuits for synthetic biology, the junction sequences might pose an obstacle when generating, for example, fusion proteins. However, for the latter purpose we have successfully modified the ID sequences to encode for rather inert amino acid spacers for fusion proteins (data not shown). Therefore, the rapid chew-back assembly method might also provide a valuable tool for fusing multiple fluorescent proteins, tags or sub-cellular localization signals to genes of interest in an expression backbone plasmid of choice. Moreover, functional amino acid sequences can be encoded in the IDs such as a His-tag (2) or a T2A ribosomal stuttering peptide (data not shown) by choosing appropriate codons for the required amino acids.
The method should be well suited to rapidly generate targeting plasmids for knock-out mice. Two PCR-amplified homology arms could be placed on either side of an antibiotic resistance gene in a single cloning step, thus providing a knock-out construct in <3 days.
The enzyme we used for the chew-back assemblies is T4 DNA polymerase, which is a standard enzyme commonly present in molecular biology laboratories.
Using the chew-back method, we were able to generate complex plasmid constructs carrying all factors necessary for the reprogramming of somatic cells to induced pluripotency (10,11). In addition, these plasmids contain a modified EBV origin of replication combined with a tet-inducible EBNA-1 gene for episomal propagation (12) intended to allow long-term expression of the reprogramming factors without using an integrating viral vector. A constitutively expressed surface marker gene for magnetic bead-based selection was further included to enrich cells that were successfully transfected or electroporated with the large plasmids. Apart from that, an embryonic stem cell-specific promoter driving an antibiotic resistance gene was bundled with the other functional moieties, providing a possible means to put positive selection pressure on the process of reprogramming.
These constructs could enable the generation of patient-derived induced pluripotent stem cells (iPS) free of exogenous DNA insertions while overcoming the relatively low efficiencies reported by Yu and colleagues (15), who used multiple episomal plasmids for reprogramming instead of only one.
In summary, the technical solution we have developed facilitates complex cloning projects by enabling the assembly of multiple PCR fragments with very low cloning background, while only requiring standard molecular biology materials and training.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S6, Supplementary Figure S1 and Supplementary References [16–19].
FUNDING
Bauer Fellows Program; ETH Zurich; ERC starting grant; National Institutes of Health (1R01CA155320-01); National Institute of General Medical Sciences Grant for Centers of Systems Biology; The undergraduate program for Molecular Biomedicine at the University of Bonn, Germany. Funding for open access charge: ETH Zurich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors want to thank Veit Hornung and Katharina Hoelle for their support and Bill Sugden for kindly providing the plasmid p3513. The plasmids pCAG2LMKOSimO and PL-SIN-EOS-C(3+)-EiP were obtained from Addgene with respective numbers 20866 and 21313.
REFERENCES
- 1.Aslanidis C, Dejong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc. Natl Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reisinger SJ, Patel KG, Santi DV. Total synthesis of multi-kilobase DNA sequences from oligonucleotides. Nat. Protocols. 2006;1:2596–2603. doi: 10.1038/nprot.2006.426. [DOI] [PubMed] [Google Scholar]
- 4.Aslanidis C, Dejong PJ, Schmitz G. Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR-Methods Appl. 1994;4:172–177. doi: 10.1101/gr.4.3.172. [DOI] [PubMed] [Google Scholar]
- 5.Donahue WF, Turczyk BM, Jarrell KA. Rapid gene cloning using terminator primers and modular vectors. Nucleic Acids Res. 2002;30:e95. doi: 10.1093/nar/gnf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 2007;35:e55. doi: 10.1093/nar/gkm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 8.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 9.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl Acad. Sci. USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu JY, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Hotta A, Cheung AYL, Farra N, Vijayaragavan K, Seguin CA, Draper JS, Pasceri P, Maksakova IA, Mager DL, Rossant J, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 13.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian-cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 14.Lindner SE, Zeller K, Schepers A, Sugden B. The affinity of EBNA1 for its origin of DNA synthesis is a determinant of the origin's replicative efficiency. J. Virol. 2008;82:5693–5702. doi: 10.1128/JVI.00332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nat. Nanotechnol. 2010;5:666–670. doi: 10.1038/nnano.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 18.Sander A, Guth A, Brenner HR, Witzemann V. Gene transfer into individual muscle fibers and conditional gene expression in living animals. Cell Tissue Res. 2000;301:397–403. doi: 10.1007/s004410000247. [DOI] [PubMed] [Google Scholar]
- 19.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.