Abstract
We introduce Grinder (http://sourceforge.net/projects/biogrinder/), an open-source bioinformatic tool to simulate amplicon and shotgun (genomic, metagenomic, transcriptomic and metatranscriptomic) datasets from reference sequences. This is the first tool to simulate amplicon datasets (e.g. 16S rRNA) widely used by microbial ecologists. Grinder can create sequence libraries with a specific community structure, α and β diversities and experimental biases (e.g. chimeras, gene copy number variation) for commonly used sequencing platforms. This versatility allows the creation of simple to complex read datasets necessary for hypothesis testing when developing bioinformatic software, benchmarking existing tools or designing sequence-based experiments. Grinder is particularly useful for simulating clinical or environmental microbial communities and complements the use of in vitro mock communities.
INTRODUCTION
The rapid development of high-throughput sequencing technologies such as 454 and Illumina has made large-scale sequencing projects both feasible and affordable. Bioinformatic tools are constantly being developed to manage and analyze data generated by these new sequencing platforms. Rigorous evaluation of the accuracy of these tools requires either the sequencing of synthetic communities of known composition created in vitro or the generation of simulated datasets in silico, which can account for both community structure and technical aspects of sequencing such as read length and errors. The construction of artificial in vitro communities and nucleic acid pools in the laboratory is both expensive and labor intensive, which limits the number of sequence libraries that can be produced (1–6). Mavromatis et al. (7) circumvented the need for in vitro manipulations when assembling the FAMES artificial metagenomes using DNA reads from existing single-genome shotgun sequencing projects. While both approaches produce realistic datasets, they are limited by confounding factors such as genome length bias (8,9), DNA amplification bias (10,11) and sequencing artifacts (12,13), the extent of which is generally unknown and can compromise interpretation of the sequence data. In contrast, bioinformatic tools that produce simulated reads based on reference sequences in silico allow users to rapidly generate large numbers of sequence libraries with controlled and predefined parameters.
Recently, characterization of microbial communities by 16S rRNA gene amplicon sequencing has experienced a renaissance, largely owing to the advent of high-throughput sequencing (14). This has spurred the development of an unprecedented number of tools and pipelines for the analysis of 16S rRNA amplicon sequences, but microbial ecologists lack a read simulator capable of generating synthetic amplicon libraries to validate existing and upcoming bioinformatic tools.
To address this limitation and also to expand upon existing shotgun sequence simulators, we present Grinder, an open-source software package that generates in silico simulated amplicon and shotgun (genomic, metagenomic, transcriptomic and metatranscriptomic) libraries from reference sequences. Grinder incorporates error models for a variety of sequencing platforms, can generate paired-end reads with variable insert size, and libraries with a user-specified species composition. Grinder libraries can also be designed based on α diversity metrics and model-based community structures, while sets of related libraries can be created by providing their β diversity. Unlike existing read simulators, Grinder can simulate the multiplexed PCR process to produce barcoded amplicon reads for any gene of interest, while also introducing experimental artifacts such as chimeras and biological biases due to variations in gene copy number between different species.
MATERIALS AND METHODS
Grinder implementation
Overview
Grinder is a platform-independent software package implemented in Perl and uses the Bioperl toolkit (15). Grinder is designed to run on a standard desktop computer and can be installed using a Perl module installer or a Debian package. Grinder includes a full test suite that automatically validates all components during installation. Grinder uses the Mersenne Twister algorithm (16) to generate random numbers because the default random number generation routines in many packages, such as Java, are below simulation grade (17).
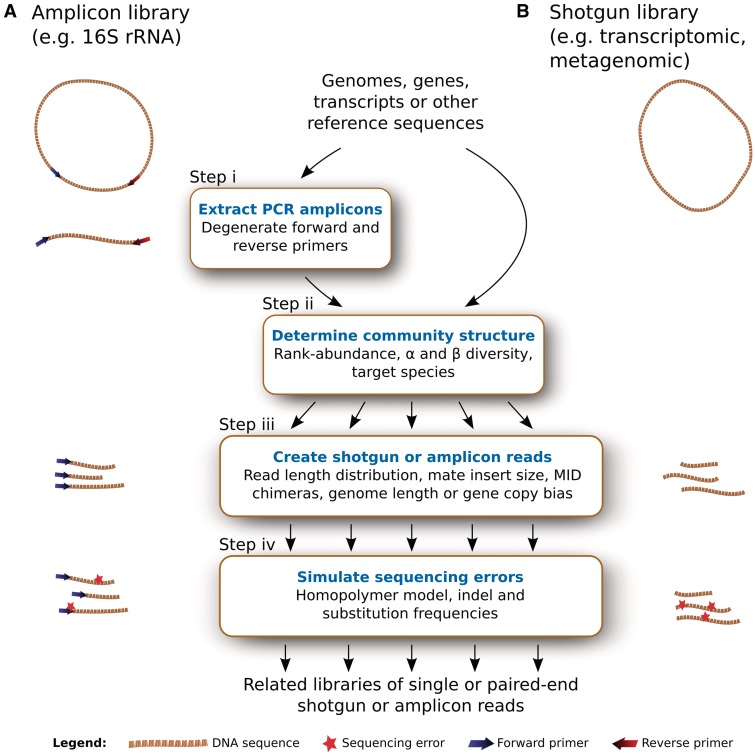
The read simulation in Grinder generates amplicon (Figure 1A), or shotgun (Figure 1B) reads. While most steps in read simulation are common to shotgun and amplicon libraries, there is an additional initial step in amplicon simulation that identifies and extracts full-length amplicons in the input reference sequences based on the provided PCR primers (Figure 1, Step i). For both amplicon and shotgun read simulations, species relative abundance (which defines community structure) is calculated from rank-abundance models, α and β diversity (Figure 1, Step ii). Reads are selected from the community either from the beginning of the full-length reference amplicon (for amplicon datasets) or randomly in the reference shotgun sequences (for shotgun datasets) (Figure 1, Step iii). Finally, sequencing errors (indels, substitutions, homopolymers) are introduced in the reads in a position-specific manner (Figure 1, Step iv). An exhaustive list of options that affect these steps can be obtained at the command line using the standard help function (Grinder–help) and all specific parameters used for a particular execution of Grinder can be put in a profile file to allow the easy reuse of complex custom configurations. A subset of the available options and features are described in detail below.
Figure 1.
Flowchart of the processes and parameters used by Grinder to generate related (A) amplicon and (B) shotgun libraries.
Input and output sequences for simulated datasets
Publicly available FASTA-formatted databases can readily be used in Grinder. For example, the curated microbial and viral genome sequences in the NCBI RefSeq collection (18) are suitable to produce artificial genomic, metagenomic or amplicon libraries. While reads can be taken from a reference sequence and its reverse complement, for example to simulate (meta)genomic data, strand-specific datasets such as some transcriptomes (19) can be put together by taking reads from only one strand, either forward or reverse, of the reference sequences. Curated gene-specific sequence databases such as Greengenes (20), Silva (21) and PseudoMLSA (22) can also be used to simulate amplicon datasets.
Simulated read libraries are output as FASTA files with optional QUAL and FASTQ files as well as accompanying text files describing library content and community rank-abundances. Grinder offers many options to adjust the read characteristics. For example, read length can have a fixed value or follow a uniform or normal distribution and insert length for mate pairs or paired-end datasets can be specified in the same way. Detailed information for each read including its source, location on the reference sequence and introduced errors are provided in its description line, making reads entirely traceable for downstream analyses and applications (Supplementary Figure S1).
PCR simulation
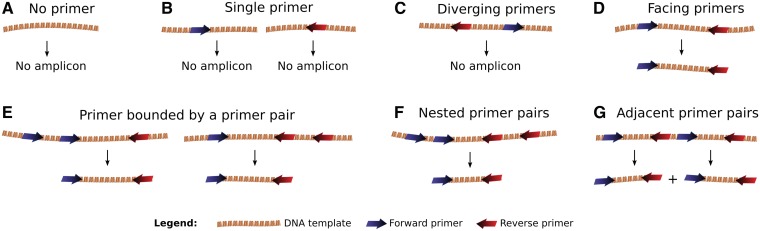
A unique feature of Grinder relative to other read simulators is that a PCR simulation is performed when an amplicon read library is requested. The forward and reverse primers provided in a FASTA file by the user can contain degenerate residues following the IUPAC convention. In cases where PCR primers match different positions of a genome, several full-length amplicons will be extracted, except if these amplicons overlap, in which case only the smallest one will be extracted to mimic the PCR process (Figure 2). In subsequent Grinder steps, simulated amplicon reads are taken from the start of each full-length PCR amplicon, forward primer included.
Figure 2.
Grinder PCR amplicon selection process. All possible combinations of degenerate primer matches on the template DNA are considered. By default, Grinder will extract the shortest amplicon.
Community structure, diversity and multiplexed identifiers
Community structure for simulated shotgun or amplicon libraries can be specified in a text file listing species and their relative abundances. Unlike most read simulators, Grinder can alternatively generate community structures based on a specified community richness (α diversity) and a deterministic rank-abundance model (uniform, linear, power law, logarithmic or exponential), with species selected randomly during library construction.
Another novel feature of Grinder is the simultaneous production of multiple read libraries (shotgun or amplicon) with related characteristics, allowing the user to vary the percentage of species shared between libraries and the percent of dominant species with different rank abundances (β diversity) (23). Multiplexed libraries consisting of individual barcoded samples pooled and sequenced on the same sequencing run can also be simulated by appending multiplexed identifiers (MIDs) given in a FASTA file to the beginning of each read. Optional MIDs are added to the reads prior to applying sequencing errors, so that MIDs may contain errors, as in real read libraries.
Simulation of biological and experimental biases
Sequencing errors such as substitutions, indels (insertions and deletions) or homopolymers can be introduced in Grinder-simulated reads by specifying position-specific models (uniform, linear or polynomial). Sanger reads can be simulated by increasing the number of substitutions and indels linearly along the reads, from 1% at its 5′ end to 2% at its 3′ end (24,25) (Supplementary Figure S2A). A fourth-degree polynomial model was implemented to reflect the accrued error rate (e) of substitutions at the 3′ end of Illumina reads (26): e = 3.10−3 + 3.3.10−8.i 4, where i is the position from the 5′ end (in bp) (Supplementary Figure S2A). Grinder also uses several deterministic models to simulate the homopolymer errors typical of 454 pyrosequencing (25,27,28). The recent empirical homopolymer model described by Balzer et al. inserts more errors as the length of the homopolymeric region increases (27). This is achieved by assigning each homopolymer a new length (n′) that is normally distributed around the actual length n, but with a standard deviation that increases linearly with homopolymer length: n′ ∼ N(n, 0.03494 + 0.06856n), for n ≥ 6 (Supplementary Figure S2B).
Quality files (FASTQ or QUAL) can be generated based on two user-specified values, one for low (e.g. 10) and one for high (e.g. 30) quality bases. Grinder assigns the low-quality score to introduced errors and the high-quality score to all other bases. Users requiring 454 pyrosequencing libraries with more realistic quality files (in native SFF format) can run Flowsim (27) on the reads generated by Grinder.
A known issue with amplicon sequencing is the formation of chimeras, spurious sequences formed during co-amplification of homologous genes (1,29). The most common type of chimera is a bimera, which results from the fusion of two amplicon template sequences. Higher order chimeras such as trimeras and quadrameras can also occur in amplicon read datasets, albeit at lower frequencies (30). In Grinder, chimeras are generated in one of two ways. In the first method, amplicon sequences and breakpoints are randomly selected in frame. The chimeras are generated by appending consecutive amplicon segments at the breakpoint. The second method is similar to that used by CHsim (31), i.e. chimeras are produced by concatenating two or more amplicon sequences, split at particular break points. The chosen breakpoints are k-mers, or short sequence stretches of k bp, shared by two amplicons and are more likely to be chosen if the amplicons are abundant and more similar to each other.
Finally, biological bias affects sequence libraries. Similar to the bias described in metagenomes arising from genome length differences (8), the presence of several gene copies in a genome may affect the composition of an amplicon library (32). When complete genomes are used as input, the effect of variable gene copy number in different genomes is modeled in Grinder by sampling species proportionally to their relative abundance and to the number of copies of the amplicon in its genome, instead of proportionally to their relative abundance only.
User interfaces
Grinder provides a command-line interface (CLI), graphical user interface (GUI) and application programming interface (API). The CLI can be used in a terminal and permits the automated generation of the many replicate datasets needed for statistical validation of bioinformatic tools. We have also implemented a GUI for Grinder on the Galaxy platform (Supplementary Figure S3) (33), which makes it possible to run Grinder through a web browser on any local desktop, remote server equipped with Galaxy or even on distributed computers (34). Unlike previous read simulators, Grinder also provides an object-oriented Perl API, which technical users can take advantage of when writing Perl pipelines. When using the API (Supplementary Figure S4), a Grinder factory has to be created first by using the new() method, which accepts the same options as the CLI. From there, the next_lib() method allows the user to proceed to the next sequence library and next_read() generates the next simulated read of that library. Each read produced is a Bio::Seq::SimulatedRead object (implemented in a Perl module written for Grinder and contributed to Bioperl) that has methods to query its nucleotide sequence, position, errors and other tracking information (Supplementary Figure S2).
I6S rRNA amplicon case study
Eight amplicon libraries, each with a unique MID sequence, were generated from the Greengenes database of named isolates (http://greengenes.lbl.gov/Download/Sequence_Data/Fasta_data_files/Isolated_named_strains_16S_aligned.fasta) using the universal primer set for 16S rRNA: 926F and 1492R (35). For each library, 454 GS-FLX Titanium pyrosequencing was simulated by requesting 5000 reads with normally distributed lengths (mean: 400 bp, standard deviation: 50 bp) and homopolymer errors (Balzer model) (27). Two additional libraries were constructed without homopolymer errors. All libraries were designed to contain 100 unique phylotypes following a power law rank-abundance curve (with parameter value of 2) and to have 80% of their phylotypes in common. The resulting Grinder files are available in Supplementary Dataset S1. FASTA and QUAL files for all libraries were concatenated prior to analysis to mimic the output of multiplexed sequencing. QIIME (36) was used to separate the libraries based on their MID, to cluster the reads at 100% and 97% identity and to assign taxonomy by comparing sequences to the Greengenes database using BLAST. A normal distribution was also fit to the empirical distribution of sequence lengths in each library by the R function fit_distr (37).
RESULTS AND DISCUSSION
Recent advances in DNA sequencing technology have allowed for the rapid generation of large sequence datasets, ushering in the age of genomics and metagenomics. Platforms and chemistries evolve quickly, engendering newer generations of sequencing that rapidly replace old methods and require the development and refinement of bioinformatic tools for analysis. Proper algorithm design and implementation requires large amounts of sequence data. However, such data may not be publicly accessible or exist in the volume necessary for rigorous testing. In silico simulated datasets overcome these limitations and also allow for optimization of study parameters, which may depend on sequencing depth and quality (e.g. sample size) in advance.
Grinder for shotgun dataset simulation
Grinder incorporates many common features of existing read simulators (Table 1) including deterministic error profiles, support for paired-end reads and the generation of sequences characteristic of particular sequencing technologies. Similar to other modern read simulators, Grinder provides sequencing errors, allowing users to flexibly specify their own error models or use preset values corresponding to known error profiles for the Sanger, 454, and Illumina platforms (Table 1). For example, Grinder was used with the Balzer error model (27) to test different short read alignment methods to improve PaPaRa (38).
Table 1.
Comparison of existing sequencing read simulators
| Name | References | Lic. | Homepage | Lang. | Interf. | Dataset types | Paired-end | Sequencing technologies | Qual. scores | Distinguishing features |
|---|---|---|---|---|---|---|---|---|---|---|
| Grinder | Angly et al. 2012 (this article) | GPL | sf.net/projects/biogrinder | Perl | CLI, API, GUI | Amplicon, (meta)genomic, (meta) transcript-omic | Yes | Sanger, 454, Illumina | Yes | Species abundance models, α and β diversity, MIDs, FASTQ output, multimeras, genome length and gene copy number bias |
| GemSIM | McElroy KE (unpublished data) | GPL | sf.net/projects/gemsim | Python | CLI | (Meta)genomic | Yes | Sanger, 454, Illumina | Yes | Haplotypes, FASTQ and SAM output |
| Mason | Holtgrewe (44) | GPL | www.seqan.de/projects/mason.html | C++ | CLI | Genomic | Yes | Sanger, 454, Illumina | Yes | Haplotypes, speed-focused |
| Flowsim | Balzer et al. (27) | GPL | biohaskell.org/Applications/FlowSim | Haskell | CLI | Genomic | No | 454 | Yes | Targets 454 simulation: SFF flowgram output, artificial replicates |
| MetaSim | Richter et al. (25) | Prop. | ab.inf.uni-tuebingende/software/metasim | Java | CLI, GUI | (Meta)genomic | Yes | Sanger, 454, Illumina | No | Genome evolution model |
| FASIM | Hur et al. (45) | Prop. | www.gem.re.kr/fasim | C | CLI | Genomic | No | Sanger | No | Biased sampling model, chimeras, chromatograms |
| CelSim | Myers (24) | Prop. | – | Awk, Perl | CLI | Genomic | No | Sanger | No | Repeat and variants generation |
| GenFrag | Engle and Burks (46,47) | Prop. | – | C | CLI | Genomic | No | Sanger | No | First read simulator |
Lic, License; Prop, proprietary; Lang, Programming language; Interf, Interfaces; Sim, Simulation; Qual, Quality.
Grinder also includes unique features such as the ability to specify a community structure based on a given richness (number of species) and ecologically-realistic species-abundance models (39). Multiple libraries representing communities with a specified structure and α and β diversity can be generated simultaneously. The β diversity feature in Grinder was recently used to establish empirical cutoffs for statistically significant differences between viral metagenomes (40). Grinder also provides parameters to introduce sampling biases inherent in metagenomic studies into sequence libraries. The development and benchmarking of GAAS (8) relied on the unique capability of Grinder to account for how the different length of genomes in a microbial or viral community affects the number of reads obtained from these genomes in a metagenome.
Grinder for amplicon dataset simulation
Grinder is the first read simulator to generate amplicon datasets (Table 1). Amplicon sequencing has most commonly been used for the characterization of bacterial and archaeal communities, but its applications are rapidly expanding to include characterization of fungal (41) and viral populations (42) as well as HLA class I genotyping (43). Amplicon libraries can be created in Grinder both with and without copy number bias, i.e. correction for the presence of multiple amplicons in a single reference sequence, and also with and without multiplex identifiers. Grinder uses an input set of PCR primers to find amplicons in reference sequences (Figure 2), and thus can be applied to any desired target gene or sequence.
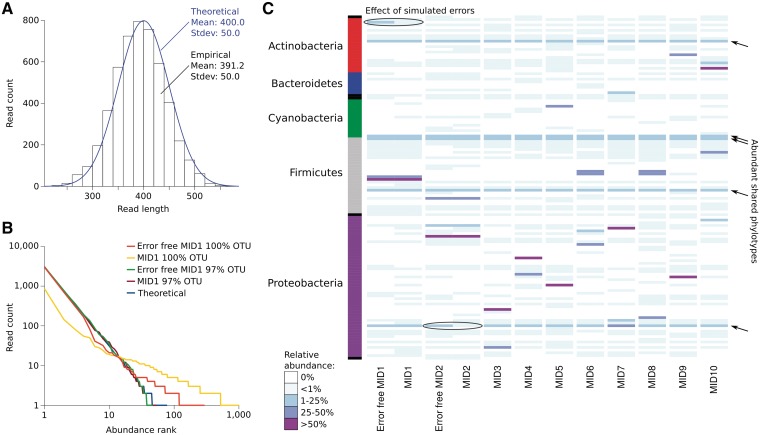
To demonstrate the use of Grinder for amplicon reads, MID-barcoded 16S rRNA libraries with and without pyrosequencing errors were simulated. Grinder faithfully produced 5000 simulated amplicon reads with MIDs in accordance with the input specifications: normal read distribution (Figure 3A), power law rank-abundance and richness (Figure 3B), β diversity (Figure 3C). All libraries were processed with QIIME and a total of 22 411 operational taxonomic units (OTUs) at 100% identity clustering, nearly 100 times the expected number. Kunin et al. (48) reported similar results for 454 amplicon pyrosequencing of Escherichia coli, demonstrating a 40- to 150-fold increase in the expected number of 100% OTUs depending on the type of quality filtering used. An approximately 100-fold increase in 100% OTUs due to homopolymer errors was also observed by Quince et al. (4). Consistent with the empirical observation of Kunin et al., 97% identity clustering reduced the number of OTUs, resulting in a rank-abundance distribution approaching the theoretical values (Figure 3B).
Figure 3.
Analysis of 10 MID-containing 16S rRNA gene amplicon libraries generated by Grinder that share 20% of their phylotypes. (A) Histogram of read lengths for the libraries and curve representing their expected normal distribution. (B) Log-log plot of phylotype rank-abundance in the MID1 amplicon library, with and without simulated sequencing errors, using 97% and 100% identity for OTU clustering in QIIME. (C) Heatmap comparison of the OTU distribution in the amplicon libraries analyzed with QIIME at 97% identity OTU clustering.
Comparison of the error-free libraries with their counterparts demonstrated changes in relative abundance for some OTUs, the introduction of 21 novel OTUs and the elimination of 15 others due to homopolymer errors (Figure 3C). While most of the discrepancies occurred for OTUs at a low abundance level (<1%), as previously reported (4,48,49), the decrease of two OTUs from a medium abundance level (1–25%) to a low abundance level (<1%) shows that care should be taken when analyzing amplicon data that contain sequencing errors. The simulated errors mostly affected low-abundance OTUs, artificially inflating the size of the rare biosphere (4,48,50). Overall, this example illustrates that Grinder is capable of creating realistic amplicon libraries and modeling the effects of 454 homopolymer errors on microbial community profiling using the 16S rRNA gene.
CONCLUSION
Grinder is a read simulator that generates shotgun and amplicon libraries for software benchmarking, algorithm development, statistical testing and educational purposes. Grinder has been used in this capacity to simulate large volumes of environmental and clinical sequence data (8,38,40,51). Grinder libraries can be given a variety of community structures by specifying an ecological species-abundance distribution and α diversity or β diversity and MIDs when multiple libraries are created simultaneously. As demonstrated here Grinder has the unique ability to generate realistic 16S rRNA amplicon reads in silico with 454 homopolymer errors. The errors of current sequencing technologies can be flexibly specified in Grinder by combining several deterministic models. Sequencing technologies evolve rapidly, but the open-source nature of Grinder will facilitate the addition of new technologies such as IonTorrent (52) as their error profiles become available. By helping test hypotheses, create better bioinformatic tools and enhance data interpretation, the more systematic use of read simulators has the potential to accelerate the rate of biological discoveries. In this context, we believe that Grinder will be a valuable tool for bioinformaticians and biologists alike.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4 and Supplementary Dataset 1.
FUNDING
QEII Fellowship from the Australian Research Council, [DP1093175 (to G.W.T.)]; University of Queensland strategic funding of the Australian Centre for Ecogenomics. Funding for open access charge: F.E.A's Discovery Early Career Research Award.
Supplementary Material
ACKNOWLEDGEMENTS
We thank people who have tested Grinder and offered comments, suggestions and support, specifically Barry Cayford, Paul Dennis, Mike Imelfort, Steve Rayhawk, Robert Schmieder, Ramzi Temanni and Albert Villela.
REFERENCES
- 1.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl Acad. Sci. USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y, Cai Y, Liu L, Yu F, Farrell ML, McKendree W, Farmerie W. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 2009;37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 5.Henn MR, Sullivan MB, Stange-Thomann N, Osburne MS, Berlin AM, Kelly L, Yandava C, Kodira C, Zeng Q, Weiand M, et al. Analysis of high-throughput sequencing and annotation strategies for phage genomes. PLoS One. 2010;5:e9083. doi: 10.1371/journal.pone.0009083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kan J, Hanson TE, Ginter JM, Wang K, Chen F. Metaproteomic analysis of Chesapeake Bay microbial communities. Saline Syst. 2005;1:7. doi: 10.1186/1746-1448-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, Land M, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 8.Angly FE, Willner D, Prieto-Davó A, Edwards RA, Schmieder R, Vega-Thurber R, Antonopoulos DA, Barott K, Cottrell MT, Desnues C, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beszteri B, Temperton B, Frickenhaus S, Giovannoni SJ. Average genome size: a potential source of bias in comparative metagenomics. ISME J. 2010;4:1075–1077. doi: 10.1038/ismej.2010.29. [DOI] [PubMed] [Google Scholar]
- 10.Yilmaz S, Allgaier M, Hugenholtz P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods. 2010;7:943–944. doi: 10.1038/nmeth1210-943. [DOI] [PubMed] [Google Scholar]
- 11.Pinard R, de Winter A, Sarkis G, Gerstein M, Tartaro K, Plant R, Egholm M, Rothberg J, Leamon J. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 13.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto M, Nishimura T. Mersenne twister: a 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans. Model Comput. Simul. 1998;8:3–30. [Google Scholar]
- 17.L’Ecuyer P, Simard R. TestU01: A C library for empirical testing of random number generators. ACM Trans. Math. Softw. 2007;33 Article 22. [Google Scholar]
- 18.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivancos AP, Güell M, Dohm JC, Serrano L, Himmelbauer H. Strand-specific deep sequencing of the transcriptome. Genome Res. 2010;20:989–999. doi: 10.1101/gr.094318.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennasar A, Mulet M, Lalucat J, García-Valdés E. PseudoMLSA: a database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol. 2010;10:118. doi: 10.1186/1471-2180-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers G. A dataset generator for whole genome shotgun sequencing. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:202–10. [PubMed] [Google Scholar]
- 25.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim: a sequencing simulator for genomics and metagenomics. PLoS One. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korbel JO, Abyzov A, Mu XJ, Carriero N, Cayting P, Zhang Z, Snyder M, Gerstein MB. PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10:R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balzer S, Malde K, Lanzén A, Sharma A, Jonassen I. Characteristics of 454 pyrosequencing data—enabling realistic simulation with flowsim. Bioinformatics. 2010;26:i420–i425. doi: 10.1093/bioinformatics/btq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang GC, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 30.Quince C, Lanzen A, Davenport R, Turnbaugh P. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosby LD, Criddle CS. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques. 2003;34:790–794, 796, 798 passim. doi: 10.2144/03344rr01. [DOI] [PubMed] [Google Scholar]
- 33.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afgan E, Baker D, Coraor N, Chapman B, Nekrutenko A, Taylor J. Galaxy CloudMan: delivering cloud compute clusters. BMC Bioinformatics. 2010;11:S4. doi: 10.1186/1471-2105-11-S12-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R development core team. R. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; [Google Scholar]
- 38.Berger SA, Stamatakis A. Aligning short reads to reference alignments and trees. Bioinformatics. 2011;27:2068–2075. doi: 10.1093/bioinformatics/btr320. [DOI] [PubMed] [Google Scholar]
- 39.Ulrich W. Models of relative abundance distributions I: model fitting by stochastic models. Pol. J. Ecol. 2001;49:145–157. [Google Scholar]
- 40.Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, Rainey PB, Schmieder R, Youle M, Conrad D, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am. J. Respir. Cell Mol. Biol. 2011 doi: 10.1165/rcmb.2011-0253OC. 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simons J, Egholm M, Lanza J, Turenchalk G, Desany B, Ronan M, Knight J, Du L, Leamon J, Rothberg J, et al. Ultra-deep sequencing of HIV from drug-resistant patients. Antivir. Ther. 2005;10:S157. [Google Scholar]
- 43.Lank SM, Wiseman RW, Dudley DM, O’Connor DH. A novel single cDNA amplicon pyrosequencing method for high-throughput, cost-effective sequence-based HLA class I genotyping. Hum. Immunol. 2010;71:1011–1017. doi: 10.1016/j.humimm.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holtgrewe M. Mason - A Read Simulator for Second Generation Sequencing Data. 2010. Institut für Mathematik und Informatik, Freie Universität Berlin. [Google Scholar]
- 45.Hur C-G, Kim S, Kim C-H, Yoon S-H, In Y-H, Kim C-M, Cho H-G. FASIM: Fragments assembly simulation using biased-sampling model and assembly simulation for microbial genome shotgun sequencing. J. Microbiol. Biotechn. 2006;16:683–688. [Google Scholar]
- 46.Engle ML, Burks C. Artificially generated data sets for testing DNA sequence assembly algorithms. Genomics. 1993;16:286–288. doi: 10.1006/geno.1993.1180. [DOI] [PubMed] [Google Scholar]
- 47.Engle ML, Burks C. GenFrag 2.1: new features for more robust fragment assembly benchmarks. Comput. Appl. Biosci. (CABIOS) 1994;10:567–568. doi: 10.1093/bioinformatics/10.5.567. [DOI] [PubMed] [Google Scholar]
- 48.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 49.Balzer S, Malde K, Jonassen I. Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics. 2011;27:i304–i309. doi: 10.1093/bioinformatics/btr251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sogin ML, Morrison HG, Huber JA, Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Treangen TJ, Sommer DD, Angly FE, Koren S, Pop M. Next generation sequence assembly with AMOS. Curr. Protoc. Bioinformatics. 2011;33:11.8.1–11.8.18. doi: 10.1002/0471250953.bi1108s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.