Kinase-inhibited FAK limits VCAM-1 production via nuclear localization and promotion of GATA4 turnover.
Abstract
Vascular cell adhesion molecule–1 (VCAM-1) plays important roles in development and inflammation. Tumor necrosis factor–α (TNF-α) and focal adhesion kinase (FAK) are key regulators of inflammatory and integrin–matrix signaling, respectively. Integrin costimulatory signals modulate inflammatory gene expression, but the important control points between these pathways remain unresolved. We report that pharmacological FAK inhibition prevented TNF-α–induced VCAM-1 expression within heart vessel–associated endothelial cells in vivo, and genetic or pharmacological FAK inhibition blocked VCAM-1 expression during development. FAK signaling facilitated TNF-α–induced, mitogen-activated protein kinase activation, and, surprisingly, FAK inhibition resulted in the loss of the GATA4 transcription factor required for TNF-α–induced VCAM-1 production. FAK inhibition also triggered FAK nuclear localization. In the nucleus, the FAK-FERM (band 4.1, ezrin, radixin, moesin homology) domain bound directly to GATA4 and enhanced its CHIP (C terminus of Hsp70-interacting protein) E3 ligase–dependent polyubiquitination and degradation. These studies reveal new developmental and anti-inflammatory roles for kinase-inhibited FAK in limiting VCAM-1 production via nuclear localization and promotion of GATA4 turnover.
Introduction
Vascular cell adhesion molecule–1 (VCAM-1) is a transmembrane protein of the immunoglobulin superfamily expressed on blood vessels after endothelial cell (EC) stimulation by inflammatory cytokines such as tumor necrosis factor–α (TNF-α; Osborn et al., 1989). VCAM-1 mediates leukocyte binding to the vascular endothelium via α4β1 or α4β7 integrins, where signaling events triggered by VCAM-1 and integrin binding contribute to the development of atherosclerosis and rheumatoid arthritis (Carter and Wicks, 2001; Libby, 2002). VCAM-1 is also expressed on the mouse embryo allantois and binds to α4β1 integrin on the chorion to facilitate chorioallantoic fusion and placental development (Gurtner et al., 1995; Kwee et al., 1995; Inman and Downs, 2007). TNF-α triggers increased VCAM-1 production via several intracellular signaling cascades, including MAPK and nuclear factor–κ light chain enhancer of activated B cells (NF-κB) pathways (Pober, 2002; Karin and Gallagher, 2009). These signaling cascades affect multiple transcription factors, including activating protein complex–1 (AP-1), NF-κB, and GATA-binding proteins that bind to the VCAM-1 promoter (Ahmad et al., 1998; Molkentin, 2000; Minami and Aird, 2001). It is a combination of transcription factor phosphorylation and stability that contributes to TNF-α–induced VCAM-1 promoter activation.
TNF-α–induced signaling is highly context dependent, triggering either cell death (cytotoxic activity) or cell activation (proinflammatory activity; MacEwan, 2002). Costimulatory signals from the ECM play an active role in preventing TNF-α cytotoxic activity and also a permissive function in facilitating TNF-α proinflammatory EC activation (Bieler et al., 2007). Transmembrane integrin receptors bind to ECM proteins and facilitate the activation of nonreceptor protein tyrosine kinases (PTKs), MAPKs, and NF-κB pathways (Schwartz, 2001). Integrin-mediated signaling supports TNF-α–induced cell survival and gene expression (Fornaro et al., 2003). This requires NF-κB activation (Beg and Baltimore, 1996) and PTKs such as Syk, and Src family PTKs have been linked to TNF-α–induced NF-κB activation (Huang et al., 2003; Takada and Aggarwal, 2004). However, integrin and FAK PTK activation are more strongly associated with TNF-α–induced MAPK activation (Short et al., 1998; Schlaepfer et al., 2007; Young et al., 2010). Although PTK activity is needed for TNF-α–induced VCAM-1 expression (Weber et al., 1995; May et al., 1996), the PTKs facilitating these signaling events remain unknown.
FAK is comprised of an N-terminal FERM (band 4.1, ezrin, radixin, moesin homology) domain, central PTK region, and a C-terminal (CT) domain that links it to integrins (Mitra et al., 2005; Schaller, 2010). FAK is activated by integrin, growth factor, and G protein–linked and cytokine stimuli that increase FAK tyrosine phosphorylation at tyrosine (Y) 397, Y576, Y861, and Y925 and link FAK to MAPK activation (Schlaepfer and Mitra, 2004). Global or EC-specific FAK knockout results in embryonic lethality associated with vascular defects (Ilic et al., 2003; Shen et al., 2005; Braren et al., 2006). A p53 tumor suppressor–dependent block in cell proliferation is associated with the FAK knockout phenotype (Lim et al., 2008). However, the presence of the FAK-related Pyk2 PTK in cells without FAK reduces effects of FAK loss on p53 and angiogenesis (Weis et al., 2008; Lim et al., 2010b). Although recent studies using conditional FAK knockout in ECs prevent tumor-induced vascular permeability (Lee et al., 2010) and angiogenesis (Tavora et al., 2010), the in vivo signaling connections for FAK in mediating these effects remains unclear.
Knockin mouse models have revealed that FAK activity is critical for developmental vasculogenesis (Lim et al., 2010a; Zhao et al., 2010). Additionally, pharmacological FAK inhibition suppresses tumor- and growth factor–stimulated angiogenesis (Roberts et al., 2008; Weis et al., 2008). FAK kinase-dead (KD) knockin mutation is embryonic lethal, and FAK-KD fibroblasts exhibit defects in cell polarity and motility but not cell proliferation (Lim et al., 2010a). FAK-KD protects primary ECs from increased apoptosis through a mechanism that involves FAK-FERM–mediated nuclear localization, binding to p53, and enhancement of murine double minute 2 (Mdm2) E3 ligase–dependent p53 ubiquitination and proteasome degradation (Lim et al., 2010b). Thus, nuclear FAK can regulate cell proliferation and survival by facilitating p53 turnover in a kinase-independent manner. As nuclear-localized FAK contributes to chromatin remodeling and gene expression (Luo et al., 2009), support for a functional role for FAK in the nucleus is growing.
Herein, we show that FAK inhibition prevents TNF-α–induced and developmental VCAM-1 expression. Although FAK activity facilitates TNF-α–induced MAPK activation, genetic or pharmacological FAK inhibition also resulted in the loss of the GATA4 transcription factor required for TNF-α–induced VCAM-1 production. We find that inhibited FAK accumulates in the nucleus, and FAK-FERM binds to the GATA4 transcription factor and enhances GATA4 ubiquitination and is degradation dependent on C terminus of Hsp70-interacting protein (CHIP) E3 ligase expression. As FAK-FERM also forms a complex with CHIP, our experiments reveal a novel regulation of GATA4 by a kinase-independent scaffolding role of nuclear FAK as part of anti-inflammatory effects of FAK inhibition.
Results
Control of TNF-α–stimulated VCAM-1 expression by FAK
ECs that line blood vessels rapidly respond to environmental changes (Weis and Cheresh, 2011). Vessel-associated changes in VCAM-1 expression occur rapidly in response to inflammatory stimuli, but the role of FAK signaling in mediating these events remains unknown. As FAK activity facilitates TNF-α–stimulated interleukin-6 (IL-6) gene expression in cultured cells (Funakoshi-Tago et al., 2003; Schlaepfer et al., 2007), in vivo signaling experiments were performed in mice to determine the effect of FAK inhibition of TNF-α–induced VCAM-1 expression. Increased VCAM-1 levels were detected within 6 h in mouse heart or lung lysates after tail vein injection of TNF-α (Figs. 1 A and S1). Pretreatment of mice with a pharmacological FAK inhibitor (FAK-I) significantly inhibited VCAM-1 expression and FAK Y397 phosphorylation but not FAK expression in heart and lung lysates (Figs. 1 B and S1). In heart lysates, oral FAK-I administration also prevented FAK pY576 phosphorylation within the kinase domain activation loop (Fig. 1 A). Staining of heart sections revealed that pY576 FAK phosphorylation is maximal in association with ECs and that EC-associated FAK pY576 is significantly inhibited by FAK-I treatment within control and TNF-α–stimulated mice (Figs. 1 [C and D] and S2 A). Analyses of heart sections revealed significantly increased EC-associated VCAM-1 expression upon TNF-α stimulation and decreased VCAM-1 staining after FAK-I administration (Figs. 1 [E and F] and S2 B). Together, these data indicate that TNF-α–mediated VCAM-1 expression within blood vessel–associated ECs is dependent on FAK activity.
Figure 1.
TNF-α–induced VCAM-1 expression within heart-associated ECs in vivo is dependent on FAK activity. PBS or TNF-α (0.02 mg/kg) was tail vein injected into mice, and, after 6 h, tissues were analyzed by immunoblotting or staining. Where indicated, FAK-I (100 mg/kg, PND-1186) was administered 3 h before starting experiments. (A) Immunoblotting of heart lysates shows increased VCAM-1 expression and FAK Y397 or FAK Y576 phosphorylation upon TNF-α stimulation in vivo. FAK-I addition prevents VCAM-1 production and FAK tyrosine phosphorylation but no change in FAK expression. Internal loading controls for each gel are shown by anti-actin immunoblotting. (B) Heart-associated VCAM-1 or FAK activation (pY397) was determined by immunoblotting (see Fig. S1) and expressed as a ratio to actin or total FAK, as determined by densitometry, respectively. Values are means (±SD) from six mice, representing two independent experiments (***, P < 0.001). (C) In vivo signaling assays were performed as in A, and heart sections were analyzed by combined staining for activated FAK (pY576) and ECs (CD31). Bar, 20 µm. (D) Mean correlation of pixel intensities from anti–pY576 FAK and anti-CD31 staining of heart sections, as shown in C. (E) Visualization of EC-associated VCAM-1 expression. Heart sections were analyzed by combined staining for VCAM-1 and ECs (CD31). A merged image is shown. Bar, 20 µm. (F) Mean correlation of pixel intensities from anti–VCAM-1 and anti-CD31 staining of heart sections, as shown in C. (D and F) 10 full-frame images were analyzed per experimental group for calculations of VCAM-1 and pY576 FAK associated with CD31 staining (±SEM; ***, P < 0.001; ****, P < 0.0001).
FAK activity controls VCAM-1 expression during mouse development
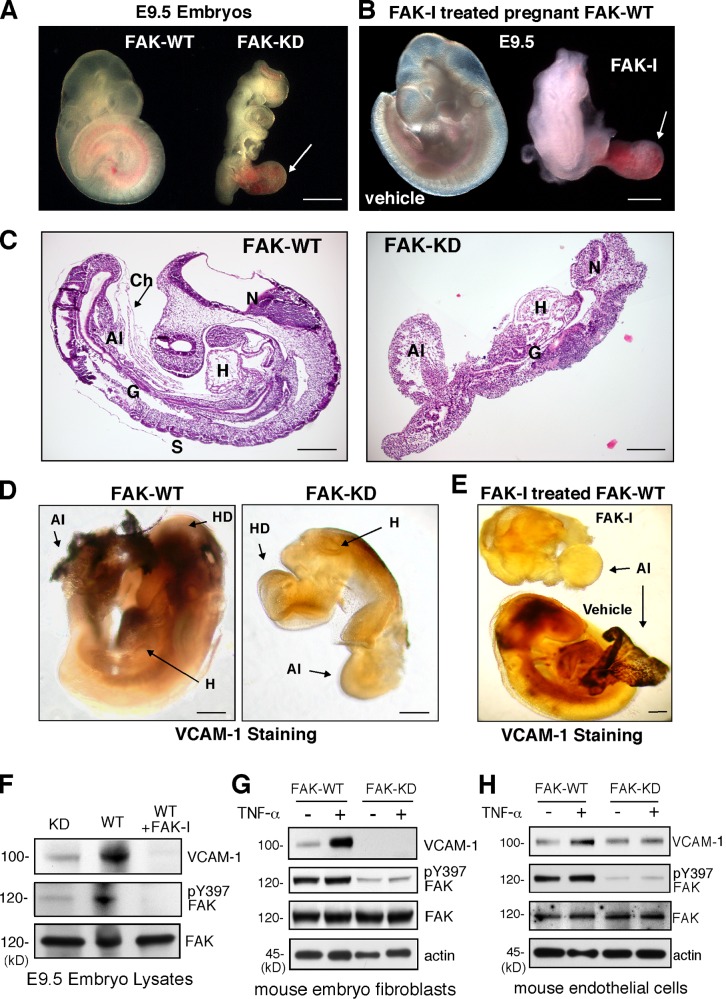
VCAM-1 expression on the developing mouse embryo allantois promotes binding to α4β1 integrin on chorion membrane, and this union facilitates chorioallantoic fusion (Inman and Downs, 2007). FAK-KD knockin mutation results in lethality at embryonic day 9.5 (E9.5; Lim et al., 2010a), with embryos exhibiting an enlarged and unfused allantois (Fig. 2 A). FAK-I administration to pregnant mice also results in E9.5 embryo lethality with defective allantois structures (Fig. 2 B). FAK-KD embryos fail to form somites, exhibit gross defects in head and heart structures, and form a rudimentary gut cavity (Fig. 2 C). Comparisons of mRNAs within FAK–wild type (WT) and FAK-KD embryos by array analyses revealed several targets that differed in expression greater than twofold. In particular, VCAM-1 mRNA levels were reduced 7.4 fold in FAK-KD embryos (Table 1). Gene ontology analyses of the mRNA array data revealed functional annotated differences that connect FAK activity to a leukocyte extravasation signaling group (Fig. S3). Other connections to endocytosis, tight junction, integrin, and Rho signaling groups were also identified. Verification of VCAM-1 protein expression differences between FAK-WT and FAK-KD embryos was obtained by immunostaining of hearts and allantois (Fig. 2 D) and immunoblotting of embryos lysates (Fig. 2 F). FAK-I treatment of pregnant mice also prevented embryo-associated VCAM-1 expression (Fig. 2 E) and FAK pY397 phosphorylation (Fig. 2 F). Notably, primary mouse embryonic fibroblasts (MEFs) or ECs isolated from FAK-WT or FAK-KD embryos showed that genetic inhibition of FAK activity prevented TNF-α–induced VCAM-1 expression (Fig. 2, G and H). Although developmental signals regulating VCAM-1 expression are complex and involve multiple transcription factors (Ferdous et al., 2011), our experiments support the importance of FAK activity in controlling both developmental and TNF-α–induced VCAM-1 expression in mice.
Figure 2.
FAK activity controls developmental VCAM-1 expression. (A) FAK-KD embryos at E9.5 exhibit an enlarged and unfused allantois (arrow) compared with normal FAK-WT embryos where chorioallantoic fusion has occurred. (B) Treatment of pregnant female mice with FAK-I (100 mg/kg, PND-1186 from E7.5 to E9.5) but not vehicle (water) results in lethality of embryos (E9.5), exhibiting defective allantois formation (arrow). (C) Sagittal sections of E9.5 WT and FAK-KD embryos. FAK-KD embryos do not form somites (S) and display gross defects in head, heart (H), and allantois (Al). The arrow indicates the chorion (Ch) membrane. G, gut; N, neural cell. (D) Whole-mount anti–VCAM-1 staining of embryos at E9.5 reveals no VCAM-1 expression in FAK-KD compared with FAK-WT embryos. HD, head. (E) Lack of VCAM-1 staining in FAK-I (100 mg/kg, PND-1186 from E7.5 to E9.5) compared with vehicle-treated embryos at E9.5. (A–E) Bars, 100 µm. (F–H) VCAM-1 protein expression and FAK activation are inhibited within FAK-KD and FAK-I–treated embryos. Immunoblotting of E9.5 embryo lysates reveals VCAM-1 levels (left) and FAK phosphorylation (pY397; right) with actin and total FAK as loading controls. Results from MEFs (G) or ECs (H) show that TNF-α (10 ng/ml, 16 h) induces VCAM-1 expression in FAK-WT but not FAK-KD cells, as determined by immunoblotting with pY397 FAK, total FAK, and actin levels, shown as controls.
Table 1.
Differences in target mRNAs from FAK-WT and FAK-KD embryos during development
| Protein name | Fold reduction (WT/KD) |
| Tropomodulin-1 | 13.1 |
| VCAM-1 | 7.4 |
| Caveolin-1 | 6.4 |
| Robo-4 | 4.9 |
| Rgs5 | 4.7 |
| CD31 | 3.6 |
| WAS | 4.3 |
| ESAM-1 | 2.7 |
| ICAM-2 | 2.6 |
| α-SMA | 2.3 |
| SEMA6B | 2.1 |
Illumina MouseWG-6 v2.0 Expression BeadChip analyses (45,200 targets) were performed. Selected mRNAs with greater than twofold change are shown. WAS, Wiskott–Aldrich syndrome; ESAM-1, endothelial selective adhesion molecule–1; ICAM-2, intercellular adhesion molecule–2; α-SMA, α–smooth muscle actin.
FAK inhibition prevents cytokine-stimulated VCAM-1 expression in human ECs
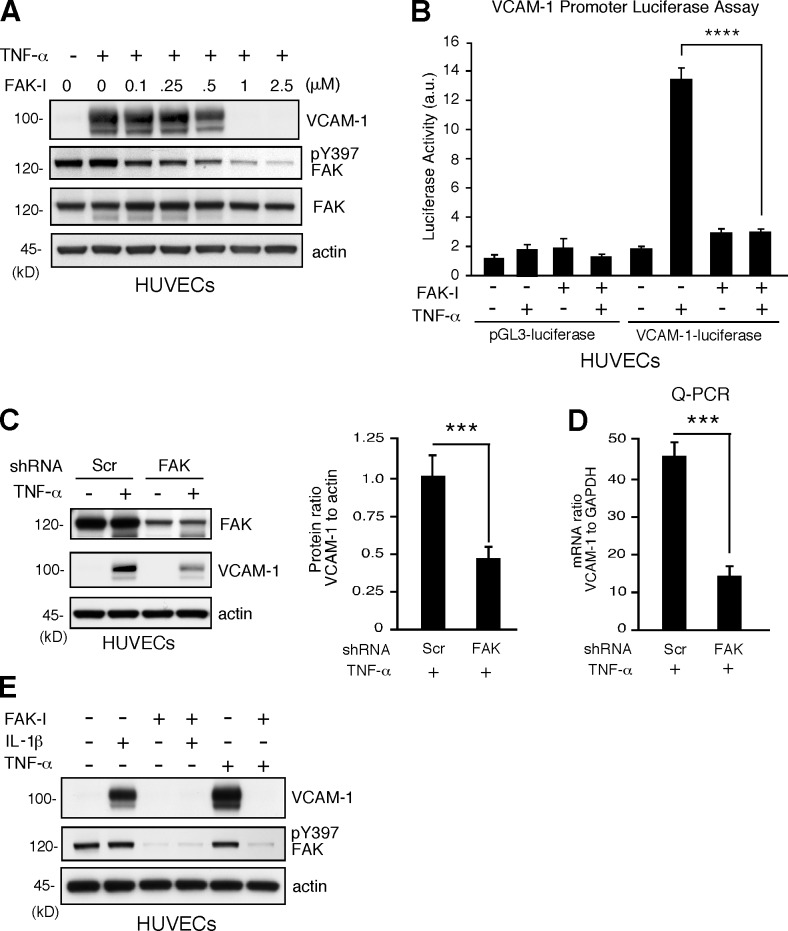
Regulation of mouse and human VCAM-1 expression share both similarities and mechanistic differences (Minami and Aird, 2001). To determine whether FAK is also important in human TNF-α–induced human VCAM-1 expression, signaling experiments were performed with human umbilical vein ECs (HUVECs). FAK-I treatment prevented TNF-α–mediated VCAM-1 expression in a dose-dependent manner with an IC50 of ∼0.5 µM that paralleled inhibition of FAK Y397 phosphorylation (Fig. 3 A). This effect was a result in part of transcriptional effects, as FAK-I significantly inhibited TNF-α–induced VCAM-1 promoter luciferase activity (Fig. 3 B). Knockdown of FAK expression also significantly inhibited TNF-α–induced VCAM-1 protein and mRNA expression (Fig. 3, C and D). As FAK-I treatment of HUVECs also prevented IL-1β–stimulated VCAM-1 expression (Fig. 3 E), our experiments show that FAK functions as a common signaling component regulating inflammatory cytokine-stimulated VCAM-1 expression.
Figure 3.
FAK activity is essential for proinflammatory cytokine-mediated VCAM-1 expression. (A) TNF-α–induced (10 ng/ml, 16 h) VCAM-1 is prevented by FAK-I (PF271) treatment of HUVECs in a dose-dependent (0.1–2.5 µM FAK-I) manner. VCAM-1, pY397 FAK, total FAK, and actin levels were determined by immunoblotting. (B) TNF-α–induced VCAM-1 promoter activity is blocked by FAK inhibition. HUVECs were transfected with pGL3 promoterless luciferase (control) or pGL3 fused with the human VCAM-1 promoter, stimulated with TNF-α, and treated with FAK-I (1 µM PF271), as indicated, and relative luciferase activity (arbitrary units [a.u.]) is shown. Values are means (±SD) from three independent experiments. (C) FAK knockdown prevents VCAM-1 expression. HUVECs were transduced with lentiviral Scr or anti-FAK short hairpin RNA (shRNA) and stimulated with TNF-α. FAK and VCAM-1 and actin expression levels were determined by immunoblotting, and densitometry values of VCAM-1 relative to actin are means (±SD; n = 3; ***, P < 0.001). (D) VCAM-1 mRNA levels to GAPDH were determined by Q-PCR (±SD; n = 3; ***, P < 0.001) in experiments, as described in C. (E) FAK activity is required for IL-1β–induced VCAM-1 expression. HUVECs were stimulated with 20 ng/ml IL-1β and 10 ng/ml TNF-α, and FAK-I (1 µM PF271) was added as indicated. After 16 h, VCAM-1, pY397 FAK, and actin levels were evaluated by immunoblotting.
FAK facilitates TNF-α–induced MAPK but not NF-κB activation
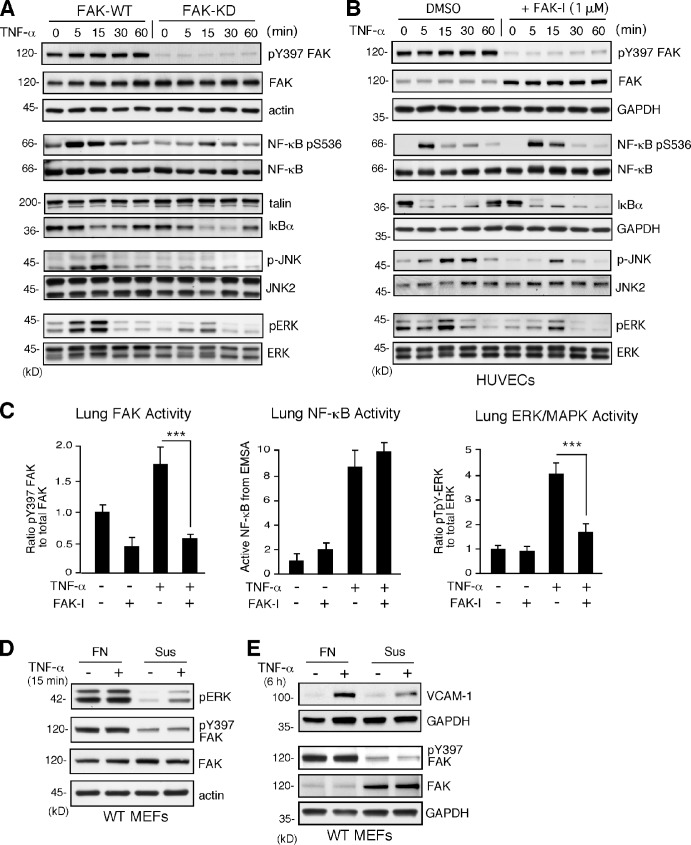
TNF-α activates extracellular-regulated kinase (ERK) MAPKs, JNK MAPKs, and p38 MAPKs as well as NF-κB to regulate gene expression (Karin and Gallagher, 2009). To determine the signaling consequences of FAK inhibition, comparisons of FAK-WT and FAK-KD MEFs or DMSO (control) and FAK-I–treated HUVECs were performed (Fig. 4). TNF-α rapidly and equally promoted NF-κB pS536 phosphorylation in both control and FAK-I–treated cells (Fig. 4, A and B). Regulatory IkBα was rapidly degraded after TNF-α addition (Fig. 4, A and B), and electrophoretic mobility shift assays (EMSAs) revealed equivalent TNF-α–stimulated NF-κB DNA binding in FAK-KD MEFs and FAK-I–treated HUVECs compared with FAK-WT and DMSO-treated controls (Fig. S4). In vivo, FAK-I significantly inhibited TNF-α–induced FAK pY397 phosphorylation but did not alter TNF-α–stimulated NF-κB DNA binding in mouse lung lysates (Figs. 4 C and S5). Despite the importance of NF-κB in promoting TNF-α–induced VCAM-1 gene transcription (Karin and Gallagher, 2009), FAK inhibition did not detectably alter NF-κB activation.
Figure 4.
TNF-α–induced MAPK but not NF-κB activation is dependent on FAK activity in vitro and in vivo. (A and B) FAK-WT or FAK-KD MEFs (A) and HUVECs pretreated with DMSO or FAK-I (1 µM PF271; B) were stimulated with 10 ng/ml TNF-α for the indicated times, and lysates were prepared for immunoblotting. Blots for activated FAK (pY397), total FAK, activated NF-κB (pS536), activated JNK (p-JNK and pT183/pY185), activated ERK (pERK and pT202/pY204), IκBα, and actin are shown. Internal loading controls for each gel are shown by anti-actin, anti-GAPDH, anti-talin, or reprobing membranes with antibodies to total NF-κB, JNK2, or total ERK1/2 immunoblotting. (C) PBS or TNF-α (0.02 mg/kg) was tail vein injected into mice, and, after 5 min (FAK and ERK activation) or 3 h (NF-κB activation), lung tissue was analyzed by immunoblotting or EMSA. Where indicated, FAK-I (100 mg/kg, PND-1186) was administered 3 h before starting experiments. Values, measured by densitometry, are means (±SD) from six mice, representing two independent experiments. ***, P < 0.001. (D and E) MEFs were replated onto FN dishes or held in suspension (Sus) for 1 h before TNF-α (10 ng/ml) addition for 15 min (D) or 6 h (E) before protein cell lysis. Blots for activated ERK (pERK and pT202/pY204), activated FAK (pY397), total FAK, and actin are shown. Anti-GAPDH blotting is shown as loading controls.
Instead, comparisons of WT and FAK-KD MEFs or FAK-I–treated HUVECs revealed that FAK-I reduced TNF-α–stimulated JNK/MAPK and ERK/MAPK activation at 5 and 15 min (Fig. 4, A and B). FAK-I treatment results in the elevation of total FAK expression but not pY397 FAK phosphorylation in HUVECs (Fig. 4 B). In lung lysates, pretreatment of mice with FAK-I significantly prevented TNF-α–induced ERK/MAPK activation in vivo (Figs. 4 C and S5), and a previous study showed that pharmacological ERK inhibition prevented TNF-α–induced VCAM-1 expression in ECs (Fitau et al., 2006). Our results support the conclusion that FAK activity facilitates TNF-α signaling to MAPKs needed for VCAM-1 expression. This role for FAK may be related to cross-talk between integrin and cytokine signaling pathways, as TNF-α signaling to ERK/MAPK (Fig. 4 D), induction of VCAM-1 expression (Fig. 4 E), and FAK Y397 phosphorylation (Fig. 4, D and E) are prevented when MEFs are deprived of matrix-mediated cell adhesion. Serum addition limited suspension-induced cell death, as determined by annexin V staining (unpublished data). As FAK−/− MEF reconstitution experiments have verified the importance of FAK activity and Y397 FAK phosphorylation in facilitating TNF-α–stimulated ERK/MAPK activation and IL-6 gene expression (Schlaepfer et al., 2007), our experiments evaluating ERK/MAPK and VCAM-1 extend this TNF-α and FAK signaling connection to HUVECs and to mice.
FAK inhibition alters GATA4 transcription factor levels
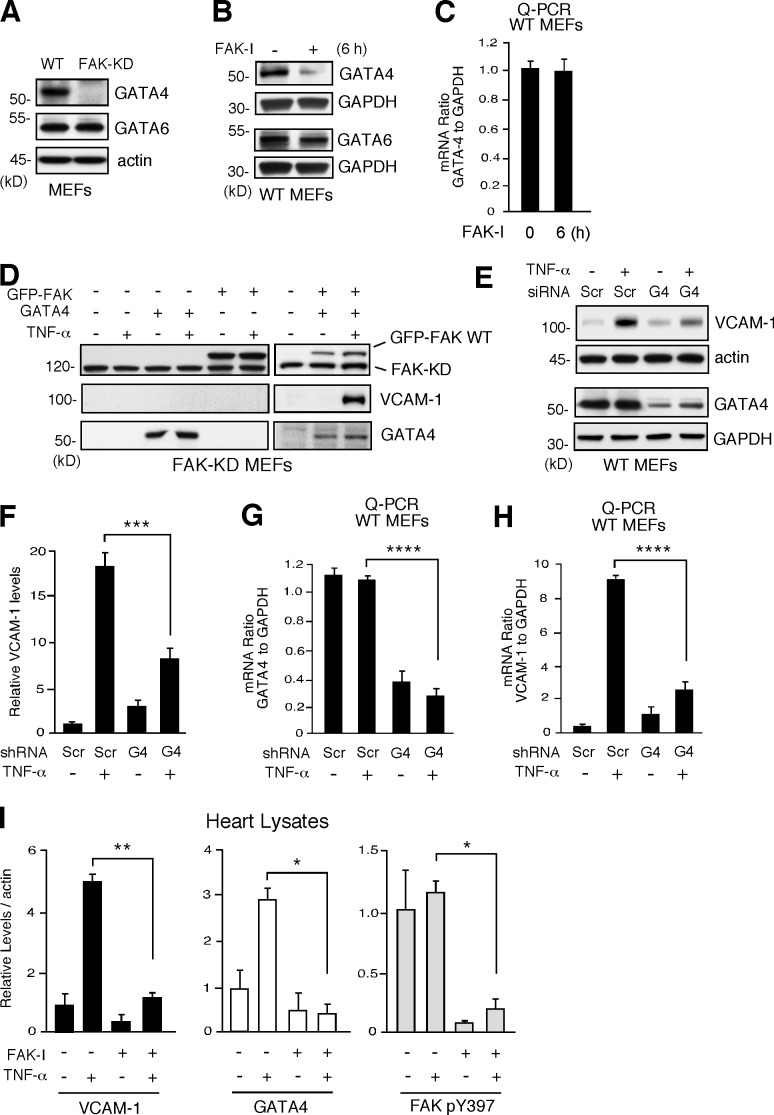
To establish the signaling linkage between FAK, MAPK, and the regulation of VCAM-1 transcription, rescue assays were performed by transient transfection of FAK-WT or activators of MAPKs (constitutively active MEK1, MKK4, or MEKK1) in FAK-KD MEFs or HUVECs treated with FAK-I. Unexpectedly, neither FAK-WT nor active MAPK alone or in combination was able to increase VCAM-1 expression upon TNF-α stimulation (unpublished data). As VCAM-1 production is transcriptionally regulated, we investigated whether FAK inhibition may alter the profile of VCAM-1–associated transcription factors in cells (Ahmad et al., 1998; Molkentin, 2000; Minami and Aird, 2001). Although genetic or pharmacological FAK inhibition did not prevent NF-κB activation upon TNF-α stimulation (Fig. 4), immunoblotting revealed the absence of GATA4 expression in FAK-KD MEFs, whereas GATA6 was equally expressed in FAK-WT and FAK-KD MEFs (Fig. 5 A). Further, FAK-I treatment of cells also resulted in the selective loss of GATA4 but not GATA6 expression by >50% within 3 h (not depicted) and resulted in an ∼90% loss of GATA4 within 6 h without effects on GATA6 (Fig. 5 B). Interestingly, these FAK-I–triggered changes in GATA4 expression occurred without alterations in GATA4 mRNA levels (Fig. 5 C). Thus, inhibition of FAK selectively affects GATA4 protein but not mRNA levels in cells.
Figure 5.
FAK inhibition decreases GATA4 levels needed for TNF-α–induced VCAM-1 expression. (A) Steady-state GATA4 and GATA6 levels in FAK-WT and FAK-KD MEFs, as determined by immunoblotting with actin as a control. (B) FAK-WT MEFs treated with DMSO or FAK-I (1 µM PF271, 6 h) and lysates blotted for GATA4 or GATA6. Anti-GAPDH blotting is shown as loading controls. (C) GATA4 mRNA levels to GAPDH were determined by Q-PCR (±SD; n = 3) in experiments, as described in B. (D) Rescue of TNF-α–induced VCAM-1 expression in FAK-KD MEFs by combined FAK-WT and GATA4 expression. Combinations of GFP-FAK and GATA4 were transfected into FAK-KD MEFs. After 24 h, cells were stimulated with 10 ng/ml TNF-α, as indicated, and FAK, VCAM-1, and GATA4 immunoblotting was performed at 40 h. (E) FAK-WT MEFs were transfected with Scr or GATA4 (G4) siRNA and, after 48 h, stimulated with TNF-α (10 ng/ml, 6 h), and immunoblotting was performed for VCAM-1 and GATA4. Anti-actin and anti-GAPDH blotting are shown as loading controls. (F) Densitometry analyses of VCAM-1 protein levels relative to actin, as described in E. (±SD; n = 2; ***, P < 0.001). (G and H) MEFs were transfected with Scr or GATA4 siRNA and stimulated with TNF-α, as described in E. GATA4 (G) or VCAM-1 (H) mRNA levels to GAPDH were determined by Q-PCR (±SD; n = 3; ****, P < 0.0001). (I) PBS or TNF-α (0.02 mg/kg) was tail vein injected into mice, and, after 6 h, heart lysates were analyzed by immunoblotting for VCAM-1, GATA4, pY397 FAK, total FAK, and actin. Where indicated, FAK-I (100 mg/kg, PND-1186) was administered 3 h before starting experiments. Data are mean densitometry values relative to actin (±SD; n = 2; *, P < 0.05; **, P < 0.001).
FAK and GATA4 rescue TNF-α–induced VCAM-1 expression in FAK-KD MEFs
To determine the significance of GATA4 loss upon FAK inhibition, FAK-WT and GATA4 alone or in combination were transfected into FAK-KD MEFs, and the effect on VCAM-1 production was evaluated in the presence or absence of TNF-α stimulation (Fig. 5 D). Notably, only the combination of FAK-WT and GATA4 with TNF-α stimulation was able to promote VCAM-1 expression. Additionally, siRNA knockdown of GATA4 in FAK-WT MEFs significantly reduced TNF-α–induced VCAM-1 protein and mRNA levels (Fig. 5, E and F). Quantitative PCR (Q-PCR) was used to confirm that GATA4 and VCAM-1 mRNA were significantly suppressed after TNF-α stimulation by siRNA to GATA4 compared with control siRNA transfection (Fig. 5, G and H). In vivo, GATA4 and VCAM-1 protein levels were increased in heart lysates after TNF-α stimulation, and this was significantly inhibited by FAK-I administration to mice (Fig. 5 I). Together, these results support the importance of GATA4 in TNF-α–induced VCAM-1 expression and suggest that loss of GATA4 associated with FAK inhibition prevents VCAM-1 production.
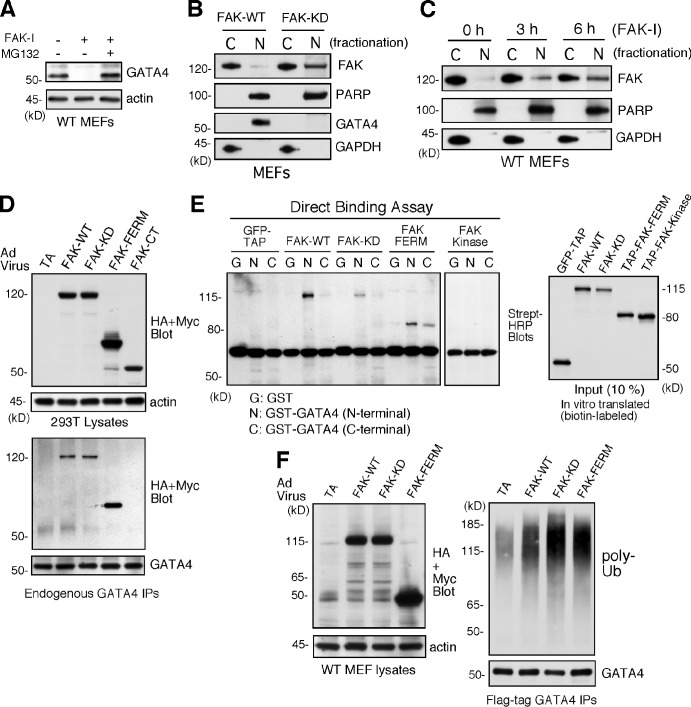
Regulation of GATA4 protein levels can occur through various posttranslational mechanisms, including phosphorylation or ubiquitination and proteasome degradation (Brewer and Pizzey, 2006). The loss of GATA4 expression by FAK-I treatment was reversed by addition of a cell-permeable proteasome inhibitor (MG132; Fig. 6 A), supporting the notion that FAK inhibition may promote enhanced GATA4 degradation. Notably, FAK can also promote p53 tumor suppressor degradation (Lim et al., 2008). Cell stress triggers FAK nuclear localization, a nuclear FAK–p53 complex, and FAK-enhanced Mdm2 E3 ligase–dependent ubiquitination and degradation of p53 (Lim et al., 2008). In FAK-KD MEFs, FAK is localized to sites of adhesion as well as to the nucleus (Lim et al., 2010a). Biochemical separation of FAK-WT and FAK-KD MEFs into cytosolic and nuclear fractions revealed elevated levels of FAK-KD in the nucleus (Fig. 6 B). Importantly, FAK-I treatment of cells could trigger enhanced FAK nuclear accumulation within 3–6 h (Fig. 6 C). Interestingly, the timing of FAK nuclear localization upon FAK-I addition parallels the time course of GATA4 loss (Figs. 5 B and 6 C). Together, these results show that FAK inhibition promotes increased FAK nuclear accumulation–correlated loss of GATA4 protein.
Figure 6.
Inhibited FAK is nuclear localized, and FAK-FERM binds GATA4 to promote GATA4 ubiquitination in cells. (A) FAK-WT MEFs treated with DMSO, FAK-I (1 µM PF271), or FAK-I with MG132 (40 µM) for 12 h and lysates blotted for GATA4 and actin. (B) FAK-WT and FAK-KD MEF lysates were separated into cytosolic (C) or nuclear (N) fractions and immunoblotted for FAK, PARP, GATA4, and GAPDH. PARP and GAPDH are nuclear and cytosolic markers, respectively. (C) FAK inhibition promotes FAK nuclear localization within 3 h. FAK-WT MEFs were treated with FAK-I (1 µM PF271) for the indicated times, and fractionated lysates were immunoblotted for FAK, PARP, and GAPDH. (D) 293T cells were transduced with Ad-TA, the indicated HA- or Myc-tagged FAK-WT, FAK-KD, FAK-FERM, and FAK-CT constructs, and association with endogenous GATA4 was determined by coimmunoprecipitation (IP). Immunoblotting shows expression of FAK constructs or actin (top) and FAK-FERM association with GATA4 (bottom). (E) GATA4 directly binds FAK. In vitro translated GFP–tandem affinity probe (TAP), FAK-WT, FAK-KD, TAP-FAK-FERM, and TAP-FAK kinase domain (386–686) were used in a direct binding assay with GST or GST fusions of GATA4 N terminus or GATA4 C terminus. Streptavidin (Strept)-HRP analyses show the amount of FAK bound (left) or 10% of input (right). (F) FAK-FERM enhances GATA4 ubiquitination. FAK-WT MEFs were transduced with Ad-TA, the indicated FAK constructs, and flag-tagged GATA4 and treated with MG132 (40 µM, 3 h). Whole-cell lysates were analyzed for expression of FAK or actin (left), and flag tag immunoprecipitates (antibody coupled to beads) were evaluated by anti-ubiquitin (Ub) and GATA4 immunoblotting.
FAK-FERM binds GATA4 and enhances GATA4 ubiquitination
Localization of FAK to integrin-enriched sites of adhesion occurs via the FAK C-terminal domain, whereas the FAK-FERM domain can target FAK to the nucleus (Lim et al., 2008). Adenoviral (Ad)-mediated overexpression experiments in human 293T cells revealed that FAK-WT, FAK-KD, and FAK-FERM but not FAK-CT associated with endogenous GATA4 by coimmunoprecipitation (Fig. 6 D). In vitro translation of various FAK constructs combined with binding assays using GST fusion proteins of GATA4 N-terminal and CT domains revealed direct binding between FAK-WT, FAK-KD, and FAK-FERM with the N-terminal domain of GATA4 (Fig. 6 E). No binding was detected between the FAK kinase domain and GATA4.
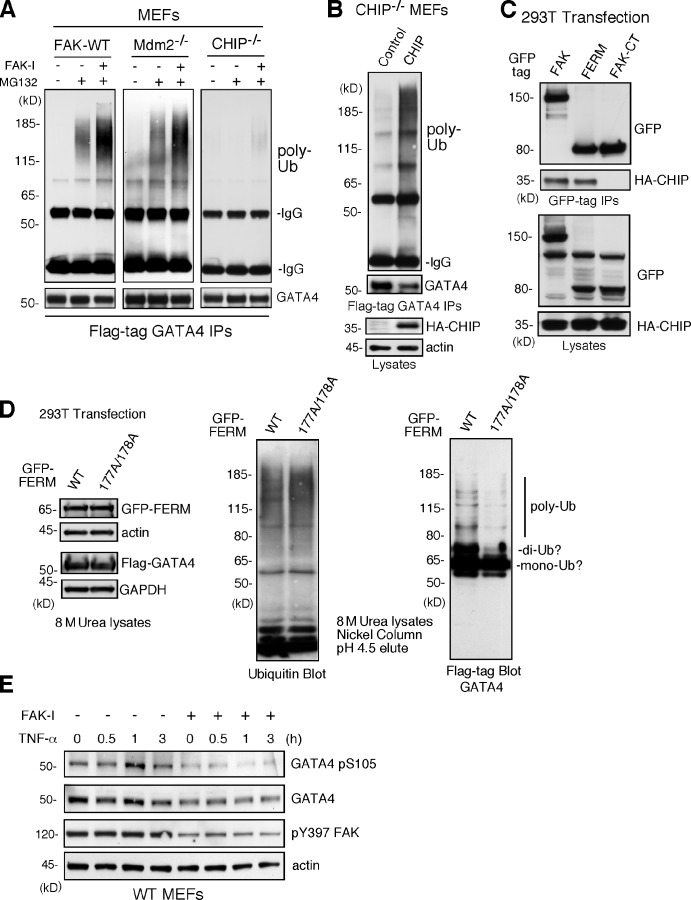
Notably, overexpression of FAK-WT, FAK-KD, or FAK-FERM increased GATA4 polyubiquitination (Fig. 6 F). These experiments were performed in the presence of MG132 proteasome inhibitor to stabilize and detect polyubiquitinylated GATA4 by immunoblotting. Interestingly, treatment of MEFs with FAK-I enhanced GATA4 ubiquitination, and this occurred equally in FAK-WT and Mdm2−/− MEFs (Fig. 7 A). As GATA4 ubiquitination is mediated in part by the cochaperone/ubiquitin E3 ligase CHIP in response to cellular stress (Dai et al., 2003; Kobayashi et al., 2007), CHIP−/− MEFs were treated with a combination of MG132 and FAK-I. No GATA ubiquitination was detected in CHIP−/− MEFs (Fig. 7 A), and this was rescued by CHIP reexpression in combination with FAK inhibition (Fig. 7 B). Transient transfection experiments revealed that full-length FAK and FAK-FERM, but not FAK-CT, formed a complex with CHIP (Fig. 7 C). As FAK-FERM forms a scaffold for p53 and Mdm2 binding to facilitate p53 ubiquitination and degradation (Lim et al., 2008), our results support a similar model whereby FAK-FERM may also form a complex with GATA4 and CHIP to facilitate GATA4 ubiquitination.
Figure 7.
FAK-enhanced GATA4 polyubiquitination is dependent on CHIP and FAK-FERM nuclear localization. (A) FAK-WT, Mdm2−/−p53−/−, and CHIP−/− MEFs were transfected with flag-tagged GATA4 and treated with MG132 (40 µM, 3 h) and FAK-I (1 µM PF271), as indicated. Flag tag immunoprecipitates (IPs) were evaluated by anti-ubiquitin (Ub) and GATA4 immunoblotting and show no ubiquitination of GATA4 in CHIP−/− MEFs. (B) CHIP−/− MEFs were transfected with HA-tagged CHIP and flag-tagged GATA4 and treated with MG132 (40 µM, 3 h) and FAK-I (1 µM PF271). Flag tag immunoprecipitates were evaluated by anti-ubiquitin and GATA4 immunoblotting and show rescue of GATA4 ubiquitination by CHIP reexpression. Lysates show HA-CHIP and actin expression. (C) 293T cells were cotransfected with GFP-tagged FAK, FAK-FERM, and FAK-CT with HA-CHIP. Coimmunoprecipitation analyses with antibodies to GFP reveal FAK-FERM and CHIP association by immunoblotting. Lysates show equal levels of GFP-FAK and HA-CHIP expression. (D) 293T cells were transfected with GFP-tagged FAK-FERM WT or FAK-FERM R177A/R178A, flag-tagged GATA4, and His-tagged ubiquitin and denatured lysates (8 M urea) purified by nickel agarose affinity binding. GFP-FERM and Flag-GATA4 (with actin and GAPDH as loading controls; left), total eluted ubiquitinated proteins (middle), and mono-, di-, and polyubiquitinated GATA4 (right), as determined by immunoblotting, are shown. (E) FAK-WT MEFs pretreated with DMSO or FAK-I (1 µM PF271, 30 min) were stimulated with 10 ng/ml TNF-α for the indicated times, and lysates were prepared for immunoblotting. Blots for activated GATA4 (pS105), total GATA4, activated FAK (pY397), and actin are shown.
FAK-FERM mutations that disrupt nuclear localization prevent efficient regulation of p53 (Lim et al., 2008). To determine whether FAK nuclear localization is important in GATA4 regulation, FAK-FERM WT or FAK-FERM R177A/R178A (point mutations that prevent nuclear localization) was evaluated for effects on GATA4 ubiquitination (Fig. 7 D). Analyses of protein lysates denatured in 8 M urea revealed equal capture and elution of ubiquitinylated proteins on a nickel affinity–binding column. Importantly, FAK-FERM WT but not FAK-FERM R177/178A overexpression increased di- and polyubiquitination of GATA4 (Fig. 7 D). These results show that FAK-FERM nuclear localization is required for efficient GATA4 polyubiquitination. Finally, as GATA4 activation is mediated in part through ERK/MAPK phosphorylation of GATA4 S105 (van Berlo et al., 2011), MEFs were treated with TNF-α in the presence or absence of FAK-I and evaluated for changes in GATA4 S105 phosphorylation (Fig. 7 E). Maximal TNF-α–induced GATA4 activation occurred at 1 h, and although FAK-I triggered loss of GATA4 protein levels, GATA4 pSer105 phosphorylation was also prevented by FAK-I addition. Collectively, these results show that FAK inhibition prevents signals leading to GATA4 phosphorylation and promotes the turnover of GATA4 protein as mechanisms limiting TNF-α–induced VCAM-1 expression.
Discussion
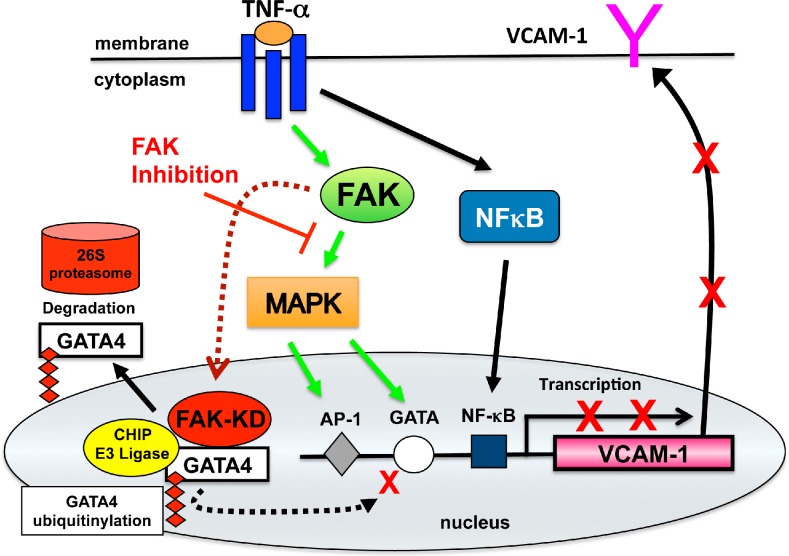
Induced VCAM-1 expression contributes to vascular inflammation (Libby, 2002). Here, we show through both genetic and pharmacological experimental results that FAK activity is essential in promoting TNF-α–induced VCAM-1 expression. These experiments define a new role for nuclear FAK in the regulation of inflammation-induced gene expression. Our results support a model (Fig. 8) whereby under normal signaling conditions, FAK functions in the cytoplasm to facilitate TNF-α–induced MAPK associated with GATA4 Ser105 phosphorylation and needed for induction of VCAM-1 expression. Upon genetic or pharmacological FAK inhibition, FAK accumulates in the nucleus and acts to enhance GATA4 polyubiquitination. Loss of GATA4 prevents TNF-α–stimulated VCAM-1 expression, and this block cannot be rescued by constitutive MAPK activation. The FAK-FERM domain promotes FAK nuclear localization and direct binding to GATA4 and acts as a scaffold to enhance CHIP E3 ligase–dependent GATA4 polyubiquitination, leading to GATA4 degradation. These experiments reveal a novel anti-inflammatory role for nuclear-localized and kinase-inhibited FAK in limiting VCAM-1 production through the modulation of GATA4 activation and turnover.
Figure 8.
Model of FAK function downstream of TNF-α in the regulation of VCAM-1 expression. TNF-α binding to cell surface receptors triggers intracellular signaling cascade activation of MAPKs and NF-κB. This leads to alterations in gene transcription of targets such as VCAM-1 that is regulated in part by combined effects of AP-1, GATA, and NF-κB transcription factors. Inhibition of FAK prevents TNF-α–induced MAPK activation and the inhibition of GATA4 Ser105 phosphorylation. Inhibited FAK (FAK-KD) accumulates in the nucleus, binds directly to GATA4, and promotes increased GATA4 ubiquitination and proteasomal degradation via interactions with the CHIP E3 ubiquitin ligase. Impairment in both FAK-mediated MAPK activation and GATA4 stability prevent cytokine-stimulated VCAM-1 transcription and reveal novel anti-inflammatory effects of FAK inhibition.
Our experiments also expand the role for FAK in mediating inflammatory signals. FAK activity promotes TNF-α–induced IL-6 expression (Funakoshi-Tago et al., 2003; Schlaepfer et al., 2007), and FAK-I treatment of mice prevents orthotopic breast tumor–associated leukocyte infiltration and tumor-associated splenomegaly (Walsh et al., 2010). As conditional deletion of FAK expression prevents macrophage or neutrophil cell migration (Owen et al., 2007), anti-inflammatory effects of FAK-I administration may result from the intrinsic inhibition of macrophage motility as well as stromal-associated mediators such as changes in VCAM-1 gene expression. As FAK-Is are being evaluated in human clinical trials as agents blocking tumor growth and angiogenesis (Schultze and Fiedler, 2010), our experiments evaluating VCAM-1 and a recent study of dominant-negative FAK inhibition preventing EC-associated E-selectin up-regulation (Hiratsuka et al., 2011) support a greater need to understand the role of FAK inhibition in altering inflammatory-associated vascular responses.
A previous study showed that FAK is catalytically activated by TNF-α stimulation of cells (Schlaepfer et al., 2007). Although other PTKs are associated with TNF-α signaling, past studies have linked PTK activity to TNF-α–induced NF-κB activation (Huang et al., 2003; Takada and Aggarwal, 2004). In contrast, FAK inhibition selectively reduced TNF-α activation of MAPKs but had no effect on NF-κB activation. Importantly, it is signaling from multiple pathways that combine to activate AP-1, NF-κB, and GATA transcription factors necessary for VCAM-1 transcriptional regulation (Karin and Gallagher, 2009). Notably, GATA4 is activated by MAPK phosphorylation at S105 (van Berlo et al., 2011), and β1 integrin signaling through MAPK contributes to GATA4 nuclear translocation and activation (Liu et al., 2009). As FAK is the major PTK activated by integrins and we find that matrix adhesion is required for efficient TNF-α–stimulated ERK/MAPK activation and VCAM-1 production, our results support the importance of FAK activity in mediating cross-talk between integrins and cytokine receptor signaling pathways.
Although FAK inhibition prevents TNF-α–induced GATA4 S105 phosphorylation, interpretations are complicated by a corresponding reduction in GATA4 protein levels. We find that genetic or pharmacological FAK inhibition triggers full-length FAK protein accumulation in the nucleus and the loss of GATA4 required for optimal TNF-α–induced VCAM-1 expression. FAK-FERM binds directly to GATA4 and enhances GATA4 polyubiquitination in cells. Expression of a FAK-FERM domain containing point mutations in the nuclear localization motif prevents efficient GATA4 di- and polyubiquitination. As FAK-I–induced loss of GATA4 in cells is blocked by proteasome inhibition, our results support the notion that nuclear-localized and kinase-inhibited FAK limits VCAM-1 production by promoting GATA4 degradation.
The role of the FAK-FERM domain in GATA4 regulation is similar to a study showing that FAK-FERM binds to and inhibits the p53 tumor suppressor protein (Golubovskaya et al., 2005). Cell stress triggers FAK nuclear localization, an FAK–p53 complex, and FAK-enhanced Mdm2 E3 ligase–dependent ubiquitination and degradation of p53 (Lim et al., 2008). FAK serves as a scaffold to limit p53 activity in an FAK kinase–independent manner. Interestingly, CHIP E3 ligase is required for FAK-FERM–enhanced GATA4 ubiquitination and not Mdm2. FAK-FERM forms a complex with CHIP, but the mechanisms for FAK-FERM–enhanced, CHIP-dependent GATA4 ubiquitination remain to be determined. As nuclear-localized FAK contributes to chromatin remodeling and gene expression (Luo et al., 2009) and FAK-I treatment of cells promotes FAK nuclear localization, this kinase-independent function of FAK deserves recognition when evaluating results of pharmacological FAK inhibition.
Moreover, the specific factors regulating FAK nuclear and cytoplasmic shuttling remain unclear. A previous study showed that FAK nuclear localization was enhanced in normal cells within 30 min of staurosporine addition or disruption of cell–matrix adhesions (Lim et al., 2008). These conditions are associated with loss of FAK tyrosine phosphorylation similar to FAK-I treatment. As treatment of cells with the nuclear export inhibitor leptomycin B results in FAK nuclear accumulation, a cytoplasmic pool of FAK likely shuttles in and out of the nucleus. Notably, a validated nuclear export sequence is located within the FAK kinase domain, and the nuclear localization sequence is within the FAK-FERM domain (Ossovskaya et al., 2008). As structural experiments show that the FAK-FERM domain makes inhibitory contacts to the FAK kinase domain and holds FAK in an inactive conformation (Lietha et al., 2007), it is tempting to speculate whether this may block FAK kinase domain nuclear export function in addition to preventing FAK catalytic activation.
In summary, our genetic and pharmacological inhibition experiments reveal that FAK activity regulates VCAM-1 gene expression during development and expand the role of FAK signaling as an essential mediator of TNF-α–induced VCAM-1 production. The regulation of GATA4 by nuclear FAK provides a novel twist to the anti-inflammatory effects of FAK inhibition.
Materials and methods
Mice
FAK-KD knockin mice containing a point mutation within the FAK kinase domain (changing the codon for Lys454 to Arg) were generated by homologous recombination, as previously described (Lim et al., 2010a), maintained as a heterozygous population, and backcrossed to a C57Bl6 background (The Jackson Laboratory) for 10 generations. Mouse experiments were approved by the University of California San Diego Institutional Animal Care and Use Committee, and mice were maintained in accordance with Association for Assessment and Accreditation of Laboratory Animal Care International–approved guidelines.
Cells and plasmids
Early-passage FAK-WT and FAK-KD MEFs were generated, immortalized by hTERT expression, and maintained on 0.1% gelatin-coated dishes, as previously described (Lim et al., 2010a). FAK-WT and FAK-KD ECs were isolated, immortalized by large T antigen expression (Addgene), and maintained as previously described (Zhao et al., 2010). HUVECs, 293T, and Mdm2−/−p53−/− fibroblasts were used as previously described (Lim et al., 2008). CHIP−/− fibroblasts were obtained from C. Patterson (University of North Carolina, Chapel Hill, NC; Dai et al., 2003). Cells were maintained in DME with 10% FBS, nonessential amino acids for MEM, 1 mM sodium pyruvate, 50 U/ml penicillin, and 50 µg/ml streptomycin. Cell culture–based experiments were performed with semiconfluent cells in growth media. Mouse GATA4 cDNA was obtained from P. Mellon (University of California San Diego, La Jolla, CA; Lawson et al., 1996) and was subcloned into p3XFlagCMV7.1 (Sigma-Aldrich). GST fusion N-terminal (aa 1–260; ends with first DNA-binding zinc finger motif) and CT (aa 255–440; starts with second zinc finger motif) GATA4 constructs (pGEX2T) were obtained from R. Viger (Centre hospitalier de l’Université Laval Research Centre, Ste-Foy, Quebec, Canada; Tremblay and Viger, 2003). Ad tetracycline transactivator (TA) was added to induce various Ad-FAK constructs and the indicated HA- or Myc-tagged full-length FAK-WT, and FAK-KD, FAK-FERM (aa 1–402, N-terminal fragment), and FAK-CT (aa 683–1,024, containing focal adhesion targeting domain) constructs were used as previously described (Lim et al., 2008; Schlaepfer et al., 2007). Lentiviral scrambled (Scr) and anti–human FAK short hairpin RNAs were used as previously described (Lim et al., 2008). Mouse CHIP cDNA was purchased (Thermo Fisher Scientific), and PCR was used to amplify the coding sequence and subcloned into pcDNA3.1 3XHA for expression as an HA-tagged protein.
Antibodies and reagents
Anti-FAK (clone 4.47), anti–mouse VCAM-1 (M/K-2), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 374) were purchased from Millipore. Anti–mouse VCAM-1 (112702) for blotting was purchased from R&D Systems. Anti–pY397 FAK (44625G), anti–pY576 FAK (70013), anti-pERK (pT202/pY204; 44654G), and anti-pGATA4 (pS105; 44–948) were purchased from Life Technologies. Anti–NF-κB pS536 (93H1) and anti–p-JNK (pT183/pY185; 9251) were purchased from Cell Signaling Technology. Anti–β-actin (AC-17), anti-talin (8d4), and anti-Flag (M4) tag were purchased from Sigma-Aldrich. Anti–human VCAM-1 (H-276), anti-IκBα (C-21), anti-GATA4 (G-4), anti-GATA6 (H-92), anti-JNK2 (D-2), and anti-ERK2 (12A4) were purchased from Santa Cruz Biotechnology, Inc. Anti-HA (16B12) and anti-Myc tag (9E10) were purchased from Covance. Anti–poly ADP ribose polymerase (PARP; clone 42), anti-CD31 (MEC13.3), and fluorescein isothiocyanate–conjugated anti-CD31 (MEC13.3) were purchased from BD. Anti-ubiquitin (Fk2) was purchased from Enzo Life Sciences, and anti-GAPDH was purchased from GeneTex Inc. (GTX239). Recombinant TNF-α (human and mouse) and IL1-β (human) were purchased from R&D Systems. Bovine plasma fibronectin (FN) was purchased from Sigma-Aldrich. MG132 was purchased from EMD. Mouse GATA4 ON-TARGETplus SMARTpool siRNA (L-040759-01-0005) and ON-TARGETplus Si Control (Scr) siRNA (D-001810-01) were purchased from Thermo Fisher Scientific. 100 pmol siRNA was used to transfect WT MEFs using Lipofectamine 2000 (Life Technologies). After 48 h, target knockdown was confirmed by immunoblotting.
FAK-Is and in vivo TNF-α signaling
FAK-I PF-271 was synthesized by Laviana Corp., as described in patent materials (Roberts et al., 2008), solubilized in DMSO, and used in experiments with cultured cells at 1 µM. FAK-I PND-1186 (Poniard Pharmaceuticals) was used for in vivo mouse experiments (Tanjoni et al., 2010; Walsh et al., 2010), solubilized in water, and administered twice daily via oral gavage (100 mg/kg) to either pregnant female C57Bl6 mice from E7.5 to E9.0 or to C57Bl6 mice 36 h before initiation of in vivo signaling assays. Recombinant TNF-α (0.02 mg/kg in 100 µl PBS) or PBS was tail vein injected into C57BL6 mice, and after 5 min or 6 h, tissues were rapidly excised and either homogenized for protein lysates or embedded in optimal cutting temperature (Tissue Tech) compound and flash frozen for sectioning.
Embryo staining
Timed matings were performed with FAKWT/KD heterozygous mice, and embryo development was visualized by ultrasound, surgically harvested at E9.0–9.5, and fixed in 3.7% PFA. For whole-mount staining, fixed embryos were permeabilized (0.1% Triton X-100 in PBS for 4 h), blocked (1% BSA and 1% goat serum in PBS for 2 h), incubated with anti–VCAM-1 (diluted at 1:100) and biotinylated goat anti–rat IgG (diluted at 1:300) using the VECTASTAIN Elite ABC kit, and visualized with a diaminobenzidine-based substrate (Vector Laboratories). Controls were performed without primary antibody addition. Bright field images of dissected and stained embryos were acquired at room temperature using a stereo microscope (M2Bio; Carl Zeiss) at 1.0× (Achromat S, FWD 69 mm) with a color charge-coupled device camera (INFINITY1; Lumenera Corporation). Embryos embedded in paraffin were thin sectioned and stained with hematoxylin and eosin. Images were acquired at room temperature using an inverted microscope (IX81; Olympus) at 4× (UPlanS Apo, NA 0.16) with a color charge-coupled device camera (INFINITY1) All images were cropped, and contrast was adjusted using Photoshop CS3 (Adobe).
Gene array and functional analysis
10 FAKWT/WT and FAKKD/KD E9.5 embryos were surgically harvested, pooled, stored in RNAlater (Life Technologies), and total RNA from tissue samples was isolated using RNeasy (QIAGEN). RNA quality was verified using the Bioanalyzer (Agilent Technologies). cDNAs were synthesized and hybridized to an Illumina mouse WG-6 v2.0 expression bead chip containing 45,200 50-mer transcripts. Data were collected and evaluated by G. Hardiman of the University of California San Diego Biomedical Genomics Core facility. Signaling pathway and functional network analyses were performed by IPA software (Ingenuity Systems).
Q-PCR
Q-PCR was used to validate changes in target mRNA level. Total RNAs were isolated by the RNeasy kit (QIAGEN), and cDNAs were synthesized using random hexamers and reverse transcription (SuperScript III; Life Technologies). Real-time PCR was performed using AmpliTaq Gold DNA polymerase and SYBR green as a substrate (Life Technologies). The primers used are mouse GATA4 forward 5′-ACCCCAATCTCGATATGTTTG-3′, GATA4 reverse 5′-CCTCGGCATTACGACGCC-3′, human VCAM-1 forward 5′-AGTTGAAGGATGCGGGAGTA-3′, human VCAM-1 reverse 5′-AGAGCACGAGAAGCTCAGGA-3′, mouse VCAM-1 forward 5′-TGAACCCAAACAGAGGCAGA-3′, and mouse VCAM-1 reverse 5′-CGGAATCGTCCCTTTTTGTAG-3′. Control GAPDH primers were obtained from Life Technologies. Quantification of each gene, relative to the calibrator, was performed with the equation within Sequence Detections software (v1.2.2; Life Technologies).
Biochemical analyses
FAK-I (1 µM PF271) was added to growing cells for 1 h before TNF-α addition (10 ng/ml) unless otherwise indicated. For replating assays, growing cells were treated with 0.06% trypsin and 2 mM EDTA in PBS (2.5 min at 37°C), collected by centrifugation, resuspended in DME with 10% FBS, enumerated (Vi-CELL XR; Beckman Coulter), and held at 37°C (2 × 105 cells/ml) for 30 min. FN-coated (10 µg/ml in PBS overnight) culture dishes were blocked with 1% BSA in PBS for 30 min and preheated to 37°C before cell replating (1 h) followed by TNF-α addition for 15 min to 6 h. Cell apoptosis was determined after 7 h by collecting cells with trypsin and flow cytometry analyses (FACSCalibur; BD) using allophycocyanin–annexin V and 7-actinomycin D staining (BD). Tissue and cultured cell protein lysates were prepared in cell extraction buffer containing 50 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 10% glycerol. For immunoprecipitation and GST binding analyses, lysates were diluted twofold in HNTG buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, and 10% glycerol) and incubated with antibodies (1 µg) or glutathione agarose beads (Sigma-Aldrich) for 3 h at 4°C. Antibodies were collected with either protein A or G Plus (Millipore) agarose beads, and beads were washed at 4°C in 1% Triton X-100–only extraction buffer followed by washes with HNTG buffer and resolved by SDS-PAGE. Densitometry of immunoblotting images was performed using ImageJ (v1.44; National Institutes of Health).
To evaluate ubiquitin incorporation into GATA4, experiments were performed in human HEK293 or normal Mdm2−/−p53−/− or CHIP−/− MEFs. MEFs were infected with 50 plaque-forming unit (pfu)/cell Ad-FAK or Ad-FAK-FERM constructs and 5 pfu/cell Ad-TA. Controls were performed with 50 pfu/cell Ad-TA. For transient transfection, 293T or MEFs using jetPRIME (PolyPlus Transfection), 1 µg Flag-GATA4, 0.5 µg HA-ubiquitin or His-ubiquitin, and 5 µg FAK-FERM plasmid DNA were used. 40 µM MG132 was added 3 h before cell lysis for ubiquitin analyses. For ubiquitin conjugate purification, transfected cells (24 h) were lysed with 8 M urea in phosphate buffer (0.1 M NaH2PO4, 10 mM Tris-HCl, pH 8.0, and 20 mM imidazole) at room temperature. His-ubiquitin–conjugated proteins were bound to a nickel agarose column (Ni–nitrilotriacetic acid; QIAGEN) and washed with 8 M urea phosphate buffer, pH 6.3, and proteins were eluted in 8 M urea phosphate buffer at pH 4.5.
For fractionation experiments, cells were lysed with Cyt buffer (10 mM Tris, pH 7.5, 0.05% NP-40, 3 mM MgCl2, 100 mM NaCl, 1 mM EGTA, 20 µg/ml aprotinin, 1 mM orthovanadate, and 10 µg/ml leupeptin), scrape loaded into tubes, incubated for 5 min at 4°C, and spun at 800 g at 4°C (5 min), and cytosolic supernatants were collected. Cell pellets were further washed with Cyt buffer, purified nuclei were resuspended in cell extraction buffer and spun at 16,000 g for 15 min, and the supernatant was collected as the nuclear fraction. Samples were separated by SDS-PAGE and immunoblotted for GAPDH and PARP as cytoplasmic and nuclear markers, respectively.
For direct binding assays, prey proteins were in vitro translated using the TNT transcription–translation system (Promega). Various pCDNA3 FAK expression constructs (1 µg) were translated in a mixture containing biotin-labeled Lys and diluted 50 fold into binding buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, and 1% Triton X-100). Purified GST-GATA4 proteins were prebound to glutathione agarose beads, incubated with in vitro translated prey for 2 h at 4°C, and washed three times in binding buffer, and the bound prey were detected by streptavidin-HRP immunoblotting.
VCAM-1 promoter assay
HUVECs were cotransfected using jetPEI-HUVEC (PolyPlus Transfection) with Renilla luciferase and promoterless pGL3 luciferase control (Promega) or pGL3 containing 1.8 kb of the human VCAM-1 promoter (−1,716 to 119) from W. Aird (Beth Israel Deaconess Medical Center, Boston, MA; Minami and Aird, 2001). After 18 h, 10 ng/ml TNF-α was added in the presence or absence of 1 µM FAK-I, and luciferase activity was measured with the dual assay kit (Promega) after 6 h.
NF-κB EMSA
Cells were scraped into 1 ml cold PBS, pelleted by centrifugation (1,500 g for 10 min), and resuspended in 160 µl of buffer A (50 mM KCl, 25 mM Hepes, pH 7.8, 10 µg/ml leupeptin, 20 µg/ml aprotinin, 125 µM DTT, and 1 mM PMSF) on ice for 15 min. 40 µl of 2.5% NP-40 was added, vortexed, and centrifuged (12,000 g for 5 min). Pellets were washed with cold buffer A, resuspended in 40 µl of buffer A with 500 mM KCl, frequently vortexed for 20 min, and centrifuged (12,000 g for 5 min). The supernatant protein concentration was determined by a Bradford assay (Bio-Rad Laboratories) and stored frozen (−70°C) as a nuclear extract. For tissue extracts, 3 g of lung tissue was minced with a razor blade, and single-cell suspensions were prepared by Dounce homogenization in 4 ml of buffer A. Nuclear extracts were prepared as previously described. Double-stranded NF-κB oligonucleotides (Promega) were labeled with γ-[32P]ATP (PerkinElmer) using DNA T4 polynucleotide kinase (New England Biolabs, Inc.), and unincorporated nucleotides were removed using a G-25 spin column (GE Healthcare). Labeled probes (∼20,000 cpm per 0.1 pmol) were incubated with 4 µg of nuclear extract for 30 min at room temperature in gel shift binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 40 mM KCl) containing 100 µg/ml poly(deoxyinosinic-deoxycytidylic) acid and 10% glycerol. Addition of 100-fold excess of unlabeled oligonucleotide probe (10 min) was used to verify binding specificity. Samples were resolved by 4% native PAGE, and binding was visualized by autoradiography.
Immunofluorescent staining
Tissues were sectioned (6 µm; CM1950; Leica), fixed in cold acetone (10 min), rehydrated in PBS containing 0.5% BSA (5 min), and blocked with 1.25% normal goat serum in PBS (30 min at room temperature). Samples were incubated with anti–FAK pY576 (1:50) or anti–VCAM-1 (M/K-2; 1:50) and anti-CD31 (1:300) overnight at 4°C followed by Alexa Fluor 488 goat anti–rat and Alexa Fluor 594 goat anti–rabbit secondary antibodies (1:500, 30 min at room temperature; Life Technologies). For VCAM-1 and CD31 staining, anti–VCAM-1 was labeled with DyLight 594 (red) by using a DyLight microscale antibody labeling kit (Thermo Fisher Scientific). Samples were incubated with fluorescein isothiocyanate CD31 (1:100) and DyLight 594–VCAM-1 (1:25) overnight. Slides were mounted in VECTASHIELD (H-1000; Vector Laboratories), and images were acquired at room temperature sequentially using a mercury lamp source, multiband dichroic, single-band exciter, and single-band emitter filter sets (Chroma Technology Corp.) on dual filter wheels, a spinning-disk confocal microscope (IX81; Olympus) at 60× (PlanApo, NA 1.42), and an OrcaER camera (Hamamatsu Photonics) controlled by SlideBook software (v5.0). Files were cropped, pseudocolored, and contrast adjusted using Photoshop CS3. The degree of association exhibited by patterns of fluorescence was measured on a pixel-by-pixel basis and calculated as a Pearson’s correlation coefficient using the Cell Profiler measure correlations module (v2.0; Broad Institute). A value of 0 indicates no overlap, and a value of 1 corresponds to 100% colocalization. Adhesion size (pixels) and number within a cell were determined using Cell Profiler using a pipeline to threshold images and reduce background fluorescent staining.
Statistical analyses
Significance between experimental groups was determined by one-way analysis of variance with a Tukey’s post hoc test using Prism software (v5.0b; GraphPad Software).
Online supplemental material
Fig. S1 shows quantitative analyses of TNF-α–stimulated VCAM-1 protein expression in mouse lung tissue and the inhibition of VCAM-1 by oral FAK-I (PND-1186) pretreatment. Fig. S2 shows visualization of TNF-α–stimulated FAK activation and VCAM-1 expression within heart ECs by indirect immunofluorescent staining. Fig. S3 shows gene ontology and signaling network analyses of mRNA array data from FAK-WT and FAK-KD embryos. Fig. S4 shows EMSAs in MEFs and HUVECs whereby FAK inhibition (FAK-I, PF271) does not block TNF-α–stimulated NF-κB activation. Fig. S5 shows analyses of lung lysates whereby pharmacological FAK inhibition (PND-1186) blocks TNF-α–induced ERK/MAPK but not NF-κB activation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201109067/DC1.
Supplementary Material
Acknowledgments
We thank Pamela Mellon, Cam Patterson, William Aird, and Robert Viger for reagents. PND-1186 was obtained from Poniard Pharmaceuticals. Array analyses were performed by the University of California San Diego Biomedical Genomics Core facility.
This work was supported by National Institutes of Health grants (HL093156 and GM087400) to D.D. Schlaepfer. N.L.G. Miller was supported by a National Research Service Award (1F32CA159558), C. Lawson was supported by a Canadian Institutes of Health Research fellowship (200810MFE-193594-139144), and I. Tancioni was supported by a grant from Susan G. Komen for the Cure (KG111237).
Footnotes
Abbreviations used in this paper:
- Ad
- adenoviral
- CHIP
- C terminus of Hsp70-interacting protein
- CT
- C terminal
- EC
- endothelial cell
- EMSA
- electrophoretic mobility shift assay
- ERK
- extracellular-regulated kinase
- FAK-I
- FAK inhibitor
- FN
- fibronectin
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- HUVEC
- human umbilical vein EC
- KD
- kinase dead
- MEF
- mouse embryonic fibroblast
- PARP
- poly ADP ribose polymerase
- pfu
- plaque-forming unit
- PTK
- protein tyrosine kinase
- Q-PCR
- quantitative PCR
- TA
- transactivator
- TNF-α
- tumor necrosis factor–α
- VCAM-1
- vascular cell adhesion molecule–1
- WT
- wild type
References
- Ahmad M., Theofanidis P., Medford R.M. 1998. Role of activating protein-1 in the regulation of the vascular cell adhesion molecule-1 gene expression by tumor necrosis factor-alpha. J. Biol. Chem. 273:4616–4621 10.1074/jbc.273.8.4616 [DOI] [PubMed] [Google Scholar]
- Beg A.A., Baltimore D. 1996. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 274:782–784 10.1126/science.274.5288.782 [DOI] [PubMed] [Google Scholar]
- Bieler G., Hasmim M., Monnier Y., Imaizumi N., Ameyar M., Bamat J., Ponsonnet L., Chouaib S., Grell M., Goodman S.L., et al. 2007. Distinctive role of integrin-mediated adhesion in TNF-induced PKB/Akt and NF-kappaB activation and endothelial cell survival. Oncogene. 26:5722–5732 10.1038/sj.onc.1210354 [DOI] [PubMed] [Google Scholar]
- Braren R., Hu H., Kim Y.H., Beggs H.E., Reichardt L.F., Wang R. 2006. Endothelial FAK is essential for vascular network stability, cell survival, and lamellipodial formation. J. Cell Biol. 172:151–162 10.1083/jcb.200506184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer A., Pizzey J. 2006. GATA factors in vertebrate heart development and disease. Expert Rev. Mol. Med. 8:1–20 10.1017/S1462399406000093 [DOI] [PubMed] [Google Scholar]
- Carter R.A., Wicks I.P. 2001. Vascular cell adhesion molecule 1 (CD106): A multifaceted regulator of joint inflammation. Arthritis Rheum. 44:985–994 [DOI] [PubMed] [Google Scholar]
- Dai Q., Zhang C., Wu Y., McDonough H., Whaley R.A., Godfrey V., Li H.H., Madamanchi N., Xu W., Neckers L., et al. 2003. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22:5446–5458 10.1093/emboj/cdg529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A., Morris J., Abedin M.J., Collins S., Richardson J.A., Hill J.A. 2011. Forkhead factor FoxO1 is essential for placental morphogenesis in the developing embryo. Proc. Natl. Acad. Sci. USA. 108:16307–16312 10.1073/pnas.1107341108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitau J., Boulday G., Coulon F., Quillard T., Charreau B. 2006. The adaptor molecule Lnk negatively regulates tumor necrosis factor-alpha-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J. Biol. Chem. 281:20148–20159 10.1074/jbc.M510997200 [DOI] [PubMed] [Google Scholar]
- Fornaro M., Plescia J., Chheang S., Tallini G., Zhu Y.M., King M., Altieri D.C., Languino L.R. 2003. Fibronectin protects prostate cancer cells from tumor necrosis factor-alpha-induced apoptosis via the AKT/survivin pathway. J. Biol. Chem. 278:50402–50411 10.1074/jbc.M307627200 [DOI] [PubMed] [Google Scholar]
- Funakoshi-Tago M., Sonoda Y., Tanaka S., Hashimoto K., Tago K., Tominaga S., Kasahara T. 2003. Tumor necrosis factor-induced nuclear factor kappaB activation is impaired in focal adhesion kinase-deficient fibroblasts. J. Biol. Chem. 278:29359–29365 10.1074/jbc.M213115200 [DOI] [PubMed] [Google Scholar]
- Golubovskaya V.M., Finch R., Cance W.G. 2005. Direct interaction of the N-terminal domain of focal adhesion kinase with the N-terminal transactivation domain of p53. J. Biol. Chem. 280:25008–25021 10.1074/jbc.M414172200 [DOI] [PubMed] [Google Scholar]
- Gurtner G.C., Davis V., Li H., McCoy M.J., Sharpe A., Cybulsky M.I. 1995. Targeted disruption of the murine VCAM1 gene: Essential role of VCAM-1 in chorioallantoic fusion and placentation. Genes Dev. 9:1–14 10.1101/gad.9.1.1 [DOI] [PubMed] [Google Scholar]
- Hiratsuka S., Goel S., Kamoun W.S., Maru Y., Fukumura D., Duda D.G., Jain R.K. 2011. Endothelial focal adhesion kinase mediates cancer cell homing to discrete regions of the lungs via E-selectin up-regulation. Proc. Natl. Acad. Sci. USA. 108:3725–3730 10.1073/pnas.1100446108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.C., Chen J.J., Chen C.C. 2003. c-Src-dependent tyrosine phosphorylation of IKKbeta is involved in tumor necrosis factor-alpha-induced intercellular adhesion molecule-1 expression. J. Biol. Chem. 278:9944–9952 10.1074/jbc.M208521200 [DOI] [PubMed] [Google Scholar]
- Ilic D., Kovacic B., McDonagh S., Jin F., Baumbusch C., Gardner D.G., Damsky C.H. 2003. Focal adhesion kinase is required for blood vessel morphogenesis. Circ. Res. 92:300–307 10.1161/01.RES.0000055016.36679.23 [DOI] [PubMed] [Google Scholar]
- Inman K.E., Downs K.M. 2007. The murine allantois: Emerging paradigms in development of the mammalian umbilical cord and its relation to the fetus. Genesis. 45:237–258 10.1002/dvg.20281 [DOI] [PubMed] [Google Scholar]
- Karin M., Gallagher E. 2009. TNFR signaling: Ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol. Rev. 228:225–240 10.1111/j.1600-065X.2008.00755.x [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Mao K., Zheng H., Wang X., Patterson C., O’Connell T.D., Liang Q. 2007. Diminished GATA4 protein levels contribute to hyperglycemia-induced cardiomyocyte injury. J. Biol. Chem. 282:21945–21952 10.1074/jbc.M703048200 [DOI] [PubMed] [Google Scholar]
- Kwee L., Baldwin H.S., Shen H.M., Stewart C.L., Buck C., Buck C.A., Labow M.A. 1995. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development. 121:489–503 [DOI] [PubMed] [Google Scholar]
- Lawson M.A., Whyte D.B., Mellon P.L. 1996. GATA factors are essential for activity of the neuron-specific enhancer of the gonadotropin-releasing hormone gene. Mol. Cell. Biol. 16:3596–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Borboa A.K., Chun H.B., Baird A., Eliceiri B.P. 2010. Conditional deletion of the focal adhesion kinase FAK alters remodeling of the blood-brain barrier in glioma. Cancer Res. 70:10131–10140 10.1158/0008-5472.CAN-10-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P. 2002. Inflammation in atherosclerosis. Nature. 420:868–874 10.1038/nature01323 [DOI] [PubMed] [Google Scholar]
- Lietha D., Cai X., Ceccarelli D.F., Li Y., Schaller M.D., Eck M.J. 2007. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 129:1177–1187 10.1016/j.cell.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.T., Chen X.L., Lim Y., Hanson D.A., Vo T.T., Howerton K., Larocque N., Fisher S.J., Schlaepfer D.D., Ilic D. 2008. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol. Cell. 29:9–22 10.1016/j.molcel.2007.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.T., Chen X.L., Tomar A., Miller N.L., Yoo J., Schlaepfer D.D. 2010a. Knock-in mutation reveals an essential role for focal adhesion kinase activity in blood vessel morphogenesis and cell motility-polarity but not cell proliferation. J. Biol. Chem. 285:21526–21536 10.1074/jbc.M110.129999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.T., Miller N.L., Nam J.O., Chen X.L., Lim Y., Schlaepfer D.D. 2010b. Pyk2 inhibition of p53 as an adaptive and intrinsic mechanism facilitating cell proliferation and survival. J. Biol. Chem. 285:1743–1753 10.1074/jbc.M109.064212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., He X., Corbett S.A., Lowry S.F., Graham A.M., Fässler R., Li S. 2009. Integrins are required for the differentiation of visceral endoderm. J. Cell Sci. 122:233–242 10.1242/jcs.037663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.W., Zhang C., Zhang B., Kim C.H., Qiu Y.Z., Du Q.S., Mei L., Xiong W.C. 2009. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. EMBO J. 28:2568–2582 10.1038/emboj.2009.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEwan D.J. 2002. TNF ligands and receptors—a matter of life and death. Br. J. Pharmacol. 135:855–875 10.1038/sj.bjp.0704549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M.J., Wheeler-Jones C.P., Pearson J.D. 1996. Effects of protein tyrosine kinase inhibitors on cytokine-induced adhesion molecule expression by human umbilical vein endothelial cells. Br. J. Pharmacol. 118:1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami T., Aird W.C. 2001. Thrombin stimulation of the vascular cell adhesion molecule-1 promoter in endothelial cells is mediated by tandem nuclear factor-kappa B and GATA motifs. J. Biol. Chem. 276:47632–47641 10.1074/jbc.M108363200 [DOI] [PubMed] [Google Scholar]
- Mitra S.K., Hanson D.A., Schlaepfer D.D. 2005. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6:56–68 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- Molkentin J.D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949–38952 10.1074/jbc.R000029200 [DOI] [PubMed] [Google Scholar]
- Osborn L., Hession C., Tizard R., Vassallo C., Luhowskyj S., Chi-Rosso G., Lobb R. 1989. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell. 59:1203–1211 10.1016/0092-8674(89)90775-7 [DOI] [PubMed] [Google Scholar]
- Ossovskaya V., Lim S.T., Ota N., Schlaepfer D.D., Ilic D. 2008. FAK nuclear export signal sequences. FEBS Lett. 582:2402–2406 10.1016/j.febslet.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen K.A., Pixley F.J., Thomas K.S., Vicente-Manzanares M., Ray B.J., Horwitz A.F., Parsons J.T., Beggs H.E., Stanley E.R., Bouton A.H. 2007. Regulation of lamellipodial persistence, adhesion turnover, and motility in macrophages by focal adhesion kinase. J. Cell Biol. 179:1275–1287 10.1083/jcb.200708093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober J.S. 2002. Endothelial activation: Intracellular signaling pathways. Arthritis Res. 4(Suppl 3):S109–S116 10.1186/ar576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts W.G., Ung E., Whalen P., Cooper B., Hulford C., Autry C., Richter D., Emerson E., Lin J., Kath J., et al. 2008. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res. 68:1935–1944 10.1158/0008-5472.CAN-07-5155 [DOI] [PubMed] [Google Scholar]
- Schaller M.D. 2010. Cellular functions of FAK kinases: Insight into molecular mechanisms and novel functions. J. Cell Sci. 123:1007–1013 10.1242/jcs.045112 [DOI] [PubMed] [Google Scholar]
- Schlaepfer D.D., Mitra S.K. 2004. Multiple connections link FAK to cell motility and invasion. Curr. Opin. Genet. Dev. 14:92–101 10.1016/j.gde.2003.12.002 [DOI] [PubMed] [Google Scholar]
- Schlaepfer D.D., Hou S., Lim S.T., Tomar A., Yu H., Lim Y., Hanson D.A., Uryu S.A., Molina J., Mitra S.K. 2007. Tumor necrosis factor-alpha stimulates focal adhesion kinase activity required for mitogen-activated kinase-associated interleukin 6 expression. J. Biol. Chem. 282:17450–17459 10.1074/jbc.M610672200 [DOI] [PubMed] [Google Scholar]
- Schultze A., Fiedler W. 2010. Therapeutic potential and limitations of new FAK inhibitors in the treatment of cancer. Expert Opin. Investig. Drugs. 19:777–788 10.1517/13543784.2010.489548 [DOI] [PubMed] [Google Scholar]
- Schwartz M.A. 2001. Integrin signaling revisited. Trends Cell Biol. 11:466–470 10.1016/S0962-8924(01)02152-3 [DOI] [PubMed] [Google Scholar]
- Shen T.-L., Park A.Y.J., Alcaraz A., Peng X., Jang I., Koni P., Flavell R.A., Gu H., Guan J.-L. 2005. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J. Cell Biol. 169:941–952 10.1083/jcb.200411155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short S.M., Talbott G.A., Juliano R.L. 1998. Integrin-mediated signaling events in human endothelial cells. Mol. Biol. Cell. 9:1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Aggarwal B.B. 2004. TNF activates Syk protein tyrosine kinase leading to TNF-induced MAPK activation, NF-kappaB activation, and apoptosis. J. Immunol. 173:1066–1077 [DOI] [PubMed] [Google Scholar]
- Tanjoni I., Walsh C., Uryu S., Tomar A., Nam J.O., Mielgo A., Lim S.T., Liang C., Koenig M., Sun C., et al. 2010. PND-1186 FAK inhibitor selectively promotes tumor cell apoptosis in three-dimensional environments. Cancer Biol. Ther. 9:764–777 10.4161/cbt.9.10.11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavora B., Batista S., Reynolds L.E., Jadeja S., Robinson S., Kostourou V., Hart I., Fruttiger M., Parsons M., Hodivala-Dilke K.M. 2010. Endothelial FAK is required for tumour angiogenesis. EMBO Mol Med. 2:516–528 10.1002/emmm.201000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J.J., Viger R.S. 2003. Transcription factor GATA-4 is activated by phosphorylation of serine 261 via the cAMP/protein kinase a signaling pathway in gonadal cells. J. Biol. Chem. 278:22128–22135 10.1074/jbc.M213149200 [DOI] [PubMed] [Google Scholar]
- van Berlo J.H., Elrod J.W., Aronow B.J., Pu W.T., Molkentin J.D. 2011. Serine 105 phosphorylation of transcription factor GATA4 is necessary for stress-induced cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. USA. 108:12331–12336 10.1073/pnas.1104499108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C., Tanjoni I., Uryu S., Tomar A., Nam J.O., Luo H., Phillips A., Patel N., Kwok C., McMahon G., et al. 2010. Oral delivery of PND-1186 FAK inhibitor decreases tumor growth and spontaneous breast to lung metastasis in pre-clinical models. Cancer Biol. Ther. 9:778–790 10.4161/cbt.9.10.11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C., Negrescu E., Erl W., Pietsch A., Frankenberger M., Ziegler-Heitbrock H.W., Siess W., Weber P.C. 1995. Inhibitors of protein tyrosine kinase suppress TNF-stimulated induction of endothelial cell adhesion molecules. J. Immunol. 155:445–451 [PubMed] [Google Scholar]
- Weis S.M., Cheresh D.A. 2011. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat. Med. 17:1359–1370 10.1038/nm.2537 [DOI] [PubMed] [Google Scholar]
- Weis S.M., Lim S.T., Lutu-Fuga K.M., Barnes L.A., Chen X.L., Göthert J.R., Shen T.L., Guan J.L., Schlaepfer D.D., Cheresh D.A. 2008. Compensatory role for Pyk2 during angiogenesis in adult mice lacking endothelial cell FAK. J. Cell Biol. 181:43–50 10.1083/jcb.200710038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S.R., Gerard-O’Riley R., Harrington M., Pavalko F.M. 2010. Activation of NF-kappaB by fluid shear stress, but not TNF-alpha, requires focal adhesion kinase in osteoblasts. Bone. 47:74–82 10.1016/j.bone.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Peng X., Sun S., Park A.Y., Guan J.L. 2010. Role of kinase-independent and -dependent functions of FAK in endothelial cell survival and barrier function during embryonic development. J. Cell Biol. 189:955–965 10.1083/jcb.200912094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.