Actin veil networks assembled by the Arp2/3 complex constrain myosin II–dependent contractility in neuronal growth cones with consequent effects on peripheral retrograde actin flow rates.
Abstract
The Arp2/3 complex nucleates actin filaments to generate networks at the leading edge of motile cells. Nonmuscle myosin II produces contractile forces involved in driving actin network translocation. We inhibited the Arp2/3 complex and/or myosin II with small molecules to investigate their respective functions in neuronal growth cone actin dynamics. Inhibition of the Arp2/3 complex with CK666 reduced barbed end actin assembly site density at the leading edge, disrupted actin veils, and resulted in veil retraction. Strikingly, retrograde actin flow rates increased with Arp2/3 complex inhibition; however, when myosin II activity was blocked, Arp2/3 complex inhibition now resulted in slowing of retrograde actin flow and veils no longer retracted. Retrograde flow rate increases induced by Arp2/3 complex inhibition were independent of Rho kinase activity. These results provide evidence that, although the Arp2/3 complex and myosin II are spatially segregated, actin networks assembled by the Arp2/3 complex can restrict myosin II–dependent contractility with consequent effects on growth cone motility.
Introduction
Growth cones are highly motile sensory structures at the tips of developing and regenerating neurites. They are composed of two distinct cytoplasmic domains: (1) the central or C-domain, characterized by fast organelle transport and high microtubule density; and (2) the peripheral or P-domain, consisting of dense networks of actin filaments (Forscher et al., 1987), which can be classified into polarized filopodial bundles and isotropic networks referred to as actin “veils” (Lewis and Bridgman, 1992). Like other migrating cells, coordinated regulation of actin filament dynamics is critical for maintaining the growth cone’s structural integrity and coherent motility.
The Arp2/3 complex has been widely investigated for its role in actin filament assembly. It is comprised of seven subunits and nucleates a branched actin filament network (Pollard and Borisy, 2003; Pollard, 2007). The Arp2/3 complex localizes to the leading edge of motile cells (Machesky et al., 1994; Welch et al., 1997; Schafer et al., 1998; Iwasa and Mullins, 2007; Le Clainche et al., 2007; Lai et al., 2008). Upon activation near the membrane by nucleation promoting factors downstream of Rac and Cdc42 and subsequent binding to the side of a preexisting filament, the Arp2/3 complex forms a daughter filament branch that grows and can generate protrusive force (Bailly et al., 1999; Prass et al., 2006; Pollard, 2007; Campellone and Welch, 2010). In neuronal growth cones, the localization and the role of the Arp2/3 complex have been questioned. Some studies reported that the Arp2/3 complex was localized to the P-domain leading edge (Mongiu et al., 2007; Korobova and Svitkina, 2008) and implicated it in veil protrusion (Mongiu et al., 2007; Tahirovic et al., 2010) and filopodium initiation (Korobova and Svitkina, 2008; Norris et al., 2009; Spillane et al., 2011). However, others reported Arp2/3 complex concentration in the C-domain and proposed a role in negative regulation of neurite outgrowth (Strasser et al., 2004; Pinyol et al., 2007).
Actin motors, particularly nonmuscle myosin II, have also been implicated in regulation of actin dynamics. Upon activation by phosphorylation of its regulatory light chains, myosin II forms bipolar mini-filaments, binds to actin filaments, and produces force on the actin filament network (Kolodney and Elson, 1995; Henson et al., 2003; Vicente-Manzanares et al., 2009). Myosin II concentrates in the lamellum and the rear of migrating cells, where it contributes to de-adherence accompanying trailing edge retraction (Ponti et al., 2004; Vicente-Manzanares et al., 2009; Wilson et al., 2010). In growth cones, myosin II is concentrated at the interface between P- and C-domains (Rochlin et al., 1995; Medeiros et al., 2006), where it contributes to retrograde flow of actin filaments and recycling of filopodial bundles (Lin et al., 1996; Medeiros et al., 2006; Wilson et al., 2010). Several lines of evidence have implicated myosin II activation in neurite retraction responses (Ahmad et al., 2000; Zhang et al., 2003; Myers et al., 2006; Brown et al., 2009).
We used small molecule inhibitors and observations by electron microscopy, immunocytochemistry, and quantitative fluorescent speckle microscopy to study the roles and interactions of Arp2/3 complex and myosin II in growth cone motility. Our observations show that the network of actin filaments comprising veils depends on the Arp2/3 complex for assembly and that retrograde flow of this network depends on myosin II activity. Our results suggest an antagonistic modulatory relationship between the Arp2/3 complex and myosin II and that spatiotemporal coordination of their respective functions regulate actin veil network integrity and dynamic behavior.
Results
Arp2/3 complex inhibition disrupts its localization in growth cones
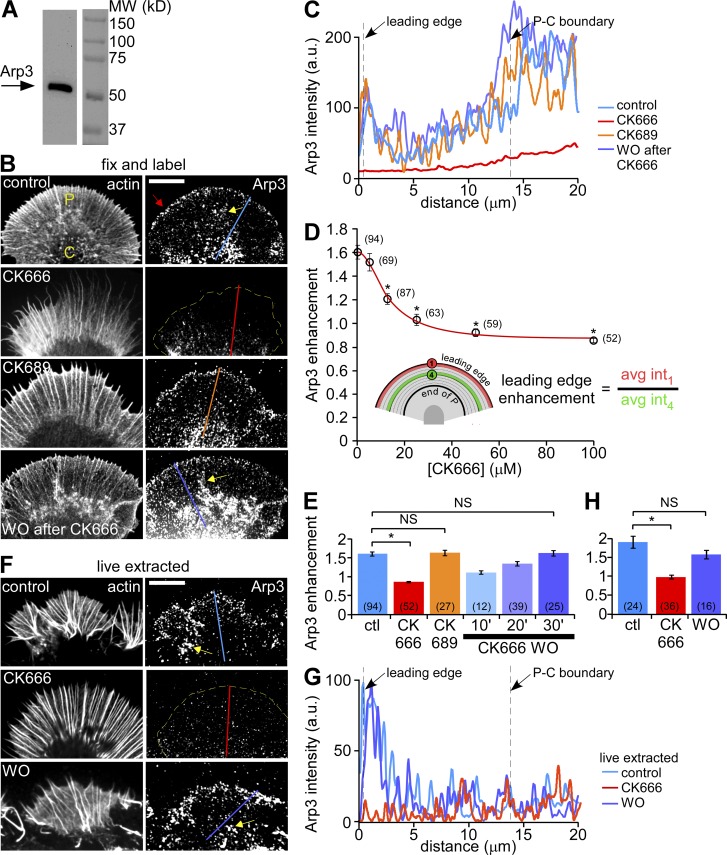
We localized Arp2/3 complex in fixed Aplysia growth cones with an antibody against the Arp3 subunit (Welch et al., 1997; Mongiu et al., 2007), which recognized a single 53-kD band on Western blots of Aplysia CNS homogenate (Fig. 1 A). Immunolabeling of Arp2/3 complex revealed a punctate pattern throughout the growth cone. Puncta density was highest in a 1–2-µm-wide band near the leading edge (Fig. 1 B, red arrow, top). Line scans spanning the P-domain (Fig. 1 C) indicated Arp2/3 complex labeling was 1.6-fold higher near the leading edge than in the adjacent region (Fig. 1, D [see inset] and E). Pre-absorbing the Arp3 antibody with purified bovine Arp2/3 complex eliminated reaction with Arp3 on Western blots and immunostaining of growth cones, confirming antibody specificity (Fig. S1, A and B). An antibody against the Arp2 subunit (Hubert et al., 2011) detected a single band at 43 kD on immunoblots of Aplysia CNS homogenate and produced labeling patterns similar to the Arp3 antibody (Fig. S1, C and D, top). Our results are consistent with previous studies of growth cones from dorsal root ganglion (Mongiu et al., 2007) and hippocampal (Korobova and Svitkina, 2008) neurons, fibroblasts (Welch et al., 1997; Schafer et al., 1998), epithelial cells (Le Clainche et al., 2007), melanoma cells (Lai et al., 2008), and Drosophila hemocytes (Iwasa and Mullins, 2007).
Figure 1.
Arp2/3 complex inhibition disrupts its localization in growth cones. (A) Western blot analysis of Aplysia CNS proteins with anti-Arp3 antibody. (B) Fluorescent labeling of Aplysia bag cell neuron growth cones with Arp3 antibody and Alexa 594 phalloidin after normal fixation. Growth cones were treated with vehicle (DMSO, top panel), CK666 (100 µM, second panel), the inactive analogue CK689 (100 µM, third panel) for 20 min, or treated with CK666 (100 µM) for 20 min followed by 30 min recovery in control medium (bottom panel). Red arrow: Arp2/3 complex enrichment. (C) Arp3 distribution profile sampled from the designated lines in B. (D) CK666 dose-dependent reduction of Arp2/3 complex enrichment at the leading edge. Red line: best fit curve using the 4-parameter nonlinear regression model (see Materials and methods), R2 = 0.9929. *, P < 0.01 with two-tailed unpaired t test versus the first data point (control). (E) Quantification of Arp2/3 complex enrichment at the leading edge for each condition in B. For images of 10- and 20-min CK666 washout, see Fig. S2 C. *, P < 0.01 with two-tailed unpaired t test. NS, not significant. (F) Fluorescent labeling of growth cones with Arp3 antibody and TRITC-phalloidin after live cell extraction. Growth cones were treated with vehicle (DMSO, top), CK666 (100 µM, middle) for 20 min, or treated with CK666 (100 µM, 20 min) followed by recovery in control medium for 30 min (bottom). (G) Arp3 distribution profiles sampled from the designated lines in F. (H) Quantification of Arp2/3 complex enrichment at the leading edge for each condition in F. *, P < 0.01 with two-tailed unpaired t test. NS, not significant. Yellow dotted lines demarcate the leading edge. Yellow arrows, intrapodia. Numbers in parentheses indicate growth cones measured. Bars, 10 µm.
We assessed effects of Arp2/3 complex inhibition on its localization using a small molecule inhibitor, CK666, which binds between Arp2 and Arp3 and blocks their movement into the active conformation (Nolen et al., 2009). Treatment of growth cones with 100 µM CK666 for 20 min delocalized Arp2/3 complex from the leading edge (Fig. 1, B [second panel], C, and E; Fig. S2 B), whereas an inactive structural analogue, 100 µM CK689 (Nolen et al., 2009), had no apparent effect (Fig. 1, B [third panel], C, and E). These effects on Arp2/3 complex localization depended on the concentration of CK666, with almost complete leading edge delocalization at concentrations above 25 µM (Fig. 1 D; Fig. S2, A and B). Delocalization effects were readily reversible (Fig. 1, B [bottom], C, and E; Figs. S1 D and S2, C and D). Treatment with CK666, but not CK689, dispersed Arp2/3 complex from the C-domain (Fig. 1, B and C), where it had also been observed previously (Strasser et al., 2004; Pinyol et al., 2007).
Arp2/3 complex in the P- and C-domains responded differently when the plasma membrane was removed by extraction in a cytoskeleton-stabilizing buffer before fixation (Fig. 1, F [top] and G). The band of enhanced Arp2/3 complex density near the leading edge was stable during extraction of control cells but was absent in cells treated with CK666 (Fig. 1, F–H). These results suggest that leading-edge Arp2/3 complex is normally tightly associated with peripheral cytoskeletal structures. In contrast, Arp2/3 complex in the C-domain appeared to be less tightly associated with the cytoskeleton as it was more readily extracted under control conditions (Fig. 1, compare controls in B vs. F).
Arp2/3 complex inhibition induces veil retraction and disrupts actin veil network
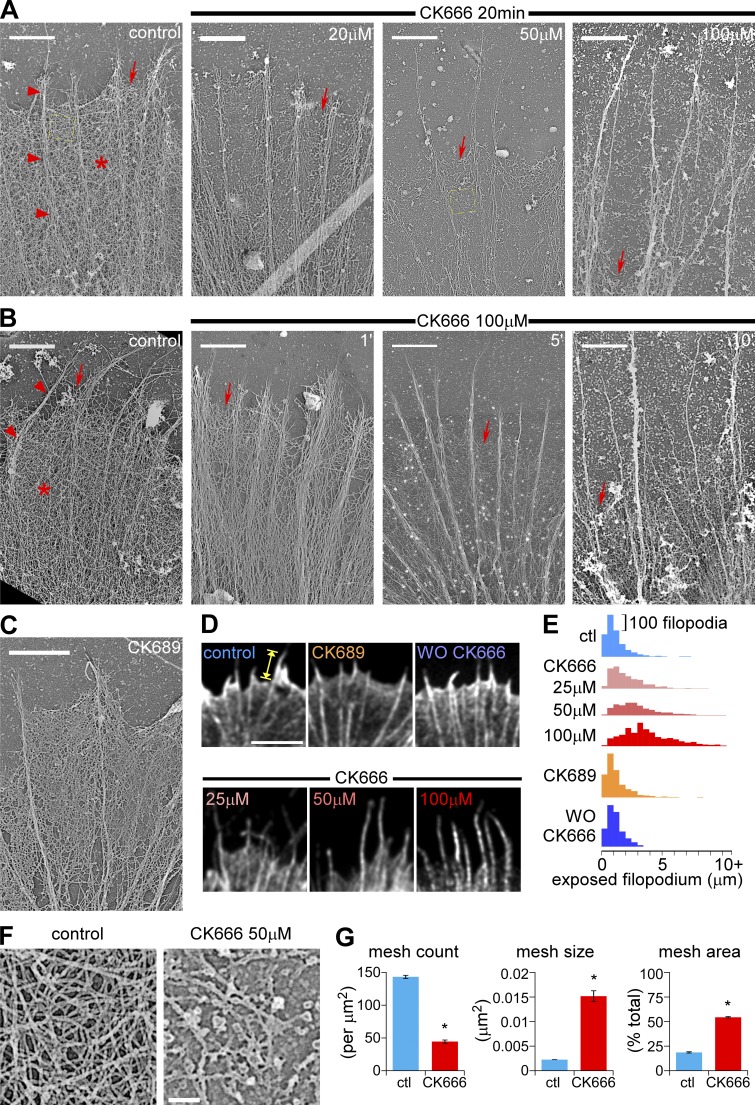
As described previously (Lewis and Bridgman, 1992; Lebrand et al., 2004), the P-domain of control growth cones contains two distinct substructures: (1) actin veils comprised of a dense isotropic network of short filaments (Fig. 2, A and B, control panels; Fig. S2 E, asterisk) and (2) filopodia comprised of bundles of longer actin filaments that span the width of the P-domain (Fig. 2 A and B, control panels; Fig. S2 E, arrowheads). Electron micrographs of platinum replicas of extracted growth cones showed that treatment with CK666 dramatically reduced the density of the actin network in veils at the leading edge in a concentration- and time-dependent manner (Fig. 2, A and B, red arrows; Fig. S2 F) and appeared to result in veil retraction. After treatment with 100 µM CK666 for 20 min, filopodial actin bundles were remarkably intact, but actin veils remained only near the base of filopodial bundles (Fig. 2 A, arrow, far right). Our results agree with a previous report showing that pharmacological inhibition of Arp2/3 complex led to veil retraction in cerebellar granule neurons (Tahirovic et al., 2010). The inactive analogue CK689 did not significantly affect actin filament organization (Fig. 2 C; Fig. S2 G).
Figure 2.
Arp2/3 complex inhibition disrupts actin veil structure and leads to veil retraction in a dose- and time-dependent manner. (A) Periphery of growth cones treated with vehicle (DMSO, left) or different concentrations of CK666 for 20 min. (B) Periphery of growth cones treated with vehicle (DMSO, 20 min, left) or CK666 (100 µM) for 1, 5, or 10 min. Arrowheads, filopodial; asterisk, actin veil; arrows, edge of the veil. Yellow dashed squares are shown in F in higher magnification. (C) Periphery of a growth cone treated with CK689 (100 µM) for 20 min. For ultrastructures of the whole growth cones, see Fig. S2, E–G. (D) Leading edge of growth cones labeled with Alexa 594 phalloidin after normal fixation. Growth cones were treated with vehicle (DMSO, 20 min, top left), CK666 at different concentrations (25, 50, or 100 µM, 20 min, bottom), CK689 (100 µM, 20 min, top middle), or CK666 (100 µM, 20 min) followed by washout for 30 min (top right). See Fig. S3 A for whole growth cones. (E) Distribution of exposed filopodium lengths (yellow caliper in D) in histograms. See Fig. S3, B and C, for statistical analysis. (F) High magnification of areas marked by the yellow boxes in A showing representative veil network ultrastructure in control (left) and CK666-treated (50 µM, 20 min, right) growth cones. (G) Quantification of actin veil network parameters from 1 × 1 µm2 regions in distal P-domain similar to those in F. n, 97 regions from 8 GCs for control; 61 regions from 6 GCs for CK666. *, P < 0.01 with two-tailed unpaired t test. Bars: (A–C) 2 µm; (D) 5 µm; (F) 200 nm.
We assessed actin veil retraction by measuring the length of exposed filopodia in growth cones treated with a range of CK666 concentrations and stained with Alexa 594 phalloidin (Fig. 2 D, yellow caliper). Under control conditions, most exposed filopodia were less than 2 µm long, consistent with our electron micrographs (Fig. 2, D [top left] and E [top histogram]). Treatment with 50 or 100 µM CK666 for 15–30 min did not change the number or length of filopodia (Fig. S3, D and E), but the lengths of filopodia exposed at the leading edge (Fig. 2 D; Fig. S3 A, bottom panels) increased with the concentration of CK666 (Fig. 2 E; Fig. S3, B and C). The length distribution of exposed filopodia was similar to controls after CK666 washout (Fig. 2, D [top right] and E; Fig. S3, A–C) or after treatment with100 µM inactive analogue CK689 (Fig. 2, D and E; Fig. S3, A–C).
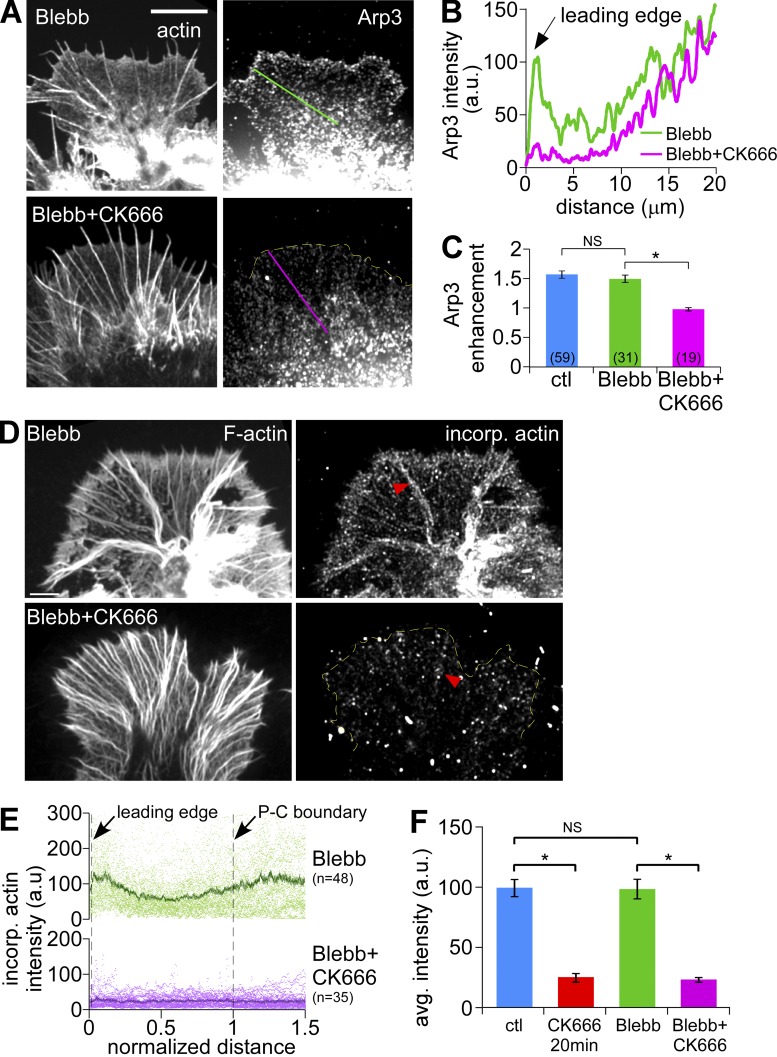
Next we assessed the effects of Arp2/3 complex inhibition on actin filament network density near the leading edge (Fig. 2, F and G). We reasoned that in the relatively isotropic actin networks typically found here, higher filament densities would be correlated with less negative (free) space. Accordingly, we measured the size, number, and sum of negative space in 1 µm2 regions of interest (ROIs) in EM images. Under control conditions, ROIs had 17.9 ± 0.7% negative space and high mesh counts (143.2 ± 2.4 /µm2, average size 0.0013 ± 0.0004 µm2), indicative of a densely packed actin filament network (Fig. 2 G, blue bars). After CK666 treatment (50 µM for 20 min), 54.3 ± 0.7% of the ROI sampled near the retracted leading edge was negative space, the mesh count significantly decreased (43.7 ± 2.4 /µm2) and average mesh size increased (0.0151 ± 0.0011 µm2). These results show that Arp2/3 complex inhibition markedly reduces actin network density (Fig. 2 G, red bars). More specifically, the Arp2/3 complex appears to regulate the integrity of actin veil networks and has modest effects, if any, on filopodial actin bundles.
Arp2/3 complex inhibition decreases the density of actin assembly sites
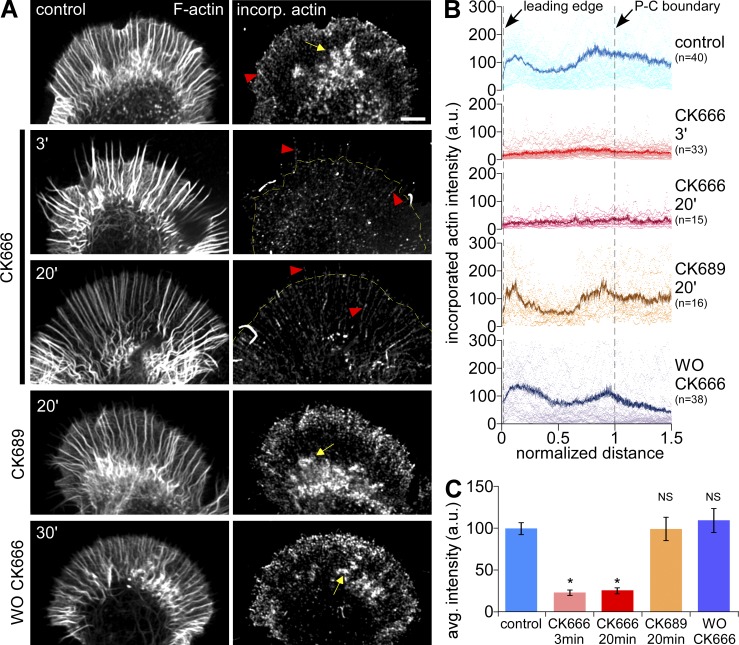
We used Alexa 488 actin monomer incorporation (Symons and Mitchison, 1991; Chan et al., 1998) to measure free actin filament barbed-end densities in permeabilized growth cones. Under control conditions, free barbed ends were concentrated in a 3–5-µm band near the leading edge (Fig. 3 A, top) and also near the peripheral–central (P–C) boundary, where they appeared to be associated with intrapodia (Fig. 3 A, yellow arrows). The density of free barbed ends decreased as early as 3 min after 50 µM CK666 addition (Fig. 3, A and B, second and third panels). Population analysis indicated that the density of barbed ends in the distal half of P-domain decreased by ∼75% after Arp2/3 complex inhibition (Fig. 3 C). Barbed-end distribution patterns recovered after CK666 washout, and the inactive analogue, CK689, had no effect on bard-end density or localization (Fig. 3, A and B [bottom two panels], and C). Note that polymerization-competent barbed ends associated with filopodial actin bundles did not appear to be affected by CK666 (Fig. 3 A, red arrowheads). These results are consistent with our electron microscopy and actin-labeling data (Fig. 2), which showed a dramatic clearance of peripheral actin veil structures after CK666 treatment. Together, these results suggest Arp2/3 complex actin nucleation significantly contributes to generation of actin veil assembly sites near the growth cone leading edge.
Figure 3.
Arp2/3 complex inhibition reduces F-actin barbed end density along the leading edge. (A) Growth cones were dual-labeled with TRITC-phalloidin to show total F-actin (left) and Alexa 488 G-actin, which incorporates at barbed ends (right). Growth cones were treated with vehicle (DMSO, 20 min, top), CK666 (50 µM, 3 or 20 min), CK689 (100 µM, 20 min), or CK666 (50 µM, 20 min) followed by washout for 30 min. Yellow dotted line demarcates the leading edge. Arrow, intrapodia; arrowheads, barbed ends on filopodia. (B) Line scan analysis of barbed-end localization in growth cones under each condition in A. Scattered dots represent dataset from individual growth cones. Solid lines represent the population average. n, growth cones measured. (C) Average barbed-end intensities in the distal half of the P-domain in each condition. *, P < 0.01 with two-tailed unpaired t test versus control; NS, not significant.
Arp2/3 complex inhibition increases retrograde actin flow rates with little effect on net actin turnover
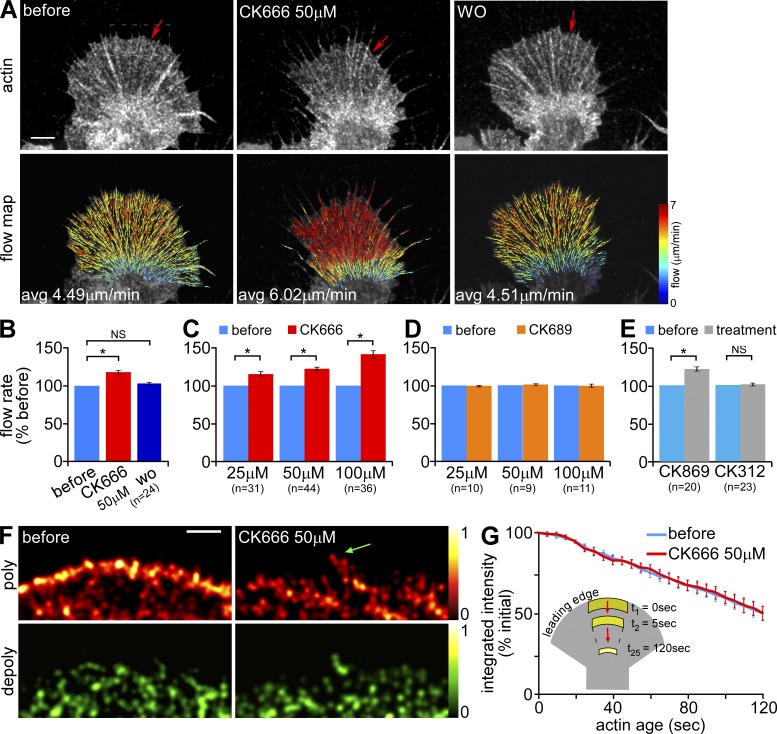
We used quantitative fluorescence speckle microscopy (Danuser and Waterman-Storer, 2006) to characterize actin filament dynamics before and after Arp2/3 complex inhibition. Neurons were injected with trace levels of either Alexa 568 actin monomers or Alexa 594 phalloidin to generate actin filament speckles for tracking over time using a quantitative cross-correlation approach (Ji and Danuser, 2005; Burnette et al., 2007; Hu et al., 2007). Calculated retrograde actin flow velocities were pseudocolor-coded and corresponding vectors overlaid on images to illustrate actin translocation (Fig. 4 A, flow map).
Figure 4.
Under control conditions, Arp2/3 complex inhibition increases peripheral retrograde actin flow but has little effect on actin turnover. (A) Representative G-actin FSM images (top) and corresponding flow maps (bottom) from a growth cone before, 20 min after treatment in 50 µM CK666, and 15 min after washout of CK666. Arrows in FSM images mark the edge of the actin veil. On the flow maps, colors encode speed (see color bar) and vectors indicate flow direction. (B–E) Summary of relative changes in retrograde actin flow rates after manipulations. (B) CK666 reversibility: before, during 50 µM CK666 treatment (15–30 min), and after washout (10–30 min). (C and D) Concentration dependence: treatment for 15–30 min with various concentrations of (C) CK666 or (D) CK689. (E) Treatment for 15–30 min with 50 µM CK869 or CK312. n, growth cones measured. (F) Map of time-averaged assembly (red) and disassembly (green) events detected near the leading edge of a growth cone before and after CK666 (50 µM, 20 min). Images were sampled from a region similar to the dotted blue box in A. Colors indicate relative assembly or disassembly rates (see color bars). Green arrow, polymerization sites on filopodia. (G) Plot of changes in integrated fluorescent intensity within the flow-displaced regions (see inset) tracked by ROI-Based Turnover Analysis before and after CK666 (50 µM, 20 min). n, 9 growth cones, 3–5 ROIs per growth cone. Images acquired every 5 s with 2 min elapsed recording time. Bars, 5 µm.
Treatment with 50 µM CK666 for 20 min significantly increased peripheral speckle flow rates and resulted in actin veil retraction (Fig. 4 A, left vs. middle panels, red arrow; Video 1). CK666 effects were reversible and retrograde flow rates returned to control levels after 10–30 min washout (Fig. 4, A and B). Individual examples (Fig. S4, A and B) and population analysis showed that after 15–30 min of exposure to CK666, flow rates increased in proportion to drug concentration: 14.9 ± 3.0% increase with 25 µM, 21.9 ± 2.3% with 50 µM, and 41.2 ± 4.7% with 100 µM (Fig. 4 C). In contrast, 15–30 min of treatment with CK689 at corresponding concentrations had no effect on flow rates (Fig. 4 D). The CK666 concentration-response trend and reversibility were consistent with antibody staining and electron microscopy results (Figs. 1 and 2).
As an alternative approach, we treated growth cones with CK869, a different class of inhibitor, which binds the Arp2/3 complex at a site distinct from CK666 (Nolen et al., 2009). Treatment with 50 µM CK869 for 15–30 min increased peripheral flow rates by 21.3 ± 2.7%; in contrast, CK312, the inactive structural analogue, had no effect (Fig. 4 E; see Fig. S4 C for CK869 flow maps). Like CK666, CK869 delocalized Arp2/3 complex from the leading edge (Fig. S1 E, bottom; see the top panel for inactive control). Taken together, these results provide strong evidence that acute Arp2/3 complex inhibition results in acceleration of retrograde actin filament flow.
We used single-speckle tracking to characterize actin filament polymerization and depolymerization rates in the P-domain (Ponti et al., 2004, 2005). Under control conditions, strong polymerization, indicated by a high density of speckle “birth” events, occurred in a narrow zone along the leading edge (Fig. 4 F, top left). Treatment with 50 µM CK666 for 20 min markedly decreased the number of speckle birth events recorded in the same sampling interval, obscuring the obvious polymerization band observed under control conditions (Fig. 4 F, top right). A punctate pattern of actin polymerization remained, some of which appeared to be associated with filopodial bundles (Fig. 4 F, green arrow).
Two lines of evidence suggest CK666 did not significantly affect actin filament turnover (life times) in the P-domain. First, speckle “death” event density, which is an indicator of filament depolymerization, appeared to be unaffected by CK666 treatment (Fig. 4 F, bottom). Second, we recently developed an algorithm (Van Goor et al., 2012) that estimates net actin turnover by tracking integrated actin speckle intensity within an ROI as it moves with retrograde flow across the P-domain. Fig. 4 G shows that the integrated actin filament intensity within the ROI steadily decreased to ∼53.0% of its initial value in 2 min either under control conditions or in the presence of 50 µM CK666 (Fig. 4 G). These results indicate that Arp2/3 complex inhibition reduces the frequency of actin assembly events near the leading edge, consistent with the actin incorporation assay results in Fig. 3, but does not appear to have a marked effect on the rate of filament depolymerization or the rate of network turnover.
Arp2/3 complex inhibition effects on retrograde flow are reversed when myosin II activity is attenuated
Previous studies in growth cones suggest retrograde actin flow depends both on pulling forces generated by myosin II localized to the transition zone and pushing forces derived from actin assembly near the leading edge (Forscher and Smith, 1988; Lin et al., 1996; Henson et al., 1999); however, maximal rates of retrograde flow depend on myosin II activity (Medeiros et al., 2006). Arp2/3 complex–dependent actin polymerization is thought to contribute to retrograde flow by generating actin assembly–dependent pushing forces at the leading edge (Mogilner and Oster, 2003; Bugyi and Carlier, 2010) and resultant actin filament array treadmilling (Pollard and Borisy, 2003). Our initial expectation was that decreased polymerization after Arp2/3 complex inhibition would decrease retrograde flow rates—the opposite of what was observed after CK666 exposure (Fig. 4, A–C and E; Fig. S4, A–C). Given this paradox, we investigated a possible role for myosin II in driving faster flow rates in CK666-treated cells. To this end, we first inhibited myosin II activity using blebbistatin, a specific myosin II ATPase inhibitor (Straight et al., 2003; Allingham et al., 2005; Medeiros et al., 2006), and then assessed residual effects of Arp2/3 complex inhibition.
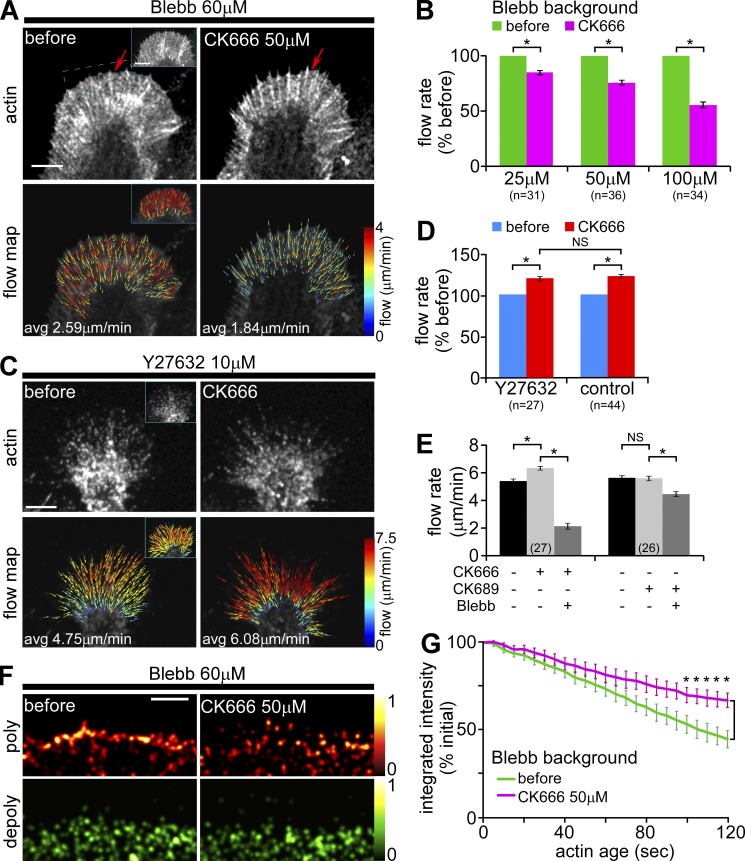
When neurons were pretreated with 60 µM blebbistatin for 10–15 min, flow rates decreased by ∼22% and were maintained by actin assembly–dependent array treadmilling as previously described (Fig. 5 A, inset vs. left; Medeiros et al., 2006). Addition of CK666 now resulted in concentration-dependent decreases in retrograde actin flow (Fig. 5 A; Fig. S4, D and E). Population analysis indicated that Arp2/3 complex–dependent networks contribute roughly 50% of the driving force for retrograde network flow when myosin II pulling force is absent (Fig. 5 B). The combined effects of CK666 plus blebbistatin on retrograde actin flow were completely reversible (unpublished data). In addition, actin veils did not retract under these conditions (Fig. 5 A, also Video 2; for comparison with controls, see Fig. 4 A and Video 1), implicating myosin II in the veil retraction process.
Figure 5.
Under conditions of low myosin II activity, Arp2/3 complex inhibition decreases peripheral retrograde actin flow rates. (A) Representative G-actin FSM images (top) and corresponding flow maps (bottom) from a growth cone before and after CK666 (50 µM, 20 min) in the presence of blebbistatin (60 µM, 10 min pretreatment). Arrows in FSM images mark the edge of the actin veil. Insets, control actin and flow map recorded before blebbistatin addition. (B) Summary of relative changes in retrograde actin flow rates in response to increasing concentrations of CK666 (15–30 min) in the presence of blebbistatin. Flow rate decreases: 14.8 ± 1.2% with 25 µM, 24.5 ± 2.3% with 50 µM, and 43.0 ± 2.6% with 100 µM CK666. n, growth cones measured. (C) Representative phalloidin-FSM images (top) and corresponding flow maps (bottom) from a growth cone before and after CK666 (50 µM, 20 min) in the presence of Y27632 (10 µM, 20 min pretreatment). Average flow rate before: 4.75 µm/min; CK666: 6.08 µm/min. Insets: control actin and flow map recorded before Y27632 addition, average flow rate 4.73 µm/min. (D) Retrograde actin flow rates still increased by 19.6 ± 2.6% in response to CK666 (50 µM, 15–30 min) in the presence of Y27632 (10 µM, 20 min pretreatment). Flow rates in response to the same concentration of CK666 under control conditions from Fig. 4 C are shown for comparison. (E) Changes in peripheral retrograde actin flow rates in response to blebbistatin after Arp2/3 complex inhibition. Growth cones were pretreated with CK666 or inactive analogue CK689 (50 µM, 15–30 min), then treated with blebbistatin (60 µM, 10–30 min) in the continued presence of either CK666 or CK689. Flow rates were assessed before drug addition, during CK pretreatment, and during cotreatment with CK and blebbistatin. Population averages are shown. CK666 treatment increased the retrograde flow by 19.3 ± 3.2%. CK689 did not alter retrograde flow. In a CK666 background, blebbistatin decreased the flow by 65.6 ± 3.6%. In a CK689 background, blebbistatin decreased the flow by 20.1 ± 2.0%. Numbers in parenthesis: growth cones measured. (F) Map of time-averaged assembly (red) and disassembly (green) events detected near the leading edge of a growth cone before and after CK666 (50 µM, 20 min) in the presence of blebbistatin. Images were sampled from a region similar to the dotted blue box in A. Colors indicate relative assembly or disassembly rates (see color bars). (G) Plot of change in integrated fluorescent intensity within the flow-displaced regions before and after CK666 (50 µM, 20 min) in the presence of blebbistatin. n, 6 growth cones, 3–5 ROIs per growth cone. *, P < 0.01 with two-tailed paired t test. Images acquired every 5 s with 2 min elapsed recording time. Bars, 5 µm.
A previous study suggested knockdown of Arp2/3 complex resulted in increased Rho activity (Korobova and Svitkina, 2008). Rho is known to increase myosin II regulatory light chain phosphorylation through Rho-kinase actions (Kimura et al., 1996; Totsukawa et al., 2000; Vicente-Manzanares et al., 2009). To test whether the increased flow rates we observed after Arp2/3 complex inhibition resulted from increased Rho-kinase activity, we inhibited Rho-kinase with Y27632 (Hirose et al., 1998). Y27632 treatment has no effect on peripheral actin flow in our Aplysia system; however, it strongly inhibits retrograde movement of actomyosin actin arcs in more central regions of the growth cone (Zhang et al., 2003). As expected, exposure to Y27632 alone did not change peripheral retrograde actin flow rates (Fig. 5 C, inset vs. left). After Y27632 pretreatment, 50 µM CK666 still increased peripheral retrograde flow rates by ∼20%—exactly the same effect we obtained with CK666 alone (Fig. 5, C and D). Additionally, in blebbistatin backgrounds, CK666 plus Y27632 decreased retrograde flow rates to the same levels as CK666 treatment alone (unpublished data). These results strongly suggest Rho-ROCK signaling is not involved in the flow rate increases we observe with Arp2/3 complex inhibition.
To further investigate how Arp2/3 complex–dependent actin networks affect myosin II function, we compared retrograde flow rates in cells exposed first to 50 µM CK666 or its inactive analogue, CK689, and then to blebbistatin (Fig. 5 E). As expected, flow rates did not change in cells pretreated with the inactive analogue, CK689 (Fig. 4 D; Fig. 5 E, right), whereas flow increased by ∼20% in cells treated with CK666 (as above; Fig. 5 E, left). Further exposure to blebbistatin decreased retrograde flow by >65% in neurons pretreated with CK666 (Fig. 5 E, left) but only decreased flow rates by ∼20% in cells pretreated with the inactive drug analogue (Fig. 5 E, right) or in control cells (not depicted). In summary, these results show that the presence of Arp2/3 complex–dependent actin networks constrains maximal retrograde flow rates under conditions of normal myosin II activity (Fig. 4, A–C and E); however, when myosin II activity is low, Arp2/3 complex–dependent actin assembly plays a significant role (∼50%; Fig. 5 B) in generating forces that drive retrograde network flow. Conversely, when Arp2/3 complex activity is low, retrograde actin flow is highly dependent on myosin II activity (Fig. 5 E).
Myosin II inhibition reveals Arp2/3 complex–dependent effects on actin turnover
We next investigated effects of Arp2/3 complex inhibition on actin turnover under conditions of low myosin II activity. After 10–15 min pretreatment with 60 µM blebbistatin, the strong band of polymerization at the leading edge observed under control conditions (Fig. 4 F) persisted (Fig. 5 F, top left). Addition of CK666 reduced the frequency of speckle birth events (Fig. 5 F, top right) as above (Fig. 4 F). Speckle analysis showed that actin depolymerization rates at the leading edge remained largely unaffected by CK666 (Fig. 5 F, bottom).
In blebbistatin alone we found integrated actin filament intensity within ROIs traversing the P-domain decreased to levels similar to controls (44.1 ± 5.0% over ∼2 min, Fig. 5 G, green line; Fig. 4 G). In contrast, after CK666 addition (50 µM) in the continued presence of blebbistatin there was a clear trend of slower actin turnover after Arp2/3 complex inhibition with integrated ROI actin intensity falling to only 66.6 ± 4.2% over the same period (Fig. 5 G, magenta line). Note that this effect was statistically significant only in the last 25 s of tracking, at which time ROIs had moved into the transition zone between P- and C-domains. More stable Arp2/3 complex–independent actin in the transition zone may be related to myosin II–dependent actin bundle turnover reported in this region (Medeiros et al., 2006). In summary, under conditions of low myosin II and Arp2/3 complex activities, residual actin flow is slower and residual actin networks appear to be more stable—especially in proximal growth cone regions.
Myosin II activity does not affect Arp2/3 complex localization and vice versa
Myosin II inhibition alone did not affect Arp2/3 complex enrichment near the leading edge (Fig. 6 A [top], B and C; for comparison with control, see Fig. 1 B, top); however, Arp2/3 complex localization was still sensitive to CK666 treatment (Fig. 6 A [bottom], B and C). Blebbistatin treatment alone did not affect the density or distribution of actin assembly sites assessed by labeled G-actin incorporation (Fig. 6, D and E [top panels] and F; Fig. 3, A and B, controls). In contrast, CK666 plus blebbistatin markedly reduced free barbed-end density in the peripheral veil and proximal transition zone regions, whereas a punctuate pattern of assembly sites remained on filopodial bundles (Fig. 6, D–F; red arrowheads). These data are in agreement with the effects of Arp2/3 complex inhibition on actin assembly in Fig. 5 F described above.
Figure 6.
Myosin II inhibition does not affect Arp2/3 complex localization or barbed-end distribution. (A) Fluorescent labeling of growth cones with Arp3 antibody (right) and Alexa 594 phalloidin (left) after normal fixation. Growth cones were treated with blebbistatin (60 µM, 20 min, top) or pretreated with blebbistatin (60 µM, 10 min) followed by blebbistatin and CK666 (100 µM, 20 min, bottom). (B) Arp3 distribution profile sampled from the designated lines in A. (C) Quantification of Arp2/3 complex enrichment at the leading edge for each condition in A. The control from Fig. 1 E is shown for comparison. Numbers in parentheses indicate growth cones measured. *, P < 0.01 with two-tailed unpaired t test. (D) Growth cones were dual-labeled with TRITC-phalloidin to show total F-actin (left) and Alexa 488 G-actin, which incorporates at barbed ends (right). Growth cones were treated with blebbistatin (60 µM, 20 min, top) or pretreated with blebbistatin (60 µM, 10 min) followed by blebbistatin and CK666 (50 µM, 20 min, bottom). Yellow dotted line demarcates leading edge. Arrowheads: filopodia. (E) Line scan analysis of barbed end localization in growth cones under each condition in D. Scattered dots represent dataset from individual growth cones. Solid lines represent the population average. n, growth cones measured. (F) Average barbed-end intensities in the distal half of the P-domain in each condition. Data for control and CK666 (50 µM, 20 min) from Fig. 3 C are shown for comparison. *, P < 0.01 with two-tailed unpaired t test. NS, not significant. Bars: (A) 10 µm; (D) 5 µm.
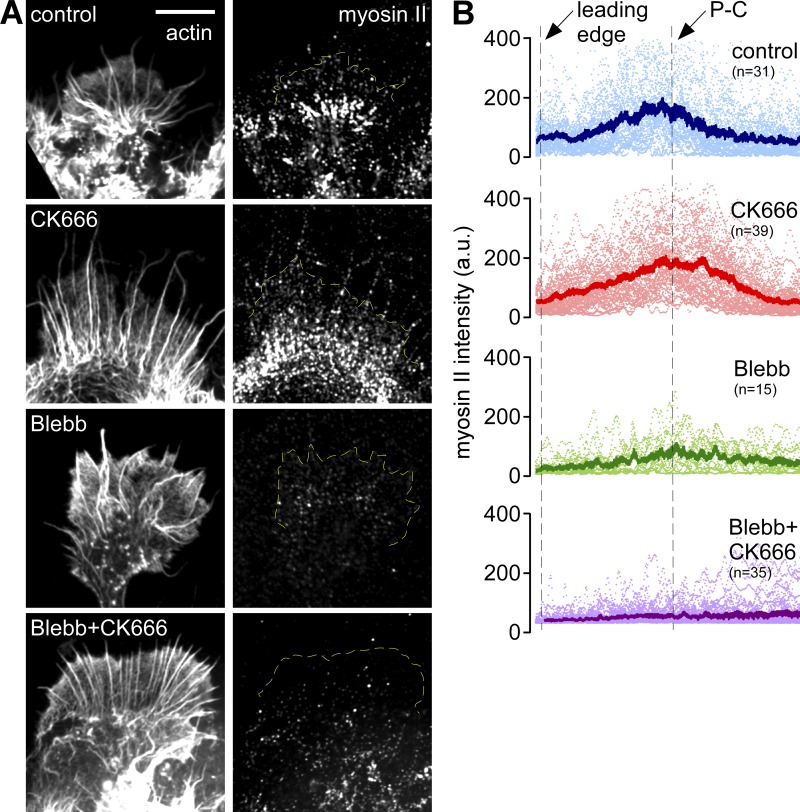
Arp2/3 complex inhibition also had little or no effect on myosin II localization, which remained concentrated in a band at the P–C boundary (Fig. 7, A and B, compare top two panels) as reported previously for growth cones under control conditions (Medeiros et al., 2006). As expected, blebbistatin delocalized myosin II from this region (Medeiros et al., 2006) and CK666 had no further effects (Fig. 7, A and B, bottom two panels). Taken together, these last two sets of results indicate that inhibition of myosin II or Arp2/3 complex alone has little or no effect on the other protein’s localization.
Figure 7.
Arp2/3 complex inhibition does not affect myosin II localization. (A) Fluorescent labeling of growth cones with myosin II antibody (right) and TRITC-phalloidin (left) after live cell extraction. Growth cones were treated with vehicle (DMSO, 20 min), CK666 (100 µM, 20 min), blebbistatin (60 µM, 20 min), or pretreated with blebbistatin (60 µM, 10 min) followed by blebbistatin and CK666 (100 µM, 20 min). (B) Line scan analysis of myosin II localization in growth cones under each condition in A. Line scans (50 pixels in width, 2x P-domain in length) were sampled along the growth axis of growth cones. Scattered dots represent dataset from individual growth cones. Solid lines represent the population average. n, growth cones measured. Yellow dotted lines demarcate leading edge. Bars, 10 µm.
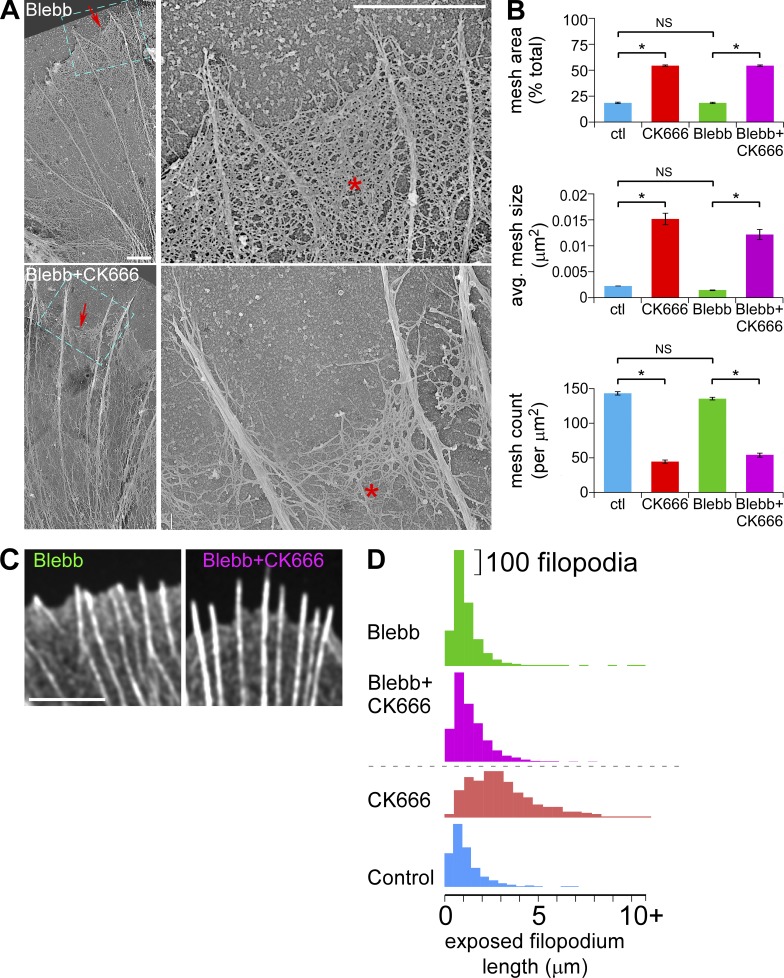
The ultrastructural changes in veil network mesh size observed after Arp2/3 complex inhibition (Fig. 2) were not significantly altered by lowering myosin II activity with blebbistatin (Fig. 8, A and B); however, veil retraction after CK666 treatment was attenuated (Fig. 8, C and D; Fig. S5, A–C). Thus, the primary effect of Arp2/3 complex inhibition is reduction of actin veil network density, whereas veil retraction appears to be a secondary effect mediated primarily by myosin II contractile activity.
Figure 8.
Myosin II inhibition attenuates veil retraction but does not affect reductions in actin veil network densities evoked by Arp2/3 complex inhibition. (A) Electron micrographs of growth cones treated with blebbistatin (60 µM, 20 min, top) or pretreated with blebbistatin (60 µM, 10 min) followed by blebbistatin and CK666 (50 µM, 20 min, bottom). Blue boxed areas on the left panels are presented in high magnification on the right. Asterisk, actin veil; arrows, edge of veils. (B) Quantification of actin veil network properties from 1 × 1 µm2 regions in distal P-domain of growth cones treated as in A. Data for control and CK666-treated growth cones from Fig. 2 G are shown for comparison. n, 45 regions from 3 GCs for blebbistatin; 42 regions from 3 GCs for blebbistatin with CK666. *, P < 0.01 with two-tailed unpaired t test versus control; NS, not significant. (C) Representative images of the leading edge of growth cones treated as in A. See Fig. S5 A for the entire growth cones. (D) Histograms of exposed filopodium lengths. CK666 and control from Fig. 2 E for comparison. See Fig. S5, B and C, for statistical analysis. Bars: (A, both panels) 2 µm; (C) 5 µm.
Discussion
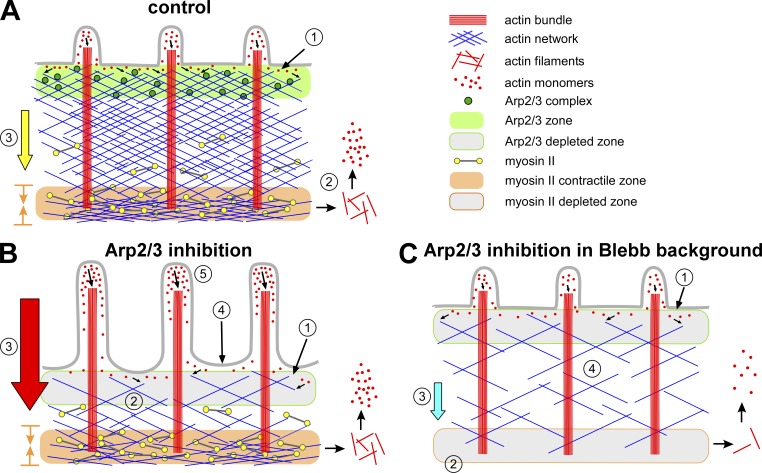
Both protrusive forces derived from actin polymerization and contractile forces produced by nonmuscle myosin II drive cell migration and growth cone advance (Cramer et al., 1994; Small et al., 1996; Borisy and Svitkina, 2000; Dent and Gertler, 2003; Lowery and Van Vactor, 2009; Vicente-Manzanares et al., 2009). The Arp2/3 complex is enriched in the P-domain near the leading edge of growth cones (Fig. 1; Mongiu et al., 2007; Korobova and Svitkina, 2008), where it generates sites for actin polymerization and is incorporated into the actin filament network (Figs. 1 and 3). Myosin II is localized in a dense band between the growth cone P- and C-domains where it is associated with actin filament structures (Fig. 7; Medeiros et al., 2006) and exerts contractile forces involved in retrograde flow (Lin et al., 1996), actin network recycling (Medeiros et al., 2006), retraction (Zhang et al., 2003), axon consolidation (Burnette et al., 2008), and engorgement (Schaefer et al., 2008). In the current study we investigated the effects of acutely inhibiting Arp2/3 complex activity in growth cones and report a role for Arp2/3 complex in regulation of actin filament veil structure and dynamics. We found that the outcome of Arp/2/3 inhibition strongly depends on preexisting levels of nonmuscle myosin II contractile activity, suggesting functional interactions between these two components. We discuss our results in the context of summary Fig. 9.
Figure 9.
Schematic summary. (A) Control. (B) Arp2/3 complex inhibition. (C) Arp2/3 complex inhibition in blebbistatin background.
Under control conditions, actin filament networks spanning the width of the growth cone P-domain assemble near the leading edge, where the Arp2/3 complex is enriched (Fig. 9 A, 1). Nonmuscle myosin II concentrates in a transition zone at the P–C domain boundary where it has been implicated in actin network recycling (2). A combination of pushing force derived from actin polymerization against the membrane and pulling force from actomyosin II contraction drives retrograde actin flow (3) (Lin et al., 1996; Forscher and Smith, 1988; Henson et al., 1999; Mogilner and Oster, 2003; Medeiros et al., 2006), which constantly moves actin networks rearward from the leading edge. Retrograde flow and actin filament turnover exist in a steady state that maintains the P-domain at relatively constant width by a process of actin network treadmilling (Van Goor et al., 2012).
Binding of CK666 to Arp2/3 complex inhibits the formation of actin filament branches on the sides of preexisting actin filaments, so Arp2/3 complex is no longer localized to the leading edge (Fig. 1; Fig. S2). As a result, actin filament barbed end production and net actin assembly rates decreased (Fig. 9 B, 1). CK666 had little or no effect on actin filament disassembly rates (Fig. 4 F). The combination of unchanged disassembly and slowed assembly rates results in a progressive decrease in P-domain actin filament density over time (Fig. 9 B, 2; Fig. 2; Fig. S2 F).
CK666 did not alter myosin II localization at the P–C boundary, where it exerts pulling forces involved in retrograde actin flow (Fig. 7; Medeiros et al., 2006). Interestingly, despite having essentially no effect on myosin II localization, inhibition of Arp2/3 complex significantly increased retrograde flow rates (Fig. 4; Fig. 9 B, 3) and induced actin veil retraction (Fig. 9 B, 4)—the latter likely resulting from the combined effect of decreased leading edge actin assembly and increased retrograde flow rates.
The increase in retrograde flow after Arp2/3 complex inhibition is interesting and somewhat unexpected. Arp2/3 complex knockdown with siRNA was reported to decrease retrograde flow levels in primary cultured hippocampal neurons and neuroblastoma cells (Korobova and Svitkina, 2008). In that study, retrograde flow rates were assessed indirectly by analysis of feature movements detected by phase microscopy, whereas we directly assessed actin filament movements by qFSM. We also note that the current study focused on acute effects of Arp2/3 complex inhibition using small molecule inhibitors, CK666 and CK869, whereas the study above analyzed long-term effects of p34-Arc or Arp3 knockdown. It is possible that the difference in flow measurement technique or the significantly different timeframe of our experiments (minutes vs. days) account for the apparent difference in our results. A more interesting alternative is that background myosin II activity was lower under their experimental conditions. Lower myosin II activity could invert the polarity of the response of retrograde flow to Arp2/3 complex inhibition and account for the slowing of actin flow estimated from their phase-contrast movies.
Arp2/3 complex inhibition has also been reported to elevate intracellular Rho activity (Korobova and Svitkina, 2008), raising the possibility that increased retrograde flow rates could result from Rho kinase–dependent elevation of nonmuscle myosin II activity. However, we observed the same CK666-dependent flow rate increases with or without Rho kinase inhibition (Fig. 5, C and D), ruling out this possibility. Moreover, increased Rho activity after Arp2/3 complex inhibition might lead to formin protein activation, resulting in increased unbranched actin filaments (Higgs, 2005; Kovar, 2006). Our results made this possibility less compelling because we obtained an overall concentration-dependent decrease in peripheral actin network density after CK666 treatments (Fig. 2). Considering that CK666 treatment decreased the density of the actin network (Fig. 2, F and G) without affecting myosin II localization (Fig. 7, A and B, second row), the scenario we favor for the increase in retrograde flow rates after Arp2/3 complex inhibition is related to the “gelation/solation model” for actomyosin network contraction (Janson et al., 1991; Kolega et al., 1991). This model suggests that decreases in actin network density tend to facilitate higher rates of myosin II–dependent network contraction as a consequence of less resistance to network compression. Alternatively, the geometry for myosin II interactions with actin filaments may be more favorable for contractile action after removal of branched Arp2/3 complex–dependent components.
In the absence of myosin II activity we found normal levels of leading edge actin assembly activity (Fig. 5 F), and no effect on Arp2/3 complex localization (Fig. 6, A and C) nor appreciable effects on actin barbed end density in P-domain (Fig. 6, D–F). This relative independence of Arp2/3 complex–dependent actin nucleation and assembly from myosin II function may help explain why growth cones are able to sustain high rates of retrograde actin network array treadmilling in the absence of myosin II activity (Medeiros et al., 2006). Indeed, we found that in the absence of myosin II activity ∼50% of residual retrograde flow depended on Arp2/3 complex activity (Fig. 9 C, 3; Fig. 5, A and B; Fig. S4, D and E). We note that when the Arp2/3 complex and myosin II were both inhibited, actin veils no longer retracted despite continued reduction in veil actin densities (Fig. 9 C, 4). These results are consistent with the system relaxing to a slower actin treadmilling steady state with reduced rates of leading edge assembly being matched by lower filament recycling rates (Fig. 9 C, 1 and 2).
It is notable that inhibition of Arp2/3 complex had little effect on actin filament bundles in filopodia (Fig. 2). Despite retraction of intervening veil networks, the length and number of filopodium bundles were unchanged (Fig. S3, D and E). These results suggest that filaments nucleated by Arp2/3 complex are not required to maintain filopodia, as proposed for other cells (Korobova and Svitkina, 2008; Norris et al., 2009; Spillane et al., 2011). Although existing filopodium bundles in mature growth cones were resistant to CK666, the current study does not formally rule out a role for Arp2/3 complex in filopodium initiation.
Note that after CK666 or CK869 treatment, retrograde flow rates increased both in filopodial actin filament bundles and in isotropic actin filament networks (Fig. 4 A, left and middle; Fig. S4, A–C). Considering that filopodia length remained constant (Fig. S3 D), these results indicate that filopodium polymerization rates increased to match the faster retrograde flow rates after Arp2/3 complex inhibition. Inhibition of Arp2/3 complex decreased the number of actin nucleation sites near the leading edge (Fig. 3), which might increase the concentration of actin monomers available at filopodium assembly sites (Fig. 9 B, 5) and account for the faster assembly rates observed. Had this kinetic compensation not occurred, filopodia would have retracted in concert with the actin veils.
In addition to filopodia, some actin filaments in the veil region persisted during Arp2/3 complex inhibition (Fig. 2). These results strongly suggest the presence of a population of actin filaments nucleated independent of the Arp2/3 complex. The obvious actin nucleation candidates are formin family proteins (Chhabra and Higgs, 2007; Pollard, 2007; Campellone and Welch, 2010), which have been reported to contribute to both lamellipodia motility by generating long filaments (Yang et al., 2007) and central contractility by making filaments that preferentially associate with myosin II (Tojkander et al., 2011). Our results suggest that actin filaments formed independent of the Arp2/3 complex depend strongly on myosin II function for translocation because retrograde flow was highly sensitive to blebbistatin treatment after inhibition of Arp2/3 complex (Fig. 5 E, left). In addition, in blebbistatin backgrounds, turnover of the Arp2/3 complex–independent filament population was significantly slower in the transition zone (Fig. 5 G) where myosin II is normally localized. These results suggest a role for myosin II in recycling actin filaments in the transition zone in line with previous observations (Medeiros et al., 2006).
In summary, we report that despite the spatial segregation of the Arp2/3 complex and myosin II in neuronal growth cones, they both contribute to regulation of actin veil dynamics and structure. Our results suggest that physiological activation of the Arp2/3 complex could promote growth cone advance by: (1) promoting leading edge actin assembly to affect veil protrusion; and (2) placing constraints on the ability of myosin II to drive retrograde flow. Conversely, Arp2/3 complex inhibition is expected to increase retrograde flow and decrease actin veil assembly rates leading to veil-specific retraction. Localized activation or inhibition of the Arp2/3 complex could meditate growth cone turning and/or avoidance in response to chemotropic guidance cues via the above effects on actin dynamics and contractility. In a broader context, it will be interesting to see if the inverse relationship between Arp2/3 complex activity and myosin II contractile efficiency described here is present in other motile cell types.
Materials and methods
Cell culture and chemicals
Primary culture of Aplysia bag cell neurons as described previously (Forscher and Smith, 1988) with the following modifications. In brief, abdominal ganglia were incubated at 22°C for ∼18 h in L-15 artificial sea water (ASW: 400 mM NaCl, 10 mM KCl, 15 mM Hepes, pH 7.8, 10 mM CaCl2, and 55 mM MgCl2), supplemented with 10 mM N-tert-butyl-α-phenylnitrone (PBN) and ∼10 mg/ml dispase. Connective tissue was manually removed and the intact bag cell cluster was triturated gently with a bent 20-µl pipette tip to separate the neurons. The isolated neurons were arranged on coverslips. These coverslips were acid washed, pretreated with 20 µg/ml poly-l-lysine for 15 min, and then covered by 50 µg/ml laminin for 2 h. A 1:10,000 dilution of fetal bovine serum was added to the culture medium 2 h after plating the neurons. CK666 was purchased from ChemDiv, Inc. CK689, CK869, CK312, blebbistatin, and Y27632 were purchased from EMD. Actin monomers purified from rabbit muscle and tagged with Alexa 568 or Alexa 488, Alexa 594 phalloidin, Alexa 488 goat anti–rabbit IgG, and Alexa 647 goat anti–rabbit IgG were from Invitrogen. Anti-Arp3 rabbit polyclonal antibody was from Millipore. Anti-Arp2 rabbit polyclonal antibody was from Abcam. DMSO and other chemicals were from Sigma-Aldrich unless otherwise stated.
Solutions
Low ionic strength artificial seawater (LIS-ASW) contained 100 mM NaCl, 10 mM KCl, 15 mM Hepes, 5 mM CaCl2, 5 mM MgCl2, and 628 mM betaine at pH 7.8, and was supplemented with 3 mg/ml BSA, 0.5 mM Vitamin E, and 1 mg/ml carnosine before experiments. Lysis buffer for homogenizing Aplysia CNS tissue contained 50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EGTA, 5 mM dithiothreitol with 0.01 mg/ml aprotinin, 0.01 mg/ml leupeptin, 5 µg/ml pepstatin, 0.4 µg/ml Pefabloc, and 1 mM Na3VO4. Tris-buffered saline (TBS) contained 25 mM Tris base, 137 mM NaCl, and 2 mM KCl. Live cell extraction buffer (LE) contained 100 mM Pipes, pH 6.9, 10 mM KCl, 100 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 4% PEG, average MW = 35,000, and 20% sucrose at ∼1,000 mOsm/L and was supplemented with 1% Triton X-100, 10 µM phalloidin, 10 µM taxol, and 10 mg/ml BAPTA immediately before use. Washing buffer for live cell extraction (WLE) contained 80 mM Pipes, pH 6.9, 5 mM EDTA, and 5 mM MgCl2. Block solution for antibody labeling was 5% BSA in PBS containing 0.1% Triton X-100 (PBS-T). MOWIOL media (EMD) for cell mounting in immunocytochemistry was prepared following the manufacturer’s protocol with 10% MOWIOL, 25% glycerol, 0.2 M Tris, pH 8.5, and 20 mM n-propyl-galate.
Western blotting and antibody preabsorption
CNS tissue from each adult Aplysia was homogenized in 1 ml ice cold lysis buffer with a Rotor Stator and SDS was added to a final concentration of 1%. Lysates were cleared by centrifugation, boiled for 3 min, resolved on 12% SDS-PAGE, and transferred to nitrocellulose membranes (GE Healthcare) by semi-dry transfer (TransBlot SD; Bio-Rad Laboratories). After probing with primary and secondary antibodies, the membrane was developed by electrochemiluminescence (Thermo Fisher Scientific) and digitally exposed using the Epi Chemi II Darkroom (UVP Laboratory Products).
For preabsorption experiments, purified bovine Arp2/3 complex (Robinson et al., 2001) was coupled to CNBr Sepharose 4B resin (Sigma-Aldrich). In brief, 0.1 g of resin was swelled in 20 ml of 1 mM HCl and washed on a sintered glass funnel. Final volume of swelled resin was 500 µl. The resin was incubated with 2.2 ml of 0.1 mg/ml Arp2/3 complex in coupling buffer (0.1 M NaHCO3 and 0.5 M NaCl, pH 8.3) overnight at 4°C. Unbound protein was removed by centrifugation and extra reactive sites were blocked by incubation with 1 M ethanolamine (pH 8). The resin was washed in a small column with 10 ml of TBS (pH 7.4) followed by three alternative washes with 5 ml 0.1 M sodium acetate (pH 4.0) and 5 ml 0.1 M Tris-HCl (pH 8.0) in 0.5 M NaCl. Purified anti-Arp3 IgG in 1.5 ml TBS were incubated with the resin overnight at 4°C. The concentration ratio of antigen to IgG was ∼10:1. Supernatant after preabsorption was used for Western blotting and immunocytochemistry.
Microinjection
Microinjection was performed using an inverted microscope (TE300; Nikon), equipped with a Quantix camera (Photometrics) and a microinjection system (Lin et al., 1996; Schaefer et al., 2002). In brief, cell bodies were impaled with a glass pipette mounted on a 3D hydraulic micromanipulator (model MMO-203; Narishige) retrofitted with a piezoelectric axial drive (model PZ-100; Buleigh Instruments). Stage position was controlled by a Newport Programmable Motion Controller (model SMC 100 series), Newport actuators (model LTA-HS), and custom control and position recording software. Neurons were microinjected with Alexa 568 actin monomers (needle concentration 0.4 mg/ml) or Alexa 594 phalloidin (needle concentration 20 µM). Reagent solution injections were typically ∼10% of cell volume. After microinjection, cells were incubated in culture medium 1 h before imaging.
Immunocytochemistry
Cells were fixed with 4% formaldehyde and 400 mM sucrose in LIS-ASW and permeabilized with 1% Triton X-100 (Forscher and Smith, 1988). Actin filaments were stained with a 1:50 dilution of Alexa 594 phalloidin stock in PBS-T. For antibody labeling, cells were blocked for 20 min with the blocking solution, incubated with 67 µM primary antibody for 30 min, washed three times with blocking solution, and incubated for 15 min with 130 µM secondary antibody. Cells were then washed three times in PBS-T and mounted in MOWIOL.
Live cell extractions were performed as described previously (Schaefer et al., 2002; Medeiros et al., 2006; Van Goor et al., 2012) with modifications. Extraction with LE for 3–5 min was followed by washing with WLE for 2 min. Free barbed ends were labeled by incubating cells with 350 nM of Alexa 488 actin monomers in WLE for 1 min and washed again (Symons and Mitchison, 1991). Cells were fixed for 20 min in 4% formaldehyde in WLE for fluorescence or 6.5% glutaraldehyde in WLE for electron microscopy. Extracted cells were labeled with antibodies as above.
Platinum/palladium replica electron microscopy
Cells were live extracted and fixed as described above followed by treatment with 6.5% glutaraldehyde in water, 0.2% tannic acid, and 0.2% uranyl acetate for 20 min each with water washes in between (Svitkina et al., 1995). Samples were dehydrated with a graded series of ethanol concentrations before critical point drying. Specimens were rotary shadowed with platinum/palladium at a 45° angle to a thickness of 2 nm followed by carbon coating. Replicas were mounted on carbon/formvar-coated EM grids and observed by transmission EM (EM-900; Carl Zeiss) at 80 kV.
To quantify actin veil network properties, 1 × 1-µm2 regions near the leading edge of cells in electron micrographs at 20,000x were subjected to an intensity threshold process until all negative spaces (meshes) among the actin filaments were selected. The resulting images were calculated with the “analyze particle” function in ImageJ (open source; National Institutes of Health). For each condition, 10–15 nonoverlapping regions from 3–8 growth cones were analyzed. Regions containing filopodial bundles or intrapodia were excluded.
Confocal microscopy
Fluorescent images were acquired using a spinning disk confocal system (Revolution XD; Andor) with a CSU-X1 confocal head (Yokogawa) mounted on an inverted microscope (TE 2000E; Nikon) with Perfect Focus, using an EMCCD camera (iXonEM +888; Andor). Transillumination was provided by a halogen lamp and controlled by a SmartShutter (Sutter Instrument). Confocal excitation was provided by an Andor laser combiner with three laser lines at 488, 561, and 647 nm. Emission wavelength was controlled using a filter wheel (LB10W-2800; Sutter Instrument) outfitted with bandpass filters from Chroma Technology Corp. A Nikon CFI Plan Apo 100x, 1.4 NA objective was used. Image acquisition and all other peripherals were controlled by Micro-Manager open source microscopy software (Edelstein et al., 2010) with a Matlab interface (MathWorks). Imaging medium was LIS-ASW at room temperature.
Quantification of actin dynamics
Fluorescent speckle microscopy images of cells injected with Alexa 488 actin monomers incorporated into filaments or low levels of Alexa 594 phalloidin, which specifically binds actin filaments, were acquired using 500–700 ms integration. Kymography (Zhang et al., 2003) and automated speckle tracking with the fsmCenter suite of custom Matlab algorithms produced by the Danuser laboratory were used to determine rates of actin filament movement. In fsmCenter, an adaptive multi-frame correlation algorithm (Ji and Danuser, 2005) was run on every five frames to determine the average flow over five frames. This information was used to initialize a single-particle tracking algorithm (Thomann et al., 2003; Vallotton et al., 2003; Ponti et al., 2004), which was run on every frame in the movie. Results of the single-particle tracking were used to generate time-averaged actin assembly–disassembly kinetic maps (Ponti et al., 2003, 2004, 2005) and analyze net population turnover with a region of interest (ROI)–based algorithm developed by D. Van Goor (Van Goor et al., 2012).
The ROI-based turnover algorithm is based upon conservation of mass principles. In brief, spatially averaged single-particle tracking vectors are used to track a user-defined ROI as it is displaced by retrograde flow from the leading edge to the peripheral–central interface. The flow-displaced ROI mimics a closed system, assuming that all assembled actin initially identified at the leading-edge moves with retrograde flow. Changes in the integrated intensity within the ROI reflect the shifting balance between assembly and disassembly over time. 3–5 regions in each growth cone were tracked for 2 min under both control and drug-treated conditions. Intensity changes were normalized to the first frame.
Image processing
Fluorescent images were subjected to background subtraction before quantitative intensity analysis. The only exception was automated speckle tracking, which was performed upon raw image data. For display only, fluorescent images were convolved with a Gaussian kernel, processed with an unsharp mask, and scaled according to a linear look-up table. For display of barbed-end images, areas outside of the growth cones were cleared with a mask. EM images were inverted and processed with pseudo-flat-field to eliminate artifacts of uneven illumination, followed by an unsharp mask. Image processing was performed in ImageJ.
Line scan analysis
Line scans were used to analyze the spatial intensity distribution of fluorescent probes. A 50-pixel-wide line was drawn either as illustrated in the figures or from the leading edge to 1.5x or 2x of peripheral domain (P-domain) width along the presumed growth axis of growth cones. The average intensity was measured with the plot profile function in ImageJ and the data exported to Excel (Microsoft). Intensity was plotted against either actual distance from the leading edge for single growth cone line scans or distance normalized to growth cone size for population line scans. When distance is normalized, the beginning (left end) of the line scan is marked as the leading edge, and 1/2 (if total length = 2x P width) or 2/3 (if total length = 1.5x P width) position is marked as the peripheral–central interface.
Quantification of Arp2/3 complex enrichment
To quantify Arp2/3 complex enrichment along the leading edge, the peripheral domain was divided into eight equal slices parallel to the leading edge, and the ratio of the average Arp3 intensity in slice 1 versus that in slice 4 was calculated as the enrichment factor (Fig. 1 D, inset).
Statistical analysis
Statistical analysis with paired or unpaired t test was performed in Excel (Microsoft) and the Kolmogorov-Smirnov test was performed in Matlab (Mathworks), with significance established at P < 0.01. Curve fitting for CK666 dose-response was done in ReaderFit (MiraiBio) according to the four-parameter nonlinear regression model: F(x) = ((a–d)/(1+((x/c)^b)))+d, where a is the minimum asymptote, b is the steepness of curve, c is the inflection point, and d is the maximum asymptote.
Online supplemental material
Fig. S1 shows verification of Arp2/3 complex localization in neuronal growth cones. Fig. S2 shows that CK666 treatment dose-dependently and reversibly delocalizes Arp2/3 complex from the leading edge and induces changes in growth cone ultrastructure. Fig. S3 shows that the Arp2/3 complex inhibitor CK666 reversibly retracts actin veil but does not affect filopodium length or number. Fig. S4 shows that Arp2/3 complex inhibition increased peripheral F-actin retrograde flow rate in a myosin II–dependent manner. Fig. S5 shows that myosin II inhibition attenuates veil retraction evoked by Arp2/3 complex inhibition. Video 1 shows that CK666 (50 µM, 20 min) increases the peripheral retrograde F-actin flow rate. Video 2 shows that CK666 (50 µM, 20 min) decreases the peripheral retrograde F-actin flow rate in blebbistatin backgrounds. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111052/DC1.
Supplementary Material
Acknowledgments
The authors thank Dr. Gaudenz Danuser (Harvard Medical School) for providing the actin flow tracking software, Dr. Barry Piekos for assisting in utilizing EM facilities, Dr. David van Goor for developing the ROI-based actin turnover algorithm, and Forscher laboratory members for insightful discussion of this work.
This work was supported by National Institutes of Health grants RO1-NS28695 and RO1-NS051786 to P. Forscher, GM-026338 to T.D. Pollard, Yale Medical Scientists Training Program TG-T32GM07205, and Nikon Partners-in-Research Program.
The authors have no commercial affiliations which would pose a conflict of interest with respect to this research.
Footnotes
Abbreviations used in this paper:
- C-domain
- central domain
- P-domain
- peripheral domain
- qFSM
- quantitative fluorescent speckle microscopy
- ROI
- region of interest
References
- Ahmad F.J., Hughey J., Wittmann T., Hyman A., Greaser M., Baas P.W. 2000. Motor proteins regulate force interactions between microtubules and microfilaments in the axon. Nat. Cell Biol. 2:276–280 10.1038/35010544 [DOI] [PubMed] [Google Scholar]
- Allingham J.S., Smith R., Rayment I. 2005. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 12:378–379 10.1038/nsmb908 [DOI] [PubMed] [Google Scholar]
- Bailly M., Macaluso F., Cammer M., Chan A., Segall J.E., Condeelis J.S. 1999. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 145:331–345 10.1083/jcb.145.2.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy G.G., Svitkina T.M. 2000. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12:104–112 10.1016/S0955-0674(99)00063-0 [DOI] [PubMed] [Google Scholar]
- Brown J.A., Wysolmerski R.B., Bridgman P.C. 2009. Dorsal root ganglion neurons react to semaphorin 3A application through a biphasic response that requires multiple myosin II isoforms. Mol. Biol. Cell. 20:1167–1179 10.1091/mbc.E08-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugyi B., Carlier M.F. 2010. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 39:449–470 10.1146/annurev-biophys-051309-103849 [DOI] [PubMed] [Google Scholar]
- Burnette D.T., Schaefer A.W., Ji L., Danuser G., Forscher P. 2007. Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat. Cell Biol. 9:1360–1369 10.1038/ncb1655 [DOI] [PubMed] [Google Scholar]
- Burnette D.T., Ji L., Schaefer A.W., Medeiros N.A., Danuser G., Forscher P. 2008. Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev. Cell. 15:163–169 10.1016/j.devcel.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K.G., Welch M.D. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11:237–251 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.Y., Raft S., Bailly M., Wyckoff J.B., Segall J.E., Condeelis J.S. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211 [DOI] [PubMed] [Google Scholar]
- Chhabra E.S., Higgs H.N. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9:1110–1121 10.1038/ncb1007-1110 [DOI] [PubMed] [Google Scholar]
- Cramer L.P., Mitchison T.J., Theriot J.A. 1994. Actin-dependent motile forces and cell motility. Curr. Opin. Cell Biol. 6:82–86 10.1016/0955-0674(94)90120-1 [DOI] [PubMed] [Google Scholar]
- Danuser G., Waterman-Storer C.M. 2006. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35:361–387 10.1146/annurev.biophys.35.040405.102114 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Gertler F.B. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 40:209–227 10.1016/S0896-6273(03)00633-0 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. 2010. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14:Unit14 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forscher P., Smith S.J. 1988. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107:1505–1516 10.1083/jcb.107.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forscher P., Kaczmarek L.K., Buchanan J.A., Smith S.J. 1987. Cyclic AMP induces changes in distribution and transport of organelles within growth cones of Aplysia bag cell neurons. J. Neurosci. 7:3600–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J.H., Svitkina T.M., Burns A.R., Hughes H.E., MacPartland K.J., Nazarian R., Borisy G.G. 1999. Two components of actin-based retrograde flow in sea urchin coelomocytes. Mol. Biol. Cell. 10:4075–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J.H., Kolnik S.E., Fried C.A., Nazarian R., McGreevy J., Schulberg K.L., Detweiler M., Trabosh V.A. 2003. Actin-based centripetal flow: phosphatase inhibition by calyculin-A alters flow pattern, actin organization, and actomyosin distribution. Cell Motil. Cytoskeleton. 56:252–266 10.1002/cm.10149 [DOI] [PubMed] [Google Scholar]
- Higgs H.N. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30:342–353 10.1016/j.tibs.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Hirose M., Ishizaki T., Watanabe N., Uehata M., Kranenburg O., Moolenaar W.H., Matsumura F., Maekawa M., Bito H., Narumiya S. 1998. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol. 141:1625–1636 10.1083/jcb.141.7.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Ji L., Applegate K.T., Danuser G., Waterman-Storer C.M. 2007. Differential transmission of actin motion within focal adhesions. Science. 315:111–115 10.1126/science.1135085 [DOI] [PubMed] [Google Scholar]
- Hubert T., Vandekerckhove J., Gettemans J. 2011. Actin and Arp2/3 localize at the centrosome of interphase cells. Biochem. Biophys. Res. Commun. 404:153–158 10.1016/j.bbrc.2010.11.084 [DOI] [PubMed] [Google Scholar]
- Iwasa J.H., Mullins R.D. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17:395–406 10.1016/j.cub.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson L.W., Kolega J., Taylor D.L. 1991. Modulation of contraction by gelation/solation in a reconstituted motile model. J. Cell Biol. 114:1005–1015 10.1083/jcb.114.5.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Danuser G. 2005. Tracking quasi-stationary flow of weak fluorescent signals by adaptive multi-frame correlation. J. Microsc. 220:150–167 10.1111/j.1365-2818.2005.01522.x [DOI] [PubMed] [Google Scholar]
- Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., et al. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 273:245–248 10.1126/science.273.5272.245 [DOI] [PubMed] [Google Scholar]
- Kolega J., Janson L.W., Taylor D.L. 1991. The role of solation-contraction coupling in regulating stress fiber dynamics in nonmuscle cells. J. Cell Biol. 114:993–1003 10.1083/jcb.114.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodney M.S., Elson E.L. 1995. Contraction due to microtubule disruption is associated with increased phosphorylation of myosin regulatory light chain. Proc. Natl. Acad. Sci. USA. 92:10252–10256 10.1073/pnas.92.22.10252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Svitkina T. 2008. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol. Biol. Cell. 19:1561–1574 10.1091/mbc.E07-09-0964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R. 2006. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18:11–17 10.1016/j.ceb.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Lai F.P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H.G., Stradal T.E., Dunn G.A., Small J.V., Rottner K. 2008. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27:982–992 10.1038/emboj.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C., Schlaepfer D., Ferrari A., Klingauf M., Grohmanova K., Veligodskiy A., Didry D., Le D., Egile C., Carlier M.F., Kroschewski R. 2007. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 282:426–435 10.1074/jbc.M607711200 [DOI] [PubMed] [Google Scholar]
- Lebrand C., Dent E.W., Strasser G.A., Lanier L.M., Krause M., Svitkina T.M., Borisy G.G., Gertler F.B. 2004. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 42:37–49 10.1016/S0896-6273(04)00108-4 [DOI] [PubMed] [Google Scholar]
- Lewis A.K., Bridgman P.C. 1992. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119:1219–1243 10.1083/jcb.119.5.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., Espreafico E.M., Mooseker M.S., Forscher P. 1996. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 16:769–782 10.1016/S0896-6273(00)80097-5 [DOI] [PubMed] [Google Scholar]
- Lowery L.A., Van Vactor D. 2009. The trip of the tip: understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10:332–343 10.1038/nrm2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M., Atkinson S.J., Ampe C., Vandekerckhove J., Pollard T.D. 1994. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 127:107–115 10.1083/jcb.127.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N.A., Burnette D.T., Forscher P. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:215–226 10.1038/ncb1367 [DOI] [PubMed] [Google Scholar]
- Mogilner A., Oster G. 2003. Polymer motors: pushing out the front and pulling up the back. Curr. Biol. 13:R721–R733 10.1016/j.cub.2003.08.050 [DOI] [PubMed] [Google Scholar]
- Mongiu A.K., Weitzke E.L., Chaga O.Y., Borisy G.G. 2007. Kinetic-structural analysis of neuronal growth cone veil motility. J. Cell Sci. 120:1113–1125 10.1242/jcs.03384 [DOI] [PubMed] [Google Scholar]
- Myers K.A., Tint I., Nadar C.V., He Y., Black M.M., Baas P.W. 2006. Antagonistic forces generated by cytoplasmic dynein and myosin-II during growth cone turning and axonal retraction. Traffic. 7:1333–1351 10.1111/j.1600-0854.2006.00476.x [DOI] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., Pollard T.D. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A.D., Dyer J.O., Lundquist E.A. 2009. The Arp2/3 complex, UNC-115/abLIM, and UNC-34/Enabled regulate axon guidance and growth cone filopodia formation in Caenorhabditis elegans. Neural Dev. 4:38 10.1186/1749-8104-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol R., Haeckel A., Ritter A., Qualmann B., Kessels M.M. 2007. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS ONE. 2:e400 10.1371/journal.pone.0000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36:451–477 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Ponti A., Vallotton P., Salmon W.C., Waterman-Storer C.M., Danuser G. 2003. Computational analysis of F-actin turnover in cortical actin meshworks using fluorescent speckle microscopy. Biophys. J. 84:3336–3352 10.1016/S0006-3495(03)70058-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti A., Machacek M., Gupton S.L., Waterman-Storer C.M., Danuser G. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786 10.1126/science.1100533 [DOI] [PubMed] [Google Scholar]
- Ponti A., Matov A., Adams M., Gupton S., Waterman-Storer C.M., Danuser G. 2005. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative Fluorescent Speckle Microscopy. Biophys. J. 89:3456–3469 10.1529/biophysj.104.058701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prass M., Jacobson K., Mogilner A., Radmacher M. 2006. Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 174:767–772 10.1083/jcb.200601159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R.C., Turbedsky K., Kaiser D.A., Marchand J.B., Higgs H.N., Choe S., Pollard T.D. 2001. Crystal structure of Arp2/3 complex. Science. 294:1679–1684 10.1126/science.1066333 [DOI] [PubMed] [Google Scholar]
- Rochlin M.W., Itoh K., Adelstein R.S., Bridgman P.C. 1995. Localization of myosin II A and B isoforms in cultured neurons. J. Cell Sci. 108:3661–3670 [DOI] [PubMed] [Google Scholar]
- Schaefer A.W., Kabir N., Forscher P. 2002. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158:139–152 10.1083/jcb.200203038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.W., Schoonderwoert V.T., Ji L., Mederios N., Danuser G., Forscher P. 2008. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell. 15:146–162 10.1016/j.devcel.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Welch M.D., Machesky L.M., Bridgman P.C., Meyer S.M., Cooper J.A. 1998. Visualization and molecular analysis of actin assembly in living cells. J. Cell Biol. 143:1919–1930 10.1083/jcb.143.7.1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J.V., Anderson K., Rottner K. 1996. Actin and the coordination of protrusion, attachment and retraction in cell crawling. Biosci. Rep. 16:351–368 10.1007/BF01207261 [DOI] [PubMed] [Google Scholar]
- Spillane M., Ketschek A., Jones S.L., Korobova F., Marsick B., Lanier L., Svitkina T., Gallo G. 2011. The actin nucleating Arp2/3 complex contributes to the formation of axonal filopodia and branches through the regulation of actin patch precursors to filopodia. Dev. Neurobiol. 71:747–758 10.1002/dneu.20907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., Mitchison T.J. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Strasser G.A., Rahim N.A., VanderWaal K.E., Gertler F.B., Lanier L.M. 2004. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 43:81–94 10.1016/j.neuron.2004.05.015 [DOI] [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., Borisy G.G. 1995. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J. Struct. Biol. 115:290–303 10.1006/jsbi.1995.1054 [DOI] [PubMed] [Google Scholar]
- Symons M.H., Mitchison T.J. 1991. Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114:503–513 10.1083/jcb.114.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S., Hellal F., Neukirchen D., Hindges R., Garvalov B.K., Flynn K.C., Stradal T.E., Chrostek-Grashoff A., Brakebusch C., Bradke F. 2010. Rac1 regulates neuronal polarization through the WAVE complex. J. Neurosci. 30:6930–6943 10.1523/JNEUROSCI.5395-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann D., Dorn J., Sorger P.K., Danuser G. 2003. Automatic fluorescent tag localization II: Improvement in super-resolution by relative tracking. J. Microsc. 211:230–248 10.1046/j.1365-2818.2003.01223.x [DOI] [PubMed] [Google Scholar]
- Tojkander S., Gateva G., Schevzov G., Hotulainen P., Naumanen P., Martin C., Gunning P.W., Lappalainen P. 2011. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21:539–550 10.1016/j.cub.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Totsukawa G., Yamakita Y., Yamashiro S., Hartshorne D.J., Sasaki Y., Matsumura F. 2000. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150:797–806 10.1083/jcb.150.4.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P., Ponti A., Waterman-Storer C.M., Salmon E.D., Danuser G. 2003. Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells: a fluorescent speckle microscopy study. Biophys. J. 85:1289–1306 10.1016/S0006-3495(03)74564-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor D., Hyland C., Schaefer A.W., Forscher P. 2012. The role of actin turnover in retrograde actin network flow in neuronal growth cones. PLoS ONE. 7:e30959 10.1371/journal.pone.0030959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., Horwitz A.R. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.D., DePace A.H., Verma S., Iwamatsu A., Mitchison T.J. 1997. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 138:375–384 10.1083/jcb.138.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.A., Tsuchida M.A., Allen G.M., Barnhart E.L., Applegate K.T., Yam P.T., Ji L., Keren K., Danuser G., Theriot J.A. 2010. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 465:373–377 10.1038/nature08994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Czech L., Gerboth S., Kojima S., Scita G., Svitkina T. 2007. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5:e317 10.1371/journal.pbio.0050317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.F., Schaefer A.W., Burnette D.T., Schoonderwoert V.T., Forscher P. 2003. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 40:931–944 10.1016/S0896-6273(03)00754-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.