Abstract
Recent studies have identified the genetic underpinnings of a growing number of diseases through targeted exome sequencing. However, this strategy ignores the large component of the genome that does not code for proteins, but is nonetheless biologically functional. To address the possible involvement of regulatory variation in congenital heart diseases (CHDs), we searched for regulatory mutations impacting the activity of TBX5, a dosage-dependent transcription factor with well-defined roles in the heart and limb development that has been associated with the Holt–Oram syndrome (heart–hand syndrome), a condition that affects 1/100 000 newborns. Using a combination of genomics, bioinformatics and mouse genetic engineering, we scanned ∼700 kb of the TBX5 locus in search of cis-regulatory elements. We uncovered three enhancers that collectively recapitulate the endogenous expression pattern of TBX5 in the developing heart. We re-sequenced these enhancer elements in a cohort of non-syndromic patients with isolated atrial and/or ventricular septal defects, the predominant cardiac defects of the Holt–Oram syndrome, and identified a patient with a homozygous mutation in an enhancer ∼90 kb downstream of TBX5. Notably, we demonstrate that this single-base-pair mutation abrogates the ability of the enhancer to drive expression within the heart in vivo using both mouse and zebrafish transgenic models. Given the population-wide frequency of this variant, we estimate that 1/100 000 individuals would be homozygous for this variant, highlighting that a significant number of CHD associated with TBX5 dysfunction might arise from non-coding mutations in TBX5 heart enhancers, effectively decoupling the heart and hand phenotypes of the Holt–Oram syndrome.
INTRODUCTION
Congenital heart diseases (CHDs) are the most prevalent neonatal disorders in humans, affecting ∼1 in every 120 live births (1). Because CHDs reflect abnormalities of heart development during embryogenesis, they are thought to result from alterations in transcription factors and other developmental pathways that coordinately control cardiac development. This hypothesis has been repeatedly reinforced by successive reports of mutations in cardiac developmental genes, resulting in CHDs (2–7). These examples, however, collectively explain but a small fraction of CHD cases.
Recent advances in DNA sequencing technology allow for the simultaneous interrogation of entire patient exomes in search of disease-causing genetic variations (8–10). This has led to the discovery of MLL2 as a causative gene for Kabuki syndrome (11), which includes congenital cardiac defects, and holds great potential as an experimental platform for the identification of new mutations and genes causing CHDs. Despite the promise of exome sequencing, its main limitation resides precisely in restricting the mutation survey area to ∼1–2% of the human genome which encodes for proteins. Indeed, the impact of genetic variants in non-protein-coding sequences in the etiology of complex diseases has become well recognized. Genome-wide association studies (GWASs) suggest that a large number of non-coding variants underlie the increased risk to various common diseases, often by disrupting cis-regulatory elements (CREs) that control the expression of nearby genes (12–19).
The potential role of non-coding variants in congenital malformations, including CHDs, has not been extensively explored. Mutations that truncate the mRNA or significantly alter the structure or amino acid composition of a transcription factor will intuitively result in severe morphological phenotypes, such as those seen in CHDs. Less clear are the potential deleterious effects of mutations in CREs of these developmental genes. The regulatory architecture of a locus spanning a transcription factor or other developmental factors often comprises various long-range CREs, each responsible for a subset of the temporal, spatial and/or quantitative aspects of the gene's expression (18). Genetic variation within one of these multiple cis-regulatory units may lead to (i) no phenotypic effect, with the remaining cis-regulatory units compensating for the dysfunction of a single enhancer; (ii) a compartmentalization of the phenotypic effects of that gene, with a subset of the global biological function of the gene being disrupted; or (iii) an altogether new phenotype, uncovering a previously unanticipated biological role for that gene. In fact, several loci encoding for transcription factors involved in embryonic development have emerged from GWASs with non-coding variation associated with adult-onset cardiac disease phenotypes (20–22), rather than congenital morphological phenotypes that were attributed to coding mutations within those same genes (2,23).
To address the potential for mutations within CREs of developmental genes to result in CHDs, we utilized the TBX5 locus. TBX5 is a transcription factor with well-defined roles in the heart and forelimb development (24,25). In addition to its demonstrated necessity for cardiogenesis (26,27), an array of coding mutations in TBX5 almost invariably leads to the Holt–Oram syndrome (2,26,28,29), indicating a strong biological sensitivity to TBX5 dosage, which has been confirmed in multiple animal models (25,30–33). Given this dosage-sensitivity, we hypothesized that sequence variations in enhancers regulating TBX5 expression during heart development are likely to perturb TBX5 expression and result in CHD.
Using a combination of bioinformatics, epigenetics, genomics and in vivo mouse transgenic reporter assays, we scanned the TBX5 locus for developmental enhancers. We subsequently sequenced three cardiac-specific enhancers in a cohort of 260 Brazilians with atrial septal defects (ASDs) and/or ventricular septal defects (VSDs), the most common forms of CHD associated with the Holt–Oram syndrome. Our results uncovered a low-frequency single-nucleotide polymorphism (SNP) segregating in the population that abrogates a cardiac-specific TBX5 enhancer, thereby decoupling the heart and hand phenotypes seen in the Holt–Oram syndrome patients. This SNP alone has the potential to lead to as many CHDs as the combined spectrum of mutations reported for the Holt–Oram syndrome, underscoring that a significant number of TBX5-related CHDs may be due to variation in CREs and are undetected by exome or other protein-coding targeted sequencing strategies.
RESULTS
Determining the regulatory landscape of the TBX5 locus
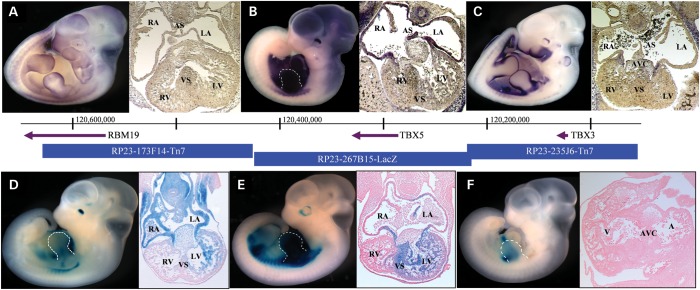
TBX5 lies amidst a nearly 700 kb gene desert, bracketed by TBX3 (261 kb upstream) and RBM19 (387 kb downstream) (Fig. 1). Although regulatory elements can lie further afield (34), we and others have had success limiting the search to the nearest neighboring genes. Using in situ hybridization (Fig. 1A–C), we established that the endogenous expression patterns of these three genes overlap minimally, thereby simplifying the assignation of enhancers within this region to their cognate gene.
Figure 1.
Regulatory landscape of the TBX5 locus. Whole mount and histological sections through the heart of embryonic day 11.5 (E11.5) embryos. Top row: In situ hybridizations showing endogenous expression (purple) of (A) RBM19, (B) TBX5 and (C) TBX3. Bottom row: β-Galactosidase staining (blue) captures the regulatory landscape of enhancers within BACs (D) RP23-173F14-Tn7, (E) RP23-267B15-LacZ and (F) RP23-235J6-Tn7. The genes probed for in A, B and C are contained, respectively, within the BACs tested in D, E and F. There is a 46 bp gap between the BACs containing RBM19 and TBX5, encompassing nucleotides 120 424 690–120 424 646. There is 1140 bp of overlap between the BACs containing TBX5 and TBX3 encompassing nucleotides 120 216 612–120 217 754. Panels B, D–F, forelimb is outlined. A–E, transverse sections; F, sagittal section. A, atrium; RA, right atrium; LA, left atrium; AS, atrial septum; V, ventricle; RV, right ventricle; LV, left ventricle; VS, ventricular septum; AVC, atrioventricular canal.
We then interrogated the entire 700 kb region for the presence of transcriptional enhancers. We identified three mouse bacterial artificial chromosomes (BACs) that tile across the region, and subsequently converted each into an enhancer trap system by recombineering lacZ onto each BAC, such that transgene expression would be driven exclusively by extant enhancers (19,35,36). We generated transgenic mice harboring each of the engineered BACs and assayed them for β-galactosidase expression in multiple independent founder embryos. Because known TBX5 mutations result in septal defects (27,30), we assayed the developing embryos after septation had begun (37,38), during their 11th day of development (E11.5) (Fig. 1D–F). Comparing β-galactosidase expression with the mRNA expression of each gene in the region, we found that BACs RP23-267B15-lacZ and RP23-173F14-Tn7 each drove cardiac expression in a manner consistent with the endogenous TBX5 expression profile. In contrast, RP23-235J6-Tn7 generated no overlapping expression with TBX5 and was therefore excluded from further investigation (Fig. 1F).
Transgenic lines (n = 6) carrying RP23-267B15-lacZ exhibited consistent expression in a pattern qualitatively similar to that of TBX5, with strong expression in the left ventricle, moderate expression in the ventricular septum and very low expression in the right ventricle (Fig. 1E). Notably, all lines exhibited weak expression in the left atrium and almost no expression in the right atrium, suggesting that the bulk of TBX5 atrial expression is regulated by enhancers outside the genomic region covered by this BAC. Sporadic expression (n = 2) was also seen in the forelimbs.
Transgenic lines (n = 4) carrying RP23-173F14-Tn7 displayed consistent expression similar to that of TBX5 in the dorsal retina and along a strong anteroposterior gradient in the heart (Fig. 1D). Both atria and the atrial septum were heavily marked, as was the mesoderm surrounding the trachea. The atrioventricular canal (AVC) and the left ventricle showed moderate expression, especially in the trabeculated myocardium, while expression in the right ventricle was minimal.
Together, these two BACs recapitulated virtually all aspects of TBX5 cardiac expression. We next sought to fine-map individual TBX5 enhancers contained within each of these BACs.
Identification of individual enhancer elements
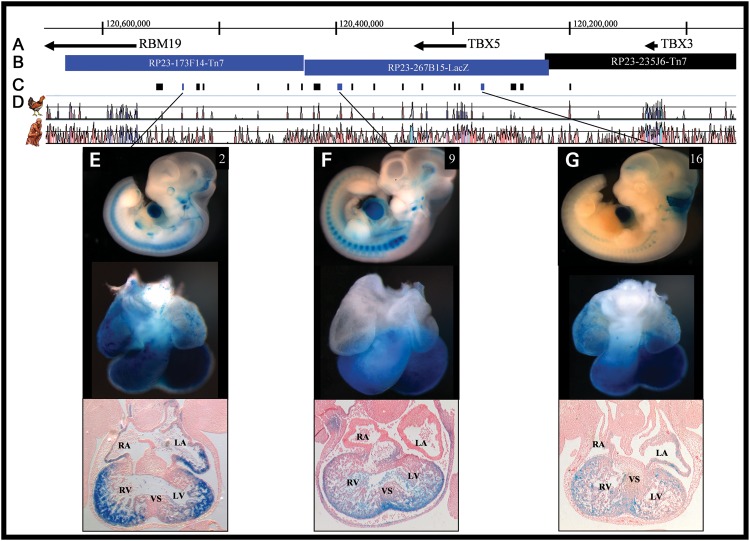
Having narrowed the TBX5 enhancer-containing region to ∼411 kb, we used an integrated approach to map the coordinates of individual enhancers. This strategy considered multiple genome-wide ChIP-seq data sets including the enhancer-associated transcriptional co-activator p300 (39,40), cardiac transcription factors Mef2a, TBX5, Gata4, Nkx2.5, SRF (40) and computationally predicted enhancers (41), as well as the enhancer-associated histone modification H3K4me1 (generated in the Nobrega lab; not shown) and evolutionary conservation (42,43) to identify and prioritize likely functional non-coding regions. In total, we identified 19 discrete (369–5026 bp) candidate human non-coding CREs (Fig. 2C and D, Supplementary Material, Table S1).
Figure 2.
Location of and β-galactosidase expression driven by TBX5 enhancers. (A and B) The TBX5 locus. (C) Individual elements tested for enhancer activity. Sequences with enhancer activity consistent with the expression pattern of TBX5 are dark blue. (D) Pairwise alignments of the mouse genome to human and chicken identify evolutionarily conserved regions (ECRs), which are displayed as peaks. (E–G) Whole mount, isolated hearts and transverse histological sections of transgenic animals carrying individual TBX5 enhancers at E11.5. Consistent expression, across multiple lines, was observed for all three enhancers in the ventricular myocardium, and although it is evident here for ECRs 2 and 16, atrial expression was not recapitulated in independent lines. Only enhancer 2 reproducibly drove expression outside the heart, in the eye. See Fig. 1 for abbreviations and Supplementary Material, Figs S1–S6 for additional lines and time points.
Each candidate CRE was tested for enhancer activity in vivo by placing it upstream of an hsp68 minimal promoter connected to lacZ (hsp68–lacZ). Using these constructs, we generated transgenic mice, each of which were assayed for β-galactosidase expression at E11.5. Enhancers exhibiting overlapping expression patterns with TBX5 in at least three independently derived embryos were considered for further analyses.
We identified three elements (CREs 2, 9, 16) with patterns of enhancer activity overlapping the endogenous expression pattern of TBX5 and the enhancer-trap BACs in which they were contained (Fig. 2E–G). Enhancer 2, located 380 kb downstream of TBX5 (hg19 chr12:114 463 712–114 464 080), drove strong expression in ventricles, but not the interventricular septum (IVS), and in both atria, but not the atrial septum or AVC. Situated 140 kb downstream of TBX5 (hg19 chr12:114 701 207–114 704 691), enhancer 9 controlled expression throughout the posterior portion of the heart, including ventricles, IVS and AVC. Enhancer 16, which is 9 kb upstream of TBX5 (hg19 chr12:114 853 271–114 858 238), imparted expression in ventricles, IVS, AVC and weakly in atria. In each case, the regulatory activity was reproducible (see Supplementary Material, Figs S1–S3 for additional time points and Supplementary Material, Figs S4–S6 for additional transgenic lines). Although these isolated enhancers gave expression in both ventricles, when in the larger context of a BAC, RV expression was not seen and supports the notion that, in vivo, the expression in the right ventricle is repressed by undefined mechanisms. Only ECR2 drove reproducible extra-cardiac expression, in the eye, which was also seen in the BAC that contains ECR2 (Fig. 1D). We also identified three elements (CREs 8, 15, 19; Supplementary Material, Figs S7–S9) that consistently yielded expression within the outflow tract (OFT), a domain in which TBX5 is excluded. TBX3, however, is critical for OFT development (44), and the most parsimonious explanation is that these are TBX3 enhancers. These enhancers were not studied further.
Interestingly, because none of the enhancers imparted both heart and limb expression, our scan suggests that the cis-regulation of this gene in the heart and forelimbs is compartmentalized, each carried out by a distinct set of modular enhancers specific to each spatial domain. These results further suggest that potentially deleterious genetic variation within these enhancers might lead to cardiac-specific phenotypes, decoupling the heart and hand phenotypes seen with TBX5 protein-coding mutations in the Holt–Oram syndrome and potentially expanding the spectrum of CHDs caused by TBX5-associated dysfunction.
Sequencing CHD patient samples
In order to establish a potential role for TBX5 cis-regulatory variation in CHD, we sequenced the three enhancers that were uncovered in a patient cohort consisting of 260 unrelated Brazilian children born with isolated, congenital heart defects, including atrial (82), ventricular (140) and atrioventricular (38) septal defects, none of whom were diagnosed with the Holt–Oram or any other syndromic CHD. We identified 19 high-quality sequence variants, 10 of which represented common polymorphisms and were discarded from further analysis (Supplementary Material, Table S2). We confirmed the remaining nine rare or novel variants via resequencing. Eight of these variants were observed in a heterozygous state, with half common to two or more individuals within our sample, while the other half were novel. In addition, most of these eight variants were devoid of or harbored minimal basewise placental mammal conservation according to PhyloP and MULTIZ (45,46), suggesting a lack of evolutionary constraint expected of functionally critical nucleotides. Furthermore, when analyzed using rVista (47), most of these lacked notable overlap with predicted conserved transcription factor binding sites (cTFBS).
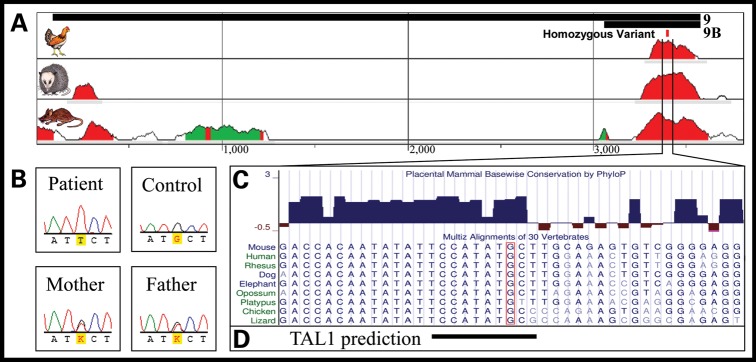
One variant, however, stood out markedly as a strong candidate for a functionally relevant sequence variant: a singleton, homozygous G→T transversion within enhancer 9 at a highly conserved nucleotide (hg19, chr12:114 704 515) inside a cTFBS (see below) (Fig. 3A–C). Notably, enhancer 9 exhibited expression within the ventricular septum (Fig. 2F), while the patient that harbored the putative functional variant was diagnosed with a VSD.
Figure 3.
Enhancer 9 genomic context and nucleotide conservation. (A) Human alignment to chicken, opossum and mouse at the enhancer 9 region (black bars, top) shows that this enhancer contains two short segments of highly conserved nucleotides (red peaks). (B) One patient possessed a homozygous G→T sequence alteration (hg19 chr12:114 704 515), with two heterozygous parents. (C) The variant nucleotide (boxed) is highly conserved and overlaps a predicted, conserved binding site for TAL1 (D).
To establish whether the variation within enhancer 9 corresponds to a functional binding site of a known transcription factor, we screened the variant nucleotide and adjacent DNA sequence for transcription factor binding sites using the TRANSFAC database (47,48) and identified a conserved, predicted binding site of TAL1 (HGNC:11556) (Fig. 3D). However, previous reports in mouse (49–51) and zebrafish (52), as well as our own RNA in situ hybridization (Supplementary Material, Fig. S10), indicate that TAL1 is confined to the endocardial aspects of the ventricular septum, and not the myocardial portion in which enhancer 9 drives expression. No other predicted binding site in TRANSFAC overlaps the variant nucleotide, suggesting that an unidentified factor binds to this DNA region in vivo. It is also possible that the variant overlaps with a binding site for a known factor that binds to DNA using alternative, non-canonical binding sites, as we have recently shown (53). The identification of such binding factor(s) will be the subject of future studies.
Upon further sequencing, we confirmed that both parents of the patient are heterozygotes at this position, and additional sequencing of 500 unrelated, unaffected Brazilians identified three additional heterozygotes, suggesting that this variation is an SNP with a minor allele frequency of 0.3% in the Brazilian population, which is slightly less common than the 2.7% frequency among Peruto Ricans, as seen in preliminary data from the 1000 Genomes Project (54), but suggests that it segregates in broader populations within the Americas. To exclude protein-coding mutations in this patient, we sequenced the exons of genes previously associated with VSD (TBX5, CITED2, GATA4 and NKX2.5). Only a common, synonymous SNP (rs2277923) in NKX2.5 was observed. All patients were karyotypically normal, and we also excluded a 22q11.2 deletion (55) as a putative underlying genetic determinant of the CHD in the patient.
Functional impact of variant in enhancer function
In vitro analysis
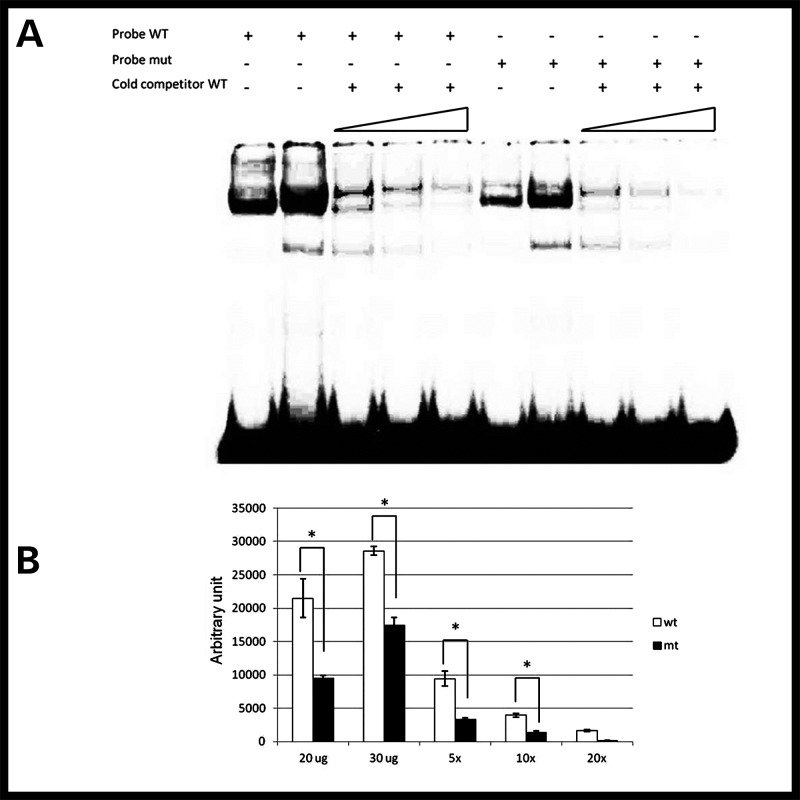
We hypothesized that the variant nucleotide disrupts transcription factor binding to enhancer 9. To test this, we performed an electrophoretic mobility shift assay using short DNA probes spanning either the wild-type (G) or variant (T) allele. Upon in vitro incubation with HeLa cell lysate, which we confirmed express TAL1 (Supplementary Material, Fig. S11), we observed that the magnitude of the shift in the variant enhancer sequence is quantitatively reduced, in a dosage-dependent effect, compared with the wild-type probe (Fig. 4, Supplementary Material, Figs S12 and S13), supporting the hypothesis that this variant does impart a functional consequence.
Figure 4.
Reduced binding to mutant DNA. Radioactive DNA probe containing wild-type G allele (WT) is bound to a greater extent than probe containing mutant T allele (mut) (A, lanes 1, 2 versus 6, 7; B, 20 and 30 μg). Cold WT probe competitively displaces mutant probe faster than WT (A, lanes 8–10 versus 3–5; B, ×5, ×10 and ×20). Statistical analyses were performed with two-way ANOVA. Each bar represents median ± standard error (n= 3). *P< 0.05 versus wild-type.
In vivo analysis
We next assayed the impact of this variant on enhancer function in vivo. For this analysis, we amplified enhancer 9 containing the putative functional variant using the patient's DNA, cloned this sequence into an hsp68–lacZ reporter construct and generated transgenic mice. While the construct harboring the wild-type (G) allele of enhancer 9 resulted in seven out of eight independent transgenic founders with reproducible β-galactosidase staining in the heart, the variant (T) allele of this enhancer resulted in 11 independent founder transgenic animals, only one of which expressed β-galactosidase, weakly in the atrial free wall of the heart (Fig. 5A, Supplementary Material, Fig. S14). These data directly demonstrate that the variation within this highly conserved DNA segment abrogates the cardiac expression potential of enhancer 9 in vivo.
Figure 5.
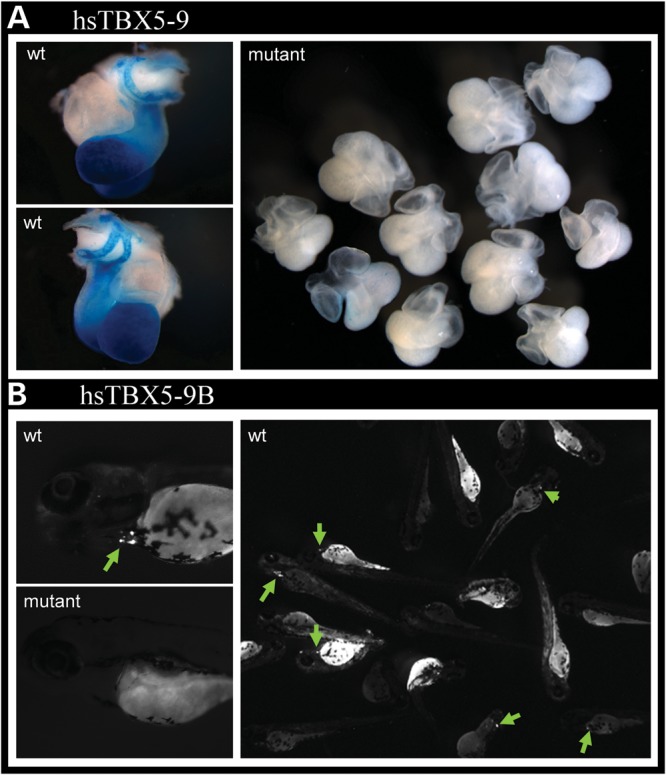
A regulatory variant abrogates enhancer function. (A) A single-base-pair mutation in enhancer 9 abrogates its ability to drive cardiac expression. (B) Enhancer 9B, containing a 515 bp ECR within enhancer 9, drives cardiac expression in zebrafish. When mutated similarly, it too loses enhancer activity.
To further narrow the critical region sufficient for enhancer 9 function, we generated a construct containing a short, highly conserved 515 bp sub-sequence of enhancer 9 (construct 9B) spanning the variant. We obtained 9B constructs harboring each allele of the variant, cloned each into a cFos-GFP reporter construct, sequence-verified them and generated transgenic zebrafish, which we assayed at 72 h for GFP expression (41). At this stage, the zebrafish heart has already developed and is undergoing rapid differentiation and proliferation, comparable to mouse embryonic stage E11.5. Of 82 fish injected with wild-type 9B constructs, 28 (34%) displayed similar expression of GFP in the heart, indicating that this shorter element alone is sufficient to retain cardiac enhancer properties (Fig. 5B). In stark contrast, of 110 fish injected with enhancer 9B containing the novel allele, only two (1.8%) displayed weak expression of GFP in the heart (Fisher's exact P-value: 3.85e−10), demonstrating a clear functional consequence of this variant, in vivo, in a second vertebrate model. Together, these results indicate that this low-frequency SNP destroys the cardiac expression properties of this enhancer.
DISCUSSION
Results from GWAS have suggested that a significant fraction of the underlying genetic variation associated with complex human diseases is non-protein coding in nature, and experimental follow-up in several of these associated regions has directly implicated regulatory variation—disrupting the function of long-range cis-regulatory sequences—as a mechanistic link to these genetic associations (12–19). Less clear, however, is the spectrum of congenital diseases that may also have regulatory variation as their genetic underpinning. Our work illustrates that a considerable number of CHD cases may be due to variation in long-range enhancers of genes that are critical in heart development. Importantly, we showed how the modularity in the cis-regulation of TBX5, comprising several cardiac enhancers, can decouple the cardiac and limb phenotypes usually associated with TBX5 protein-coding mutations and the Holt–Oram syndrome. This example highlights that regulatory variations leading to CHDs may present with compartmentalized phenotypes compared with the broader phenotypes associated with protein-coding mutations in their cognate gene.
Our results show that the enhancer variant tested in vitro and in vivo represents a low-frequency SNP in the population studied. It also implies that ∼1/100 000 individuals will be homozygous for this variant. The penetrance of this variant is unknown, but its frequency raises the possibility that this SNP alone may result in as many cases of CHD as the population-wide incidence of the Holt–Oram syndrome, significantly increasing the number of cases of CHD connected to TBX5 dysfunction. Because the patient's parents and the three unrelated Brazilians appear to be unaffected carriers of the variant, we speculate that a single allele of this variant is not sufficient to significantly change the expression of TBX5, which we show is under the control of at least three independent cardiac enhancers. Moreover, while we were successful in uncovering multiple cardiac enhancers within the TBX5 locus, this was not a comprehensive enhancer screen, and it is likely that we are missing other regulatory sequences that could harbor additional variants leading to additional cases of CHD.
Regulatory variation in long-range enhancers such as the one studied here will be missed by exome sequencing strategies. With the advent of whole-genome sequencing, such variants will finally be captured by routine resequencing efforts. Nevertheless, two major hurdles limit the interpretation of sequencing data. First, a complete catalog of functional non-coding sequences will be necessary in order to prioritize among the non-coding fraction of the genome those regions in which variation may result in phenotypes. Technological developments in high throughput genomics and large-scale experimental efforts such as the ENCODE project have made this an addressable challenge (56,57). Second, the functional interpretation of sequence variation in non-protein-coding sequences remains elusive. As demonstrated in our work, we were not able to establish which transcription factor(s) binds differentially to the allelic variants of the novel SNP within the TBX5 enhancer that we studied. Given the rapidly increasing repertoire of examples of non-coding variation as genetic underpinning for both developmental and complex, adult-onset diseases, the development of new, high throughput, systematic experimental tools to evaluate the functional implications of such variants will likely take center stage.
MATERIALS AND METHODS
BAC modification
Mouse BACs were engineered in vitro in two ways: through targeted insertion and through transposon-mediated, random insertion of a cassette containing an antibiotic resistance gene, for selection of recombinant bacteria, and a β-galactosidase (lacZ) gene which is expressed only in the presence of cis-regulatory elements within the respective BACs.
BAC RP23-267B15 had a lacZ–ampicillin cassette inserted, in frame, replacing the first exon of TBX5 using a Red/ET recombination kit and protocol from Gene Bridges (catalog #K001). Ampicillin-resistant colonies were polymerase chain reaction (PCR)-screened for homologous recombination using primers in TBX5 and the vector cassette (Supplementary Material, Table S1), and the insertion junction was verified by sequencing. Recombinant BACs were confirmed intact by restriction digest with MluI (NEB).
BACs RP23-173F14 and RP23-235J6 were modified using a Tn7 transposon-based system (19,35,36). Using a Nucleobond AX Kit (Macherey-Nagel), BAC DNA was extracted, 40 ng of which were combined with 20 ng of Tn7 vector carrying a β-globin minimal promoter adjacent to a lacZ gene and a downstream kanamycin resistance gene, GPS buffer and TnsABC transposase (NEB), and incubated at 37°C for 10 min. Start solution was added and incubation continued for 1 h, followed by a heat inactivation 75°C for 10 min and a 1 h dialysis against ultra-pure H20. Electro-competent DH10B cells were transformed with 2 μl of the transposition reaction and plated on LB agar containing 20 μg/ml kanamycin and 12.5 μg/ml chloramphenicol. Recombinant colonies were identified by PCR. Modified BAC clones were then digested with NotI and separated by pulsed field gel electrophoresis (PFGE) overnight on a 1% agarose gel to determine the number of copies and approximate position(s) of the integrated Tn7β-lacZ cassette.
Finally, BAC DNA from both systems was extracted from fresh overnight culture, an aliquot linearized and checked for length and degradation via PFGE, and the remainder dialyzed against PBS and submitted for pronuclear injection.
Enhancer reporter constructs
DNA was PCR-amplified and, depending on amplicon size, cloned into either of the Gateway entry vectors pENTR/D-TOPO (<2 kb) or pDONR 221 (Invitrogen), and then transferred to a Gateway-hsp68-lacZ reporter vector through LR recombination (58). Constructs were restriction digest and sequence-verified, linearized with SalI (NEB) and the vector backbone excised to leave only the putative enhancer, the hsp68 minimal promoter and the lacZ gene. After a final ethanol precipitation, the DNA was submitted to the University of Chicago Transgenic Core for pronuclear injection.
Mouse in vivo transgenic reporter assay
Embryos were harvested into cold 100 mm phosphate-buffered saline, pH 7.3 (PBS), followed by, depending on the embryonic stage, 20–60 min incubation in 4% paraformaldehyde at 4°C. Embryos were washed twice for 20 min in wash buffer (2 mm MgCl2, 0.01% deoxycholate, 0.02% NP-40, 100 mm phosphate buffer, pH 7.3), and stained for 16–20 h at room temperature with freshly made staining solution (0.8 mg/ml X-gal, 4 mm potassium ferrocyanide, 4 mm potassium ferricyanide, 20 mm Tris, pH 7.5 in wash buffer). After staining, embryos were rinsed at least five times in PBS and post-fixed in 4% paraformaldehyde. Embryonic yolk sacs were digested overnight at 55°C in 50 μl lysis buffer (100 mm Tris–HCl pH8.5, 5 mm ethylenediaminetetraacetic acid, 0.2% sodium dodecyl sulphate, 200 mm NaCl, 20 μg Proteinase K) and genomic DNA was extracted and purified using a Gentra Puregene kit (Qiagen).
Zebrafish reporter construction and in vivo transgenic assay
DNA from Patient 3403 and a control were PCR-amplified and cloned into pDONR 221 (Invitrogen) Gateway entry vector and then shuttled via LR reaction to a reporter vector containing eGFP under the control of a minimal c-fos promoter, all of which were flanked by tol2 transposon sites. Each construct was coinjected into one- to two-cell stage zebrafish embryos as 25 ng/μl circular plasmid with 35 ng/μl to l2 transposase mRNA transcribed in vitro using the mMessage mMachine Sp6 kit (Ambion). Zebrafish were maintained in accordance with standard protocols (59,60), and embryos were obtained from natural crosses of wild-type mating pairs, incubated at 28.5°C, and staged (61). Developing fish were observed daily for enhancer activity beginning 24 h after injection.
Histological processing and sectioning
Embryos were trimmed of excess tissue and submitted for dehydration and paraffin infusion to the University of Chicago Human Tissue Resource Center. Embryos were then embedded in paraffin blocks, sectioned and lightly stained with eosin at the University of Chicago Cancer Research Center Immunohistochemistry Core facility.
Enhancer sequencing in humans
Patient DNA was collected at the Instituto do Coracao in San Paulo, Brazil, under local IRB regulatory approval. Patients were ascertained by heart defect: 82 possessed ASDs, 140 possessed VSDs and 38 had complex atrioventricular septal defects. Enhancers were PCR-amplified (primers in Supplementary Material, Table S1) and then sequenced with BigDye Terminator chemistry (Applied Biosystems) according to standard protocols. Sequences were analyzed using Mutation Surveyor (SoftGenetics) and putative variants sequenced a second time in the opposite direction.
In situ hybridization
Whole mount and section in situ hybridizations against TBX5, Tbx3 and Rbm19 mRNA using digoxigenin-labeled antisense and sense (control) riboprobes were performed according to standard protocols (62). Probes were generated from full-length mouse cDNA clones: TBX5 (plasmid TBX5 w35), kindly provided by S. Evans; TBX3 (IMAGE ID 30547736); and RBM19 (IMAGE ID 3673396). Tissues were stained overnight, whole mount embryos transferred to 10% buffered formalin phosphate prior to imaging and sections mounted in Fluoro-Gel (Electron Microscopy Sciences).
Electrophoretic mobility shift assay
Assays were performed as previously reported (28). Probe sequences are available upon request. HeLa cells were shown to significantly express TAL1 compared with HEK 293T cells transiently transfected to overexpress TAL1 and endothelial cells that endogenously express TAL1 by western blot. Immunoblot membranes were probed first with TAL1 antiserum M258C (kindly provided by R. Baer) and subsequently with mouse anti-rabbit IgG/horseradish peroxidase conjugate (Invitrogen). Data were expressed as mean ± SEM. Statistical analyses were performed using two-way analysis of variance (ANOVA) and/or Student's t-test. Values of P < 0.05 were considered significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (grant numbers HL088393, HG004428, DK078871) to M.A.N., and the Genetics and Regulation Training Grant (grant number T32GM007197) to S.S.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ivy Aneas for technical help, Miles Tracy and Fabio Arimura for assistance in cloning and dissection and the rest of the Nobrega lab for troubleshooting and discussion. The TBX5 plasmid was kindly provided by Sylvia Evans, and the TAL1 antiserum graciously donated by Richard Baer.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hoffman J.I., Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 2.Basson C.T., Bachinsky D.R., Lin R.C., Levi T., Elkins J.A., Soults J., Grayzel D., Kroumpouzou E., Traill T.A., Leblanc-Straceski J., et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt–Oram syndrome. Nat. Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 3.Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A., Rothrock C.R., Eapen R.S., Hirayama-Yamada K., Joo K., et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 4.Schott J.J., Benson D.W., Basson C.T., Pease W., Silberbach G.M., Moak J.P., Maron B.J., Seidman C.E., Seidman J.G. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 5.Lopes Floro K., Artap S.T., Preis J.I., Fatkin D., Chapman G., Furtado M.B., Harvey R.P., Hamada H., Sparrow D.B., Dunwoodie S.L. Loss of Cited2 causes congenital heart disease by perturbing left-right patterning of the body axis. Hum. Mol. Genet. 2011;20:1097–1110. doi: 10.1093/hmg/ddq554. [DOI] [PubMed] [Google Scholar]
- 6.Reamon-Buettner S.M., Ciribilli Y., Inga A., Borlak J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum. Mol. Genet. 2008;17:1397–1405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- 7.Bruneau B.G. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 8.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worthey E.A., Mayer A.N., Syverson G.D., Helbling D., Bonacci B.B., Decker B., Serpe J.M., Dasu T., Tschannen M.R., Veith R.L., et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 10.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 11.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubbe S.J., Pittman A.M., Olver B., Lloyd A., Vijayakrishnan J., Naranjo S., Dobbins S., Broderick P., Gomez-Skarmeta J.L., Houlston R.S. The 14q22.2 colorectal cancer variant rs4444235 shows cis-acting regulation of BMP4. Oncogene. 2011 doi: 10.1038/onc.2011.564. doi:10.1038/onc.2011.564. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 14.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M., et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pittman A.M., Naranjo S., Jalava S.E., Twiss P., Ma Y., Olver B., Lloyd A., Vijayakrishnan J., Qureshi M., Broderick P., et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 2010;6:e1001126. doi: 10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittman A.M., Naranjo S., Webb E., Broderick P., Lips E.H., van Wezel T., Morreau H., Sullivan K., Fielding S., Twiss P., et al. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome. Res. 2009;19:987–993. doi: 10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visel A., Rubin E.M., Pennacchio L.A. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakabe N.J., Savic D., Nobrega M.A. Transcriptional enhancers in development and disease. Genome. Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasserman N.F., Aneas I., Nobrega M.A. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome. Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm H., Gudbjartsson D.F., Arnar D.O., Thorleifsson G., Thorgeirsson G., Stefansdottir H., Gudjonsson S.A., Jonasdottir A., Mathiesen E.B., Njolstad I., et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 21.Pfeufer A., van Noord C., Marciante K.D., Arking D.E., Larson M.G., Smith A.V., Tarasov K.V., Muller M., Sotoodehnia N., Sinner M.F., et al. Genome-wide association study of PR interval. Nat. Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sotoodehnia N., Isaacs A., de Bakker P.I., Dorr M., Newton-Cheh C., Nolte I.M., van der Harst P., Muller M., Eijgelsheim M., Alonso A., et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirk E.P., Sunde M., Costa M.W., Rankin S.A., Wolstein O., Castro M.L., Butler T.L., Hyun C., Guo G., Otway R., et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am. J. Hum. Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberatore C.M., Searcy-Schrick R.D., Yutzey K.E. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev. Biol. 2000;223:169–180. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi J.K., Koshiba-Takeuchi K., Suzuki T., Kamimura M., Ogura K., Ogura T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130:2729–2739. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- 26.Bruneau B.G., Logan M., Davis N., Levi T., Tabin C.J., Seidman J.G., Seidman C.E. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt–Oram syndrome. Dev. Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 27.Bruneau B.G., Nemer G., Schmitt J.P., Charron F., Robitaille L., Caron S., Conner D.A., Gessler M., Nemer M., Seidman C.E., et al. A murine model of Holt–Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 28.Li Q.Y., Newbury-Ecob R.A., Terrett J.A., Wilson D.I., Curtis A.R., Yi C.H., Gebuhr T., Bullen P.J., Robson S.C., Strachan T., et al. Holt–Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 29.Holt M., Oram S. Familial heart disease with skeletal malformations. Br. Heart. J. 1960;22:236–242. doi: 10.1136/hrt.22.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori A.D., Zhu Y., Vahora I., Nieman B., Koshiba-Takeuchi K., Davidson L., Pizard A., Seidman J.G., Seidman C.E., Chen X.J., et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 31.Albalat R., Baquero M., Minguillon C. Identification and characterisation of the developmental expression pattern of tbx5b, a novel tbx5 gene in zebrafish. Gene. Expr. Patterns. 2010;10:24–30. doi: 10.1016/j.gep.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Garrity D.M., Childs S., Fishman M.C. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129:4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 33.Horb M.E., Thomsen G.H. Tbx5 is essential for heart development. Development. 1999;126:1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- 34.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 35.Savic D., Ye H., Aneas I., Park S.Y., Bell G.I., Nobrega M.A. Alterations in TCF7L2 expression define its role as a key regulator of glucose metabolism. Genome Res. 2011;21:1417–1425. doi: 10.1101/gr.123745.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitz F., Gonzalez F., Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 37.Dalgleish A.E. The development of the septum primum relative to atrial septation in the mouse heart. J. Morphol. 1976;149:369–382. doi: 10.1002/jmor.1051490306. [DOI] [PubMed] [Google Scholar]
- 38.Rentschler S., Vaidya D.M., Tamaddon H., Degenhardt K., Sassoon D., Morley G.E., Jalife J., Fishman G.I. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blow M.J., McCulley D.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42: 806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He A., Kong S.W., Ma Q., Pu W.T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl Acad. Sci. USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narlikar L., Sakabe N.J., Blanski A.A., Arimura F.E., Westlund J.M., Nobrega M.A., Ovcharenko I. Genome-wide discovery of human heart enhancers. Genome Res. 2010;20:381–392. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mesbah K., Harrelson Z., Theveniau-Ruissy M., Papaioannou V.E., Kelly R.G. Tbx3 is required for outflow tract development. Circ. Res. 2008;103:743–750. doi: 10.1161/CIRCRESAHA.108.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loots G.G., Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drake C.J., Fleming P.A. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- 50.Giroux S., Kaushik A.L., Capron C., Jalil A., Kelaidi C., Sablitzky F., Dumenil D., Albagli O., Godin I. lyl-1 and tal-1/scl, two genes encoding closely related bHLH transcription factors, display highly overlapping expression patterns during cardiovascular and hematopoietic ontogeny. Gene Expr. Patterns. 2007;7:215–226. doi: 10.1016/j.modgep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez M., Gottgens B., Sinclair A.M., Stanley M., Begley C.G., Hunter S., Green A.R. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development. 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- 52.Bussmann J., Bakkers J., Schulte-Merker S. Early endocardial morphogenesis requires Scl/Tal1. PLoS Genet. 2007;3:e140. doi: 10.1371/journal.pgen.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen T., Aneas I., Sakabe N., Dirschinger R.J., Wang G., Smemo S., Westlund J.M., Cheng H., Dalton N., Gu Y., et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J. Clin. Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium T.G.P. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Driscoll D.A., Spinner N.B., Budarf M.L., McDonald-McGinn D.M., Zackai E.H., Goldberg R.B., Shprintzen R.J., Saal H.M., Zonana J., Jones M.C., et al. Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am. J. Med. Genet. 1992;44:261–268. doi: 10.1002/ajmg.1320440237. [DOI] [PubMed] [Google Scholar]
- 56.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myers R.M., Stamatoyannopoulos J., Snyder M., Dunham I., Hardison R.C., Bernstein B.E., Gingeras T.R., Kent W.J., Birney E., Wold B., et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poulin F., Nobrega M.A., Plajzer-Frick I., Holt A., Afzal V., Rubin E.M., Pennacchio L.A. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 60.Whitlock K.E., Westerfield M. The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development. 2000;127:3645–3653. doi: 10.1242/dev.127.17.3645. [DOI] [PubMed] [Google Scholar]
- 61.Warga R.M., Kimmel C.B. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108:569–580. doi: 10.1242/dev.108.4.569. [DOI] [PubMed] [Google Scholar]
- 62.Wilkinson D.G., Nieto M.A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.