EGF signaling activates Rab5c and promotes the intracellular association of AMAP1 and PRKD2 to enhance β1 integrin recycling and promote the invasiveness of breast cancer cells.
Abstract
Epidermal growth factor receptor (EGFR) signaling is one of the crucial factors in breast cancer malignancy. Breast cancer cells often overexpress Arf6 and its effector, AMAP1/ASAP1/DDEF1; in these cells, EGFR signaling may activate the Arf6 pathway to induce invasion and metastasis. Active recycling of some integrins is crucial for invasion and metastasis. Here, we show that the Arf6–AMAP1 pathway links to the machinery that recycles β1 integrins, such as α3β1, to promote cell invasion upon EGFR stimulation. We found that AMAP1 had the ability to bind directly to PRKD2 and hence to make a complex with the cytoplasmic tail of the β1 subunit. Moreover, GTP-Rab5c also bound to AMAP1, and activation of Rab5c by EGFR signaling was necessary to promote the intracellular association of AMAP1 and PRKD2. Our results suggest a novel mechanism by which EGFR signaling promotes the invasiveness of some breast cancer cells via integrin recycling.
Introduction
Invasive phenotypes and mechanisms are highly diverse among different types of tumors (Friedl and Wolf, 2003), and are also significantly diverse even among different breast cancer cell lines (Bowden et al., 2001). However, loss of sedentary epithelial phenotypes, i.e., loss of E-cadherin–based cell–cell adhesion and activation of some integrins, is a hallmark characteristic of the development of malignancy of most tumor cells with epithelial origin. Thus, identification of the signaling pathways and mechanisms that regulate the activities of integrins and E-cadherin is important for cancer cell biology.
Integrins are αβ heterodimeric receptors for extracellular matrices, and their downstream signaling pathways play pivotal roles in proliferation, survival, migration, and invasion of tumor cells as well as normal cells (Hynes, 2002). Among the different types of integrins, β1 integrins, such as α3β1, have been highly implicated in the development of the malignancy of many primary breast cancers (Coopman et al., 1996; Morini et al., 2000). Moreover, in a mouse model, the β1 subunit has been shown to be pivotal for the induction and progression of mammary tumors, and its absence results in a state of tumor cell dormancy (White et al., 2004).
Several integrins, including β1 integrins, recycle between endosomal compartments and the plasma membrane (Molnar et al., 1987; Bretscher, 1989, 1992). The recycling of integrins, which involves their endocytosis from the cell surface and recycling back to the plasma membrane, is important for their activities, such as cell migration (Bretscher, 1992), as well as for the regulation of their cell surface levels. Moreover, cancer cell invasion into the extracellular matrices generally involves active phagocytosis of the degraded matrix components by integrins (Coopman et al., 1996). Although different small GTPases and their regulators, as well as serine/threonine kinases, have been implicated in the intracellular trafficking and recycling of integrins (Ng et al., 1999; Roberts et al., 2001; Ivaska et al., 2002; Powelka et al., 2004; Woods et al., 2004; Li et al., 2005; Caswell et al., 2007, 2008; Caswell and Norman, 2008), the actual mechanisms by which such signaling molecules are involved in the intracellular dynamics of integrins still largely remain elusive. Moreover, mechanisms and factors involved in the intracellular trafficking of integrins appear to be diverse, and may be cell type– and context-dependent (also see Discussion).
Human breast cancer MDA-MB-231 cells provide an excellent model to study cancer invasion and metastasis (Bowden et al., 1999). α3β1 integrin is localized to invadopodia of MDA-MB-231 cells, which are specific protrusive structures invading into basement membranes (Coopman et al., 1996). Bowden et al. (1999) have shown that protein kinase Cµ (PKCµ), which has now been renamed protein kinase D1 (PKD1/PRKD1), and also several other proteins, such as cortactin and paxillin, localize to the invadopodia of MDA-MB-231 cells. Moreover, these proteins form a complex in MDA-MB-231 cells, and this complex formation appears to be necessary for the invasive activity (Bowden et al., 1999). PRKD family members consist of three isoforms, PRKD1–3, and are involved in intracellular trafficking as well as cell proliferation and apoptosis (Van Lint et al., 2002). PRKD1 has been shown to bind to the β3 integrin subunit, and this binding is involved in the PDGF-induced recycling of αvβ3 integrin from endosomes to the plasma membrane in NIH3T3 fibroblasts (Roberts et al., 2001; Woods et al., 2004). PRKD1 and PRKD2 have moreover been implicated in the basolateral sorting of β1 integrin and E-cadherin in polarized MDCK epithelial cells (Yeaman et al., 2004).
A small GTPase, Arf6, primarily regulates the recycling of plasma membrane components (Donaldson, 2003; D’Souza-Schorey and Chavrier, 2006). Arf6 activity has been implicated in β1 integrin recycling, although the precise mechanisms are unknown (Brown et al., 2001; Powelka et al., 2004; Dunphy et al., 2006). We have shown previously that Arf6 and one of its downstream effectors, AMAP1/DDEF1/ASAP1 (we call this protein AMAP1 here), are abnormally overexpressed in highly invasive breast cancer cells, including MDA-MB-231 cells, and that this contributes greatly to their invasive and metastatic activities (Sabe, 2003; Hashimoto et al., 2004; Onodera et al., 2005). Such overexpression of Arf6 and AMAP1 proteins in malignant cancer cells occurs at levels >10–20 fold higher than that in normal mammary epithelial cells and weakly or noninvasive breast cancer cells. However, EGF receptor (EGFR) is also frequently overexpressed in breast cancer and is highly implicated in malignancy (Blume-Jensen and Hunter, 2001; Hynes and Lane, 2005). We have shown that ligand-activated EGFR directly binds to GEP100 (Someya et al., 2001) to activate Arf6 (Morishige et al., 2008). Activated Arf6 (i.e., the GTP-bound form of Arf6) then recruits AMAP1 by direct binding (Hashimoto et al., 2005). Immunohistochemical analyses indicated that overexpression of AMAP1, as well as coexpression of EGFR and GEP100, correlate, with statistical significance, with the malignancy of primary ductal carcinomas of the human breast (Onodera et al., 2005; Morishige et al., 2008). Therefore, the EGFR–GEP100–Arf6–AMAP1 signaling pathway appears to be up-regulated in significant populations of malignant breast cancers, and used for their invasion and metastasis when EGFR is activated. The GEP100–Arf6–AMAP1 pathway is also activated by overexpressed Her2/ErbB2/Neu, one of the other members of the EGFR family, and this activation appears to be crucial for the distant metastases of lung adenocarcinomas (Menju et al., 2011). Moreover, angiogenesis also involves cell protrusion and invasion. Arf6 and AMAP1 are also highly expressed in vascular endothelial cells, and the GEP100–Arf6–AMAP1 pathway is directly activated by vascular endothelial cell growth factor receptor 2 (VEGFR2) as an integral part of angiogenesis (Hashimoto et al., 2011). However, the molecular mechanism by which the GEP100–Arf6–AMAP1 pathway functions to evoke invasive and metastatic activities, when activated by receptor tyrosine kinases, has not yet been identified.
AMAP1 is an integral component of invadopodia in MDA-MB-231 cells (Onodera et al., 2005). Here, we analyzed the mechanisms by which AMAP1 functions in cancer invasion by using MDA-MB-231 cells as a model. We show that AMAP1 plays a crucial role in EGF-induced recycling of β1 integrins, such as α2β1 and α3β1, through its binding to PRKD2 and hence complex formation with the cytoplasmic tail of the β1 subunit. We moreover show that activation of Rab5c by EGF stimulation, which is another binding partner of AMAP1, is crucial to promote the intracellular association of AMAP1 and PRKD2.
Results
AMAP1 forms a complex with PRKD2 in highly invasive breast cancer cells
Since the original study by Bowden et al. (1999), the primary structures of the PRKD family members have been characterized in detail (Sturany et al., 2001), and the anti-PKCµ antibody used in the original study was later found to be reactive against both PRKD1 and PRKD2. We found that MDA-MB-231 cells express PRKD2, but not PRKD1 (Fig. 1 A). Other breast cancer cell lines we examined all expressed PRKD2, but not necessarily PRKD1 (Fig. 1 A).
Figure 1.
Association of PRKD2 with AMAP1 in highly invasive breast cancer cells. (A) Lysates from different breast cancer cells and HMECs were probed with the indicated antibodies. (B) Lysates from the indicated cells were immunoprecipitated with an anti-AMAP1 antibody or an anti-AMAP2 antibody, then subjected to blotting with the indicated antibodies. Preimmune serum of the anti-AMAP1 antibody was used as a control (Pre-imm). WCL, whole cell lysate (5 µg). (C and D) MDA-MB-231 cells, expressing mVenus-PKD2 and cultured on Alexa Fluor–labeled gelatin film, were fixed and stained with an anti-AMAP1 antibody. Original colors were: PRKD2, green; AMAP1, red; and sites of gelatin degradation, blue (C). Bar, 10 µm. Red intensities (AMAP1, arbitrary units) and green intensities (PRKD2, arbitrary units) at the sites of gelatin degradation were measured and are shown as a scatter plot (D). (E–H) MDA-MB-231 cells were transfected with PRKD2 siRNA or a dsRNA with a nontargeting, irrelevant sequence (Irr), together with pcDNA3.1 HisC-resPRKD2 (rescue PRKD2) or control pcDNA3 empty vector (−), as indicated (E and F). Hs578T and MDA-MB-435s cells were transfected with PRKD2 siRNA or a dsRNA with an irrelevant sequence (Irr), as indicated (G and H). Total lysates were subjected to blotting with the indicated antibodies (E and G). Activities of cell adhesion to collagen (Adhesion) and Matrigel chemoinvasion (Invasion) are shown (F and H). In F and H, data are presented as percentages calculated by normalizing the values obtained for the control cells as 100%. 4,213 ± 516 (∼4.21%), 2,562 ± 309 (∼2.56%), and 1,790 ± 271 (∼1.79%) control MDA-MB-231, Hs578T, and MDA-MB-435s cells, respectively, were calculated to have transmigrated per 6.4-mm-diam Matrigel-coated Boyden chamber filter under these conditions. Data are shown as mean ± SEM of triplicate experiments (error bars).
PRKD2 was well colocalized with AMAP1 at the invadopodia of MDA-MB-231 cells (Fig. 1, C and D), and we moreover found that PRKD2 is coprecipitated with an anti-AMAP1 antibody from their cell lysates (Fig. 1 B). We also confirmed coprecipitation of AMAP1 by an anti-PRKD2 antibody (Fig. S1 A). Such coprecipitation of PRKD2 with AMAP1 was also observed in other highly invasive breast cancer cells, such as Hs578T and MDA-MB-435s, which express AMAP1 at high levels (Fig. 1 B). In contrast, coprecipitation of PRKD2 with AMAP1 was not detected in weakly or noninvasive breast cancer cell lines, such as MCF7 and MDA-MB-468, nor in a primary culture of human mammary epithelial cells (HMECs; Fig. 1 B), which express much lower basal levels of AMAP1 (Onodera et al., 2005). We also confirmed that PRKD2 is not coprecipitated with AMAP2 (Hashimoto et al., 2005), a close isoform of AMAP1, although these highly invasive cells seem to express AMAP2 at levels comparable to AMAP1 (Figs. 1 B and S1 B; Onodera et al., 2005).
PRKD2 is required for invasive activity
Like in the case of AMAP1 (Onodera et al., 2005), siRNA-mediated knockdown of PRKD2 efficiently inhibited Matrigel invasion activity of MDA-MB-231 cells, without notably inhibiting their adhesion to collagen (Fig. 1, E and F). This inhibition was restored by a rescue construct of PRKD2 cDNA, conjugated with an Xpress tag (Fig. 1, E and F). We have previously shown that AMAP1 knockdown is also effective in inhibiting the invasion of MDA-MB-435s cells, but less effective in Hs578T cells (Onodera et al., 2005), perhaps suggesting different levels of contribution of AMAP1 to the invasive activities in these cell lines. Similarly, knockdown of PRKD2 was very effective in blocking the Matrigel invasion activity of MDA-MB-435s, and less effective in Hs578T cells (Fig. 1, G and H).
Properties of AMAP1 binding to PRKD2
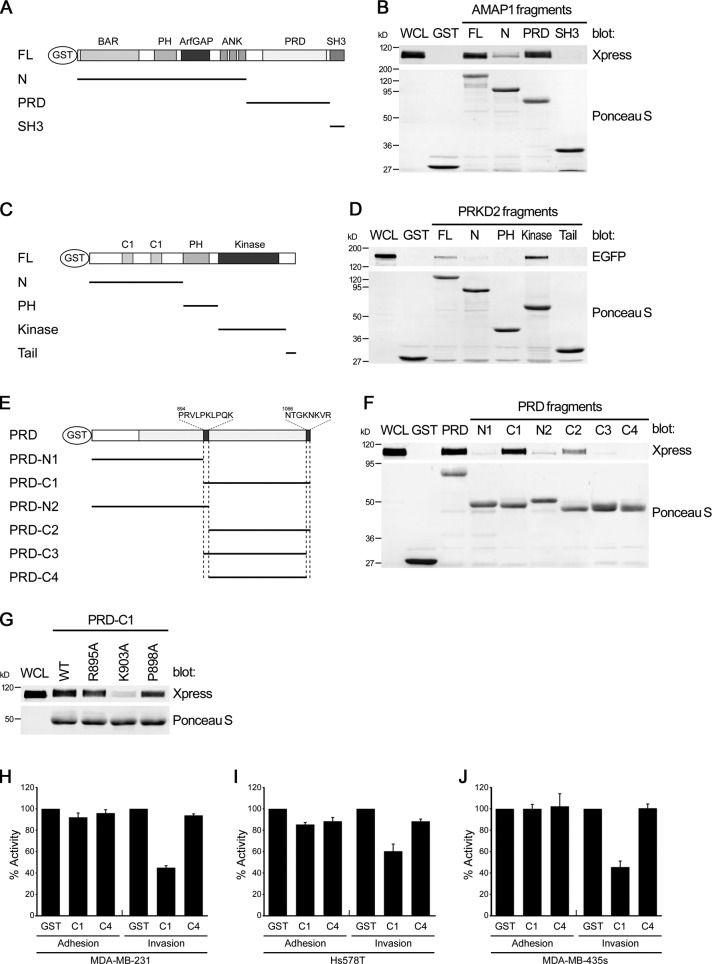
To confirm whether AMAP1 binds to PRKD2 directly, we then identified the sites of these proteins necessary for their binding. For this, we first divided AMAP1 into several fragments based on its functional modules (N, PRD, and SH3; see Fig. 2 A) and expressed them in mammalian cells, each tagged with GST. After purification on glutathione beads, these GST fusion proteins were incubated with cell lysates overexpressing full-length PRKD2 conjugated with an Xpress tag. We found that the proline-rich domain (PRD), but not other regions of AMAP1, binds to PRKD2 (Fig. 2 B). We then examined which region of PRKD2 is essential for its binding to AMAP1 by expressing GST-tagged fragments of PRKD2 (N, PH, Kinase, and Tail; Fig. 2 C) in mammalian cells, and incubating them with cell lysates overexpressing full-length AMAP1 conjugated with EGFP; we found that the kinase domain of PRKD2 binds to AMAP1 (Fig. 2 D). Therefore, binding of AMAP1 with PRKD2 appears to be primarily mediated by the PRD of AMAP1 and the kinase domain of PRKD2.
Figure 2.
Properties of AMAP1 and PRKD2 binding. (A–G) GST-tagged AMAP1 fragments (A and E) or AMAP1 PRD fragments bearing the indicated mutations (G) were each incubated in vitro with lysates of Cos7 cells expressing full-length Xpress-PRKD2. GST-tagged PRKD2 fragments (C) were each incubated in vitro with lysates of Cos7 cells expressing full-length EGFP-AMAP1. Coprecipitation of GST fusion proteins with PRKD2 or AMAP1 was analyzed by immunoblotting using anti-Xpress (B, F, and G) or anti-EGFP (D) antibodies, respectively. In B, D, F, and G, amounts of GST fusion proteins used were analyzed by Ponceau S staining. WCL, whole cell lysate (5 µg); GST, GST alone used as a control. (H–J) The GST-C1 fragment was overexpressed in MDA-MB-231 (H), Hs578T (I), and MDA-MB-435s cells (J), and their adhesion to collagen (Adhesion) and Matrigel chemoinvasion (Invasion) was measured. Cells overexpressing GST alone or the GST-C4 fragment were included as controls. Data are presented as percentages calculated by normalizing the values obtained for the GST-expressing control cells as 100%. 1,617 ± 212 (∼5.39%), 836 ± 96 (∼3.34%), and 457 ± 58 (∼2.17%) MDA-MB-231, Hs578T, and MDA-MB-435s cells expressing GST (and mVenus as a marker), respectively, were calculated to have transmigrated per 6.4-mm-diam Matrigel-coated Boyden chamber filter under these conditions. Data are shown as mean ± SEM of triplicate experiments (error bars).
The AMAP1 PRD is 373 aa long (aa 704–1,076 of AMAP1), and contains more than a dozen repeats of proline-rich sequences with some diversity, most of which conform to the Pro-X-X-Pro motif of the Src homology 3 (SH3)-binding regions (Onodera et al., 2005). The AMAP1 PRD also contains other aa sequences. We next sought to identify the region of the PRD essential for its binding to PRKD2. After testing several different fragments, we found that a fragment containing aa 894–1076 (C1 fragment; Fig. 2 E), but not the rest of the PRD region (aa 704–893, N1 fragment; Fig. 2 E), binds to PRKD2 (Fig. 2 F). The binding of this C1 fragment to PRKD2 seemed to be as strong as that of the entire region of the PRD (Fig. 2 F).
We then found that a deletion of several amino acids from the N terminus, Pro894-Arg-Val-Leu-Pro-Lys-Leu-Pro-Gln-Lys903 (resulting in the C2 fragment; Fig. 2 E), almost completely abolishes its binding to PRKD2 (Fig. 2 F). This peptide is rich in prolines and basic residues, and we found that mutation of Lys903 into alanine (K903A) almost completely abolishes the binding of C1 to PRKD2, while mutation of Arg895 or Pro898 into Ala (R895A and P898A, respectively) did not (Fig. 2 G). However, we found that deletion of the C-terminal amino acids Gln1066-Thr-Gly-Lys-Asn-Lys-Val-Arg1073 from the C1 fragment (resulting in the C3 fragment; Fig. 2 E) also abolishes its binding to PRKD2 (Fig. 2 F). Addition of Pro894-Arg-Val-Leu-Pro-Lys-Leu-Pro-Gln-Lys903 to the N1 fragment (resulting in the N2 fragment; Fig. 2 E) did not result in notable binding toward PRKD2 (Fig. 2 F). We also confirmed that deletion of both of these N-terminal and C-terminal peptides from the C1 fragment (resulting in the C4 fragment; Fig. 2 E) abolishes the binding to PRKD2 (Fig. 2 F). Therefore, the mode of binding between AMAP1 and PRKD2 appears to be very unusual, in which at least two separate regions within the PRD domain of AMAP1, namely the N-terminal and the C-terminal peptides of the C1 fragment, are simultaneously necessary for the binding of AMAP1 to the kinase domain of PRKD2.
Because the kinase domain of PRKD2 appeared to be used for its binding to AMAP1, we were interested in whether the kinase activity of PRKD2 is necessary for its binding to AMAP1 and/or for invasion. We generated a kinase-dead form of PRKD2 by substituting the crucial amino acid Asp695 into alanine (D695A; Mihailovic et al., 2004), and found that this kinase-dead mutant binds to AMAP1 (Fig. S2 A). This kinase-dead cDNA construct was designed to be unaffected by the PRKD2 siRNA used above, and we then expressed it in MDA-MB-231 cells, pretreated with the PRKD2 siRNA. We found that this construct of the kinase-dead mutant only partially, but not fully, restores the Matrigel invasion activity, whereas the wild-type rescue construct of the PRKD2 cDNA fully restores the invasion activity (Fig. S2, B and C). Expression of this kinase-dead mutant did not interfere with the adhesion of cells to collagen (Fig. S2 C). These results suggest that the kinase activity of PRKD2 is dispensable for its binding to AMAP1, but may be independently required for cell invasion activity.
Involvement of the AMAP1–PRKD2 complex in invasion activity
We then sought to obtain evidence supporting the finding that AMAP1–PRKD2 binding is involved in invasion. Expression of GST-C1, but not GST-C4, in MDA-MB-231 cells blocked the endogenous binding of AMAP1 with PRKD2 (Fig. 3 B). We found that expression of GST-C1, but not GST-C4, blocks the Matrigel invasive activity of MDA-MB-231 cells without affecting its adhesion to collagen (Fig. 2, H–J). Again, blockage of invasion activity by GST-C1 was similarly effective in MDA-MB-435s cells, but less effective in Hs578T cells (Fig. 2 I).
Figure 3.
AMAP1 associates with the β1 integrin subunit through PRKD2. (A) MDA-MB-231 cells plated onto collagen I–coated dishes were prestarved for serum for 2 h and then treated or untreated with 10 ng/ml EGF (5 min). Cells were lysed and anti-AMAP1 immunoprecipitates were analyzed for coprecipitation of PRKD2 and several integrin subunits (β1, β3, α2, α3, and α5) by immunoblotting, as indicated. Pre-immune serum of the anti-AMAP1 antibody was used as a control (Pre-imm). (B) Anti-AMAP1 immunoprecipitates from MDA-MB-231 cells overexpressing GST-C1 or GST-C4, prestarved and treated with EGF, were analyzed as above. Coprecipitation of PRKD2 and β1 integrin with AMAP1 was analyzed by immunoblotting using the indicated antibodies. In A and B, blots of whole cell lysates (WCL, 10 µg) are also shown. (C and D) Lysates from 293T cells expressing V5-PRKD2 or AMAP1-HA were mixed in different ratios, as indicated, and incubated in vitro with the GST-tagged cytoplasmic region of β1 integrin (GST-β1T) bound to glutathione beads. Precipitates were analyzed by immunoblotting using anti-V5, HA, and GST antibodies. GST alone was used as a control. (E–G) MDA-MB-231 cells expressing V5-PRKD2 were cultured on collagen I–coated dishes. Cells were serum-starved, and then treated (F) or untreated (E) with 10 ng/ml EGF (5 min) before fixation and immunolabeling. In the merged picture, V5-PRKD2, β1 integrin, and AMAP1 are shown as green, red, and blue, respectively. Bar, 10 μm. Colocalization of these proteins in serum-starved cells (open bars) and EGF-stimulated cells (closed bars) was measured, in which 1.0 and −1.0 indicate perfect colocalization and exclusion, respectively. Results represent mean ± SEM of >10 measurements (error bars).
AMAP1 forms a complex with β1 integrin via PRKD2
PRKD1 has been shown to bind directly to the β1 subunit of integrin (Woods et al., 2004). We hence next investigated whether PRKD2, complexed with AMAP1, binds to some integrins. MDA-MB-231 cells express several integrins (Fig. 3 A; Morini et al., 2000). We found that the β1 subunit, but not the β3 subunit, is coprecipitated by anti-AMAP1 immunoprecipitation, along with PRKD2, from MDA-MB-231 cell lysates (Fig. 3 A). The α2 and α3 subunits of integrins, but not the α5 subunit, were also coprecipitated (Fig. 3 A). Moreover, GST-C1, but not GST-C4, blocked the coprecipitation of integrins with AMAP1 (Fig. 3 B).
We also investigated how the β1 subunit associates with the AMAP1–PRKD2 complex. For this, we first found that the cytoplasmic tail of the β1 subunit, fused to GST (GST-β1T), pulls down HA-tagged AMAP1 (AMAP1-HA) and V5-tagged PRKD2 (V5-PRKD2) proteins expressed in 293T cells (Fig. 3 C). Moreover, the amounts of AMAP1-HA pulled down by GST-β1T were increased by the further addition of V5-PRKD2, whereas the amount of V5-PRKD2 pulled down by GST-β1T was not notably changed by the further addition of AMAP1-HA (Fig. 3 D). Together with the results described above, these results indicate that through binding to PRKD2, AMAP1 forms a complex with the cytoplasmic tail of the β1 subunit of integrins in which the subunit compositions are likely to be α2β1 and α3β1. However, α5β1 integrin does not seem to be a binding partner of the AMAP1–PRKD2 complex, although the α5 subunit is expressed at a high level in MDA-MB-231 cells.
EGFR signaling facilitates AMAP1–PRKD2–β1 integrin complex formation
MDA-MB-231 cells express EGFR and respond to EGF to become highly invasive and metastatic (Price et al., 1999; Morishige et al., 2008). We found that the amounts of PRKD2 and β1 integrins coprecipitated with AMAP1 are increased several-fold upon stimulation of MDA-MB-231 cells by EGF (Fig. 3 A), in which cells are prestarved for serum to minimize their invasive activity (Morishige et al., 2008). Therefore, EGFR signaling appears to facilitate formation of the AMAP1–PRKD2–β1 integrin complex. It should be noted that these cells used in previous experiments were cultured in the presence of serum and not prestarved.
AMAP1 resides mostly on intracellular vesicles and tubulovesicular structures in unstimulated epithelial cells, and a subfraction of AMAP1 becomes recruited to the plasma membrane upon EGF stimulation (Hashimoto et al., 2005). PRKDs are also known to localize at endosomal compartments and at the Golgi apparatus (Prestle et al., 1996; Jamora et al., 1999; Liljedahl et al., 2001). We found that AMAP1 and PRKD2 are both localized to tubulovesicular structures in serum-starved, EGF-unstimulated MDA-MB-231 cells (Fig. 3 E). In this analysis, we used cells cultured on collagen, and hence not forming invadopodia, to observe the morphologies of their endosomes and tubulovesicular structures more clearly.
In the serum-starved cells, a small fraction of AMAP1 was already found to be colocalized with PRKD2 (Fig. 3 E), which is consistent with the above described biochemical results. We then found that AMAP1 becomes better colocalized with PRKD2 upon their EGF stimulation, particularly at the plasma membrane (Fig. 3, F and G). A fraction of β1 integrin was also found to reside at the structures labeled with both AMAP1 and PRKD2 after EGF stimulation (Fig. 3, E–G). These results further support the finding that association of AMAP1, PRKD2, and β1 integrin is regulated by EGFR signaling.
Activation of Rab5c by EGFR signaling is necessary for intracellular complex formation of AMAP1 and PRKD2
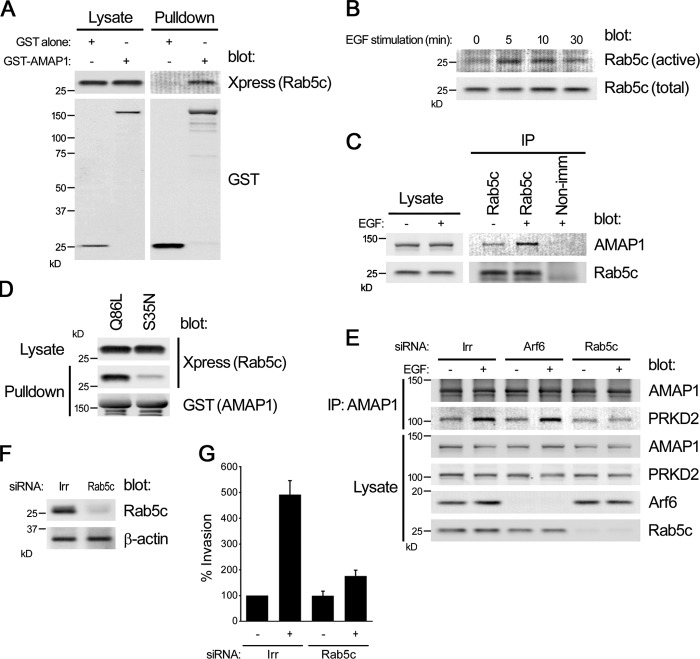
We then addressed the mechanism of how EGFR signaling facilitates AMAP1-mediated protein complex formation. Rab family GTPases are pivotal for intracellular membrane traffic and vesicle fusion (Stenmark, 2009; Hutagalung and Novick, 2011). We previously performed a yeast two-hybridization assay using AMAP1 as bait (Onodera et al., 2005; Nam et al., 2007) and identified Rab5c as one of its potential binding partners (unpublished data). We confirmed coprecipitation of Rab5c with AMAP1 by expressing GST-AMAP1 and Xpress-tagged Rab5c in 293T cells (Fig. 4 A). EGF stimulation is known to activate Rab5a (Chen et al., 2009). We found that EGF stimulation of serum-starved MDA-MB-231 cells causes enhanced activation of endogenous Rab5c (Fig. 4 B), and that the amounts of Rab5c, coprecipitated with AMAP1, are increased several-fold upon EGF stimulation (Fig. 4 C). Consistently, AMAP1 bound preferentially to the GTP hydrolysis-deficient form of Rab5c, Rab5cQ86L, rather than the GTP binding–deficient form, Rab5cS35N (Fig. 4 D). Therefore, AMAP1 appears to bind to Rab5c preferentially in its active form, which is generated upon EGFR signaling.
Figure 4.
Rab5c activation by EGF stimuli facilitates AMAP1–PRKD2 interaction. (A) GST-AMAP1 and Xpress-Rab5c were overexpressed in 293T cells. GST-AMAP1 was precipitated by glutathione beads, and coprecipitation of Xpress-Rab5c was analyzed by immunoblotting. GST alone was also used as a control. (B and C) MDA-MB-231 cells cultured on collagen I–coated dishes were serum-starved for 2 h and stimulated with 10 ng/ml EGF (5 min). The GTP-bound form of endogenous Rab5c was precipitated by GST-R5BD (B). Endogenous Rab5c was immunoprecipitated, and coprecipitation of AMAP1 was analyzed by immunoblotting. Nonimmune rabbit IgG was used as a control (Non-imm). (D) GST-AMAP1, immobilized on glutathione beads, was incubated with either Xpress-tagged Rab5c-Q86L or -S35N. Coprecipitation of Rab5c mutants was analyzed by immunoblotting. (E) MDA-MB-231 cells were transfected with Arf6 or Rab5c siRNAs or a dsRNA with a nontargeting, irrelevant sequence (Irr), and cultured on collagen I-coated dishes. Cells were serum-starved for 2 h and treated (+) or untreated (−) with 10 ng/ml EGF (5 min). Cells were lysed and AMAP1 was immunoprecipitated. Co-precipitation of PRKD2 was analyzed by immunoblotting. (F and G) MDA-MB-231 cells were transfected with Rab5c siRNA or a dsRNA with a nontargeting, irrelevant sequence (Irr). Total lysates were subjected to blotting with the indicated antibodies (F). Matrigel chemoinvasion activities in the presence or absence of 10 ng/ml EGF are shown (G). Data are presented as percentages calculated by normalizing the values obtained for the irrelevant dsRNA-treated, EGF-untreated control cells as 100%. 732 ± 71 (∼0.73%) control cells were calculated to have transmigrated per 6.4-mm-diam Matrigel-coated Boyden chamber filter under this condition. Data are shown as mean ± SEM of triplicate experiments (error bars).
We then found that knockdown of Rab5c by the siRNA method abolishes the increased coprecipitation of PRKD2 with AMAP1 in response to EGF stimulation of serum-starved MDA-MB-231 cells (Fig. 4 E). Rab5c knockdown also substantially blocked the EGF-induced Matrigel invasion activity (Fig. 4, F and G). Arf6 is also activated by EGFR signaling (Morishige et al., 2008), and binds to AMAP1 preferentially in its GTP form (Hashimoto et al., 2005). We found, however, that knockdown of Arf6 does not notably affect the EGF-induced coprecipitation of PRKD2 with AMAP1 (Fig. 4 E). Therefore, activation of Rab5c by EGFR signaling appears to promote the intracellular association of AMAP1 with PRKD2, and this association appears to be independent of Arf6 activation.
EGF induces the colocalization of AMAP1 with PRKD2 via Rab5c
The next target for investigation was the mechanism by which GTP-Rab5c facilitates intracellular complex formation of AMAP1 with PRKD2. For this, we first tested whether GTP-Rab5c is necessary for the binding of AMAP1 to PRKD2 in vitro. Purified GST-AMAP1 was incubated with cell lysates overexpressing V5-tagged PRKD2, together with increasing amounts of cell lysates overexpressing Xpress-Rab5cQ86L. We found, however, that the amount of PRKD2 coprecipitated with AMAP1 is not notably changed by the addition of Rab5cQ86L (Fig. S3). Therefore, GTP-Rab5c is not likely to mediate the physical association of AMAP1 with PRKD2.
Nevertheless, Rab5c knockdown abolished the EGF-induced enhancement of the colocalization of AMAP1 and PRKD2 (Fig. 5, A–C), which is consistent with the above described biochemical results. We were also interested in subcellular localization of Rab5c. Because antibodies against Rab5c that are applicable to cell labeling are not available, we used Xpress-tagged Rab5c. We found that Xpress-Rab5c is not appreciably colocalized with AMAP1 in serum-starved cells, although a fraction of PRKD2 staining seems to be merged with that of Rab5c (Fig. 5, E and F). It is well known that only a small fraction of intracellular small GTPases, such as Ras and Rho, becomes activated upon growth factor stimulation (Satoh et al., 1990), which may also be true for Rab5c activation upon EGFR signaling (Fig. 4 B and Materials and methods). We found that a small but significant fraction of Rab5c becomes colocalized with AMAP1 and PRKD2 upon EGF stimulation (Fig. 5, D–F), which is consistent with our biochemical results indicating that the activated form of Rab5c preferentially binds to AMAP1. These results collectively suggest that Rab5c and its activation are a key step to promote the intracellular association of AMAP1 with PRKD2 in response to EGFR signaling,
Figure 5.
EGF induces the colocalization of AMAP1 with PRKD2 via Rab5c. (A–F) MDA-MB-231 cells were cultured on collagen I–coated dishes, then transfected with Rab5c siRNA and pCMV-V5-PRKD2 one after another (A–C) or with pCMV-V5-PRKD2 and pcDNA3.1 HisC Rab5c simultaneously. Cells were serum-starved for 2 h, then treated (A and D) or left untreated (B and E) with 10 ng/ml EGF (5 min) before fixation. AMAP1, Xpress, and V5 epitopes were immunostained. In the merged image, V5-PRKD2 and AMAP1 are shown as green and red, respectively, in A and B; V5-PRKD2, Xpress-Rab5c, and AMAP1 are shown as green, red, and blue, respectively, in D and E. Colocalization indices of each protein were quantified, and the results represent mean ±SEM of >10 measurements (error bars; C and F). The rightmost panels in D and E show enlarged views of the boxed regions. Bars, 10 µm.
AMAP1 regulates β1 integrin recycling by binding to PRKD2
We finally investigated the roles of complex formation between AMAP1, PRKD2, andβ1 integrins with regard to integrin function and invasion. Arf6 activity plays crucial roles in the outward flow of plasma membrane components from recycling endosomes, but perhaps not for the recruitment of de novo synthesized proteins to the plasma membrane (Radhakrishna et al., 1996; Radhakrishna and Donaldson, 1997; D’Souza-Schorey et al., 1998). We have shown that GTP-Arf6 acts to recruit AMAP1 to the plasma membrane (Hashimoto et al., 2005). We then tested whether AMAP1 and its complex formation with PRKD2 is involved in plasma membrane recruitment of recycling β1 integrins.
For this, we tracked β1 integrins after their internalization from the cell surface in MDA-MB-231 cells treated with siRNAs specific to AMAP1, PRKD2, or Rab5c. Cells were first incubated with an anti–β1 antibody at 4°C to label surface pools of the β1 subunit and then incubated at 37°C for 2 h to induce their internalization. Cells were acid-washed to remove the antibodies remaining on the cell surface, then further incubated at 37°C to induce recycling of the internalized β1 integrins back to the cell surface. Anti–β1 antibody molecules then appearing on the cell surface were detected by immunofluorescence. We found that there is a basal level of recycling activity of β1 integrins in serum-starved MDA-MB-231 cells, and the recycling activity is enhanced several-fold upon their EGF stimulation (Fig. 6 C). We then found that knockdown of AMAP1 or PRKD2 abolishes the EGF-induced recycling activity of β1 integrins (Fig. 6, A and C). Consistently, cell surface levels of β1 integrins were significantly reduced in these knockdown cells cultured in the presence of serum (Fig. 6 B). Blocking AMAP1–PRKD2 association by knockdown of Rab5c or overexpression of GST-C1 (but not GST-C4) also abolished the EGF-induced recycling activity of β1 integrins (Fig. 6, E, F, and H). However, it has been shown that β1 integrin is internalized at almost similar rates in HeLa cells in the presence and absence of extracellular stimulation (Li et al., 2005). Likewise, we found that rates of β1 integrin internalization are not notably affected in the presence or the absence of EGF in MDA-MB-231 cells (not depicted), and that knockdown of AMAP1, PRKD2, or expression of GST-C1 did not affect the rates of β1 integrin internalization, whereas Rab5c knockdown partially inhibited it (Fig. 6, D and I). Therefore, AMAP1, PRKD2, and Rab5c, as well as the physical association of AMAP1 and PRKD2, appear to be essential for the EGF-induced recycling of β1 integrins by primarily mediating their plasma membrane recruitment.
Figure 6.
Association of AMAP1 and PRKD2 is required for the recycling back of β1 integrin. (A–D) MDA-MB-231 cells, treated with siRNAs specific to AMAP1 or PRKD2, were analyzed for the total cellular amounts (A), surface expression (B), activities of recycling back (C), and internalization (D) of β1 integrin. Percentages of β1 integrin molecules recycled back to the plasma membrane or internalized, as compared with those internalized or initially present at the plasma membrane, respectively, are shown. siRNA-treated cells were serum-starved for 2 h and processed for each assay as described in Materials and methods, and then treated (+) or left untreated (−) with 10 ng/ml EGF (C), or cultured in the absence of serum or EGF for the indicated times (D). An RNA duplex with an irrelevant sequence (Irr or Irrelevant) was used as a control. (E–I) MDA-MB-231 cells, treated with Rab5c siRNA (E–G) or overexpressing GST, GST-C1, or GST-C4 (H and I), were analyzed for their recycling-back (F and H) and internalization (G and I) of β1 integrin as above. Data are shown as mean ± SEM of triplicate experiments (error bars).
To further investigate the possible role of Rab5c in EGF-induced recycling of β1 integrins, we examined the subcellular localization of the internalized β1 integrins in MDA-MB-231 cells, treated with control or Rab5c siRNAs. Cell surface β1 integrins were labeled and induced to be internalized in the absence of serum and EGF, as above. These cells were then either fixed immediately or stimulated with EGF before fixation. Rab4 is a representative marker for recycling endosomes, whereas Rab7 is a marker for late endosomes. We found that the internalized β1 integrins exhibit appreciable colocalization with Rab4, but not with Rab7 (Fig. S4, and not depicted). Such colocalization of β1 integrins and Rab4 disappeared when cells were treated with EGF (Fig. S4), which induced the recycling of β1 integrins to the cell surface. However, we found that when Rab5c was silenced, β1 integrins stay colocalized with Rab4 at intracellular endosomes even after EGF stimulation (Fig. S4). Therefore, it is likely that Rab5c primarily acts at a site in which recycling β1 integrins move en route to the cell surface from the Rab4-positive endosomes in response to EGF stimulation.
Discussion
EGFR signaling is one of the major factors crucial for the development of malignancy in breast cancer. We have shown previously that AMAP1, as well as Arf6, is aberrantly overexpressed in a large population of malignant cancers of the human breast (40–80%), and in such cancer cells EGFR may use the Arf6–AMAP1 pathway to induce invasion and metastasis (Hashimoto et al., 2004; Onodera et al., 2005; Morishige et al., 2008). In this paper, we showed that AMAP1 directly binds to PRKD2 and hence forms a complex with certain types of β1 integrins, such as α2β1 and α3β1. Activated Arf6 recruits AMAP1 to the plasma membrane by its direct binding to AMAP1 (Hashimoto et al., 2005; Morishige et al., 2008). We hence propose that Arf6, activated upon EGFR signaling, recruits β1 integrins associated with AMAP1 via PRKD2 to the plasma membrane. We demonstrated that silencing of AMAP1 and PRKD2, as well as interference of AMAP1–PRKD2 binding, all effectively inhibit the EGF-induced recycling of β1 integrins back to the plasma membrane and invasion activities of different breast cancer cells. β1 integrins are crucial for the malignant development of breast cancer cells (White et al., 2004; Huck et al., 2010), and α3β1 is essential for the phagocytosis of degraded ECM fragments as well as cancer outgrowth (Coopman et al., 1996; Morini et al., 2000). Our results moreover suggest that α5β1 integrin, the major fibronectin receptor, may be regulated independently of α2β1 and α3β1 in EGFR signaling. Because the aberrant overexpression of AMAP1 seen in breast cancer cells is necessary for its efficient complex formation with PRKD2, this AMAP1 pathway may provide an excellent molecular target for cancer therapeutics.
AMAP1 on its own has the biochemical property to bind directly to PRKD2. We found that EGFR signaling is necessary to promote intracellular complex formation of AMAP1 and PRKD2. As a mechanism for this phenomenon, we showed that Rab5c and its activation by EGFR signaling are crucial to promote their intracellular association (also see below). Silencing of Rab5c abolished the EGF-enhanced association of these two proteins, and blocked the EGF-induced recycling of β1 integrins, as well as the invasive activities of breast cancer cells. We moreover showed that the intracellular association of AMAP1 and PRKD2 was independent of Arf6. Therefore, Rab5c activation upon EGFR signaling appears to be another step regulating activation of the AMAP1 pathway for cancer invasion, in addition to the activation of Arf6.
Rab5c was dispensable for the physical binding of AMAP1 with PRKD2 in vitro. However, we found that GTP-Rab5c, but not GDP-Rab5c, binds to AMAP1 both in vivo and in vitro. Rab family GTPases consist of >60 members in humans, and have crucial roles in intracellular trafficking, including vesicle budding and fusion (Stenmark, 2009; Hutagalung and Novick, 2011). Our microscopic observations indicated that only small fractions of AMAP1 and PRKD2 colocalize with each other in starved cells, and that EGF stimulation enhances their colocalization, for which Rab5c is necessary. Moreover, Rab5c appeared to colocalize more with PRKD2 than with AMAP1 in starved cells. Collectively, one possible scenario is that GTP-Rab5c employs AMAP1 as its “target protein” to mediate the fusion of AMAP1-containing vesicles and PRKD2-containing vesicles. However, AMAP1 interacts with several different proteins and exhibits different functions (Hashimoto et al., 2005; Onodera et al., 2005; Nam et al., 2007), and hence does not seem to use only Rab5c for its intracellular trafficking. Consistent with this notion, only limited fractions of AMAP1 were colocalized with Rab5c and with PRKD2 even in stimulated cells. Rab5a has been shown to be activated by Rin1 upon EGFR signaling (Chen et al., 2009). The precise mechanisms as to how Rab5c is activated by EGFR signaling, and also as to how Rab5c mediates vesicle fusion, deserve further investigation. Likewise, the mechanisms by which the intracellular association of PRKD2 and β1 integrins is regulated also remain to be elucidated.
We provide a line of evidence that the AMAP1–Rab5c–PRKD2 pathway is central to EGFR-induced recycling of β1 integrins in MDA-MB-231 cells. However, this pathway may not be the sole pathway regulating the recycling of β1 integrins among the different types of cells and tumors. For example, Arf6 has been shown to colocalize with β1 integrin at endosomes in HeLa cells and is implicated in the serum-induced recycling-back of β1 integrin, in which Rab11, but not Rab4, appears to be involved (Powelka et al., 2004). EGFR may also bind directly to an ArfGAP, ACAP4, to control recycling of β1 integrins, which affects the migration of HeLa cells on fibronectin (Yu et al., 2011). In T cells, β1 integrin also associates with PRKD1 together with Rap1 upon T cell receptor activation (Medeiros et al., 2005). In the case of other types of integrins, PRKD1 was shown to mediate the recycling of β3 integrin in NIH3T3 cells; this was dependent on Rab4 but not Rab11 (Roberts et al., 2001; Woods et al., 2004). Rab11 is involved in the trafficking of β4 integrin in the hypoxia-induced invasion of MDA-MB-231 cells (Yoon et al., 2005). Moreover, epigenetic inactivation of PRKD1 expression is frequently found in primary gastric cancer, and is implicated in the acquisition of invasiveness (Kim et al., 2008). However, in these studies, precise mechanisms as to how PRKD1, Rap1, and Rabs are involved in the integrin recycling and function still remain largely elusive.
In conclusion, we show here that AMAP1 functions to recycle certain types of β1 integrins to induce the invasion activity of some breast cancer cells, together with the detailed molecular mechanisms. MDA-MB-231 cells are categorized as “triple-negative” type in which patients show a very bad prognosis. The question of what populations of “triple-negative” breast cancers express Arf6, AMAP1, PRKD2, and Rab5c, together with α2β1 and α3β1 integrins, remains to be studied with clinical samples. In this regard, it should be noted that overexpression of Arf6 and AMAP1 proteins in breast cancer cells are primarily controlled posttranscriptionally (Hashimoto et al., 2004; Onodera et al., 2005).
Materials and methods
Cells
Breast cancer cell lines were purchased from the American Type Culture Collection (ATCC). MDA-MB-231 cells were cultured in a 1:1 mixture of DME and RPMI 1640 supplemented with 10% FCS (HyClone) and 5% NuSerum (BD), as described previously (Bowden et al., 1999). Other cells were cultured according to the ATCC instructions. Cells were cultured using collagen type I–coated (10 µg/ml) plates for each assay, unless otherwise indicated. A primary preparation of normal HMECs (Cambrex) was cultured according to the manufacturer’s instructions. FCS was purchased from Hyclone. For stimulation with EGF, cells were prestarved for FCS for 2 h. Cells were then stimulated with EGF at a final concentration of 10 ng/ml and incubated for the indicated periods before being subjected to analyses.
Antibodies
Rabbit polyclonal antibodies against AMAP1 and AMAP2 were raised using GST-fused peptides corresponding to aa 935–1,002 (AMAP1) and 871–929 (AMAP2), respectively, as described previously (Hashimoto et al., 2005; Onodera et al., 2005). Other antibodies were purchased from commercial sources; rabbit polyclonal antibodies against PRKD1; β3 integrin (Cell Signaling Technology); PRKD2 (EMD); α2, α3, and α5 integerins (Millipore); Rab5c (Sigma-Aldrich); mouse monoclonal antibodies against β-actin (Millipore), Xpress, V5 (Invitrogen), β1 integrin for immunoblotting (BD) and for immunofluorescence (Millipore), AMAP1/DDEF1 (Abnova), myc epitope (Covance) and GST (Millipore); and chicken polyclonal antibody against the V5 epitope (Abcam). Secondary polyclonal antibodies to mouse IgG, rabbit IgG, or chicken IgY, each conjugated with peroxidase, Alexa Fluor dyes (488, 555, or 647), or DyLight488, were from Jackson ImmunoResearch Laboratories, Inc., Invitrogen, and Abcam, respectively.
cDNAs and transfection
AMAP1 cDNA (Onodera et al., 2005) was cloned into pEGFP C1 (Takara Bio Inc.). cDNA for PRKD2, amplified by PCR from the first-strand cDNA of MDA-MB-231 cells, was cloned into pmVenus C1 (see below), pcDNA3.1 HisC (Invitrogen), or pCMV-V5 (see below). Full-length Rab5c cDNA was amplified by PCR from a pACT2 (Takara Bio Inc.) clone isolated from a human cDNA library by yeast two-hybrid screening. AMAP1 cDNA fragments corresponding to the N terminus (aa 1–703), PRD (aa 704–1,076), PRD-N1 (aa 704–893), PRD-N2 (aa 704–903), PRD-C1 (aa 894–1,076), PRD-C2 (aa 904–1,076), PRD-C3 (aa 894–1,065), PRD-C4 (aa 904–1,065), and SH3 domain (aa 1,074–1,133) were amplified by PCR and ligated into pEBG (Mayer et al., 1995). PRKD2 cDNA fragments encoding the N terminus (aa 1–397), PH domain (aa 398–539), kinase domain (aa 540–838), and tail region (aa 839–878) were amplified by PCR and cloned into pEBG. The rescue cDNA for PRKD2 (resPRKD2) was constructed by substituting the nucleotides (underlined) within the siRNA target to 5′-TTATGGCAATTGAATTTACAA-3′, without changing the coding amino acids. pmVenus C1 was generated by substituting a cDNA fragment encoding EGFP in the pEGFP vector with that of Venus, which bears the A206K “monomeric” mutation (Zacharias et al., 2002). pCMV-V5 was generated as follows: the NdeI–BamHI fragment of pcDNA3.1 HisA (Invitrogen) was replaced with an NdeI–NheI fragment containing the cytomegalovirus (CMV) promoter excised from pEGFP C1 and a NheI–BamHI fragment encoding the V5 epitope generated by PCR.
For inhibition of the endogenous association of AMAP1 and PRKD2, Matrigel invasion, and integrin recycling, PRD-C1 and PRD-C4 bearing P898A and/or R1063A mutations were used to avoid inhibition of the binding of several SH3 proteins known to interact with AMAP1 PRD. The mutants of AMAP1 PRD or resPRKD2 were generated by a PCR-based method using the following primers: PRD R895A, 5′-CCGGATCCCCAGCAGTTCTTCCTAAACTACCTCAG-3′; PRD K903A, 5′-CCGGATCCCCAAGAGTTCTTCCTAAACTACCTCAGGCAGTGGCACTAAGG-3′; PRD P898A, 5′-CCGGATCCCCAAGAGTTCTTGCTAAACTACCTCAGAAAGT-3′; PRD R1063A, 5′-AAGCGGCCGCTAGATTTTTGCGGGCAGTGG-3′; and resPRKD2 D695A, 5′-TTCCTCAGGTGAAGCTGTGTGCCTTTGGCTTTGCTCGC-3′.
For the in vitro binding assay, 5 × 105 COS-7 cells were transfected with 3 µg of pEBG, pEGFP C1-AMAP1, or pcDNA3.1 HisC-PRKD2, using FuGENE 6 (Roche) according to the manufacturer’s instructions. For assays using MDA-MB-231 cells, 106 cells were cotransfected with 3.2 µg of pEBG vectors and 0.3 µg of pmVenus C1 using Lipofectamine LTX (Invitrogen).
Protein knockdown by siRNA
Cells were transfected with siRNA duplexes using Lipofectamine RNAi MAX (Invitrogen) according to the reverse transfection method provided by the manufacturer.
Duplex oligonucleotides were chemically synthesized and purified by Japan BioService. The siRNA duplexes used were: AMAP1, 5′-GACCUGACAAAAGCCAUUAdTdT-3′ and 5′-UAAUGGCUUUUGUCAGGUCdTdT-3′; and PRKD2, 5′-CUGCAAGUUUAACUGUCACAAdTdT-3′ and 5′-UUGUGACAGUUAAACUUGCAGdTdT-3′.
In vitro binding assay and immunoprecipitation
Cells were lysed with Nonidet P-40 buffer (1% Nonidet P-40, 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 5 mM EDTA, 1 mM Na3VO4, 1 mM PMSF, 5 µg/ml aprotinin, 2 µg/ml leupeptin, and 3 µg/ml pepstatin A). For in vitro binding assays, GST fusion proteins were first purified on glutathione beads. 5 µg of GST fusion proteins were then incubated with 300 µg of cell lysates. For coprecipitation assays, 500 µg of cell lysates were subjected to immunoprecipitation, and proteins precipitated were detected by immunoblotting after separation by SDS-PAGE, as described previously (Onodera et al., 2005). Antibodies against AMAP1, PRKD2, and Rab5c were used at dilutions of 1:200, 1:100, and 1:50, respectively.
Measurement of Rab5c activity
The Rab5-binding domain (R5BD) of Rabaptin 5 (aa 739–862) was amplified by PCR and ligated into pGEX4T-1 (GE Healthcare) using EcoRI–SalI sites. GST-R5BD was expressed in bacteria and purified by glutathione beads. MDA-MB-231 cells were serum-starved for 2 h and treated or untreated with 10 ng/ml EGF (5–30 min), then lysed in Rab5-binding buffer (25 mM Hepes, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 10% glycerol, and 1 mM DTT) supplemented with protease inhibitors (1 mM PMSF, 5 µg/ml aprotinin, 2 µg/ml leupeptin, and 3 µg/ml pepstatin A). 30 µg of GST-R5BD was incubated with 1 mg lysates from each condition for 30 min at 4°C, then washed four times with Rab5-binding buffer. GTP-Rab5c bound to GST-R5BD was detected by immunoblotting using an anti-Rab5c antibody.
Immunofluorescence microscopy
For analyzing the colocalization of PRKD2 with AMAP1 in invadopodia, MDA-MB-231 cells expressing mVenus-PRKD2 were cultured on cross-linked, Alexa Fluor 594–labeled gelatin films for 8 h and then subjected to immunolabeling with an anti-AMAP1 antibody, coupled with an Alexa Fluor 647–labeled anti–rabbit IgG antibody, after fixation in 4% paraformaldehyde and 0.1% Triton X-100 in PBS for 10 min, as described previously (Onodera et al., 2005).
For analyzing the colocalization of PRKD2, AMAP1, Rab5c, and β1 integrin, cells were cultured on plastic dishes coated with collagen I for 24 h and then starved for serum for 2 h, followed by stimulation with 10 ng/ml EGF (Sigma-Aldrich) for 5 min, before fixation with methanol/acetone and immunostaining. Cells were labeled with anti-AMAP1, V5 epitope (for V5-tagged PRKD2), Xpress epitope (for Xpress-tagged Rab5c), or β1 integrin antibodies, coupled with DyLight 488 or Alexa Fluor 488–, 555–, or 647–conjugated secondary antibodies.
Stained cells were mounted with 50% glycerol in PBS for imaging by an LSM 510 confocal laser scanning microscope (Carl Zeiss; 100× oil-immersion lens, 1.40 NA, LSM 510 software) or an A1R (Nikon; 60× oil-immersion lens, 1.40 NA, NIS-Elements software) at room temperature. For measurement of the colocalization of proteins, z sections of cells were also taken. The Pearson’s correlation coefficient between two proteins was measured in each image by the ImageJ software (National Institutes of Health, version 1.44p). Images were handled using Photoshop version 7 (Adobe).
Matrigel invasion assay
In vitro invasive activity was examined using Biocoat Matrigel chambers (BD), as described previously (Hashimoto et al., 2004; Onodera et al., 2005). In brief, 105 cells were seeded on the upper wells of 24-well chambers in the absence of serum, and lower wells were filled with conditioned medium of NIH3T3 cells cultured for 24 h in the absence of serum. After incubation for 12 h, the number of cells that migrated out onto the lower surface of the membranes was scored by staining with 1% crystal violet (for siRNA-treated cells) or by identifying Venus-positive cells by fluorescent microscopy (for AMAP1 PRD-expressing cells) after fixing the cells in 4% paraformaldehyde. Data were collected from three independent experiments, each performed in triplicate, and are presented as percentages calculated by normalizing the values obtained for the control cells as 100%.
Adhesion assay
Cell adhesion to collagen type I was examined as follows: 3 × 104 cells were seeded on collagen type I–coated 8-well chamber slides (10 µg/ml) in the absence of serum. The cells were fixed with 4% paraformaldehyde after incubation for 30 min, and the number of attached cells was counted.
Surface expression, internalization, and recycling of β1 integrin
Trafficking of β1 integrin in MDA-MB-231 cells was examined by an antibody labeling–based method, as described previously (Powelka et al., 2004), with minor modifications. In brief, for the internalization assay, cells were incubated with 1 µg/ml of an anti–β1 integrin antibody (clone MEM-101A [Santa Cruz Biotechnology, Inc.], dialyzed with PBS) in PBS for 30 min on ice. After washing twice with ice-cold PBS, cells were incubated with serum-depleted culture media for the indicated times at 37°C in a CO2 incubator, followed by washing three times with an acidic buffer (0.5% glacial acetic acid, pH 3.0, and 0.5 M NaCl) for 2 min. Cells were then washed twice with ice-cold PBS, fixed in 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature before immunostaining. For the recycling assay, following the acid wash, cells were incubated with serum-depleted culture media with or without 10 ng/ml EGF for the indicated times at 37°C in a CO2 incubator, then subjected to immunolabeling after fixation without permeabilization. For surface labeling, cells were fixed immediately after washing in PBS at room temperature and subjected to immunolabeling fixation in the absence of detergents.
Cells were then incubated with an IRdye 800–conjugated anti–mouse IgG antibody (LI-COR Biosciences; 1:800 dilution) and DRAQ5 (Biostatus Limited; 1:20,000 dilution), a cell-permeable nuclear staining dye, for 1 h at room temperature. Staining intensity was measured with the Odyssey Infrared Imaging System (LI-COR Biosciences) using both 700 and 800 nm channels. Nonimmune mouse IgG was used to measure nonspecific labeling, and the value was subtracted from each measurement. Data were collected from three independent experiments, each performed in duplicate.
Tracking of internalized β1 integrin
To examine the localization of internalized β1 integrin in the cells treated with Rab5c siRNA, cells were cultured on plastic dishes coated with collagen I with siRNA complexes (see above). For Rab4 staining, cells were further transfected with pEF-BOS myc-Rab4 after 24 h. 48 h after the initial transfection with siRNA duplexes, β1 integrin on the cell surface was labeled with a β1 integrin antibody as above. For internalization of β1 integrin, cells were washed twice with ice-cold PBS, then incubated with serum-depleted culture media for 2 h at 37°C in a CO2 incubator. Cells were washed three times with acidic buffer and fixed with 4% paraformaldehyde. To examine the localization of internalized β1 integrin after EGF stimulation, cells were further incubated for 1 h with serum-depleted culture media supplemented with 10 ng/ml EGF after the internalization. Cells were then washed three times with acidic buffer and fixed with 4% paraformaldehyde. Fixed cells were permeabilized and labeled with an anti-myc antibody, then stained with fluorophore-labeled secondary antibodies.
Online supplemental material
Fig. S1 shows specificity of AMAP1–PRKD2 binding. Fig. S2 shows roles of PRKD2 kinase activity. Fig. S3 shows the effect of Rab5c binding on AMAP1–PRKD2 complex formation. Fig. S4 shows localization of internalized β1 integrin in the cells treated with Rab5c siRNA. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201065/DC1.
Supplementary Material
Acknowledgments
We are grateful to E. Hayashi for her secretarial work and H. Yoshino for her technical assistance. We also thank M. Bissell and C. Park for support, and H. A. Popiel for her critical reading of the manuscript.
J.-M. Nam was supported by a grant from the Matching Program for Innovations in Future Drug Discovery and Medical Care from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT). This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas from MEXT, a research grant for life science from Takeda Science Foundation, a research grant in the natural sciences from the Mitsubishi Foundation, and a research grant from Suhara Memorial Foundation.
Footnotes
Abbreviations used in this paper:
- EGFR
- EGF receptor
- HMEC
- human mammary epithelial cell
- PRD
- proline-rich domain
References
- Blume-Jensen P., Hunter T. 2001. Oncogenic kinase signalling. Nature. 411:355–365 10.1038/35077225 [DOI] [PubMed] [Google Scholar]
- Bowden E.T., Barth M., Thomas D., Glazer R.I., Mueller S.C. 1999. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 18:4440–4449 10.1038/sj.onc.1202827 [DOI] [PubMed] [Google Scholar]
- Bowden E.T., Coopman P.J., Mueller S.C. 2001. Invadopodia: unique methods for measurement of extracellular matrix degradation in vitro. Methods Cell Biol. 63:613–627 10.1016/S0091-679X(01)63033-4 [DOI] [PubMed] [Google Scholar]
- Bretscher M.S. 1989. Endocytosis and recycling of the fibronectin receptor in CHO cells. EMBO J. 8:1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M.S. 1992. Circulating integrins: alpha 5 beta 1, alpha 6 beta 4 and Mac-1, but not alpha 3 beta 1, alpha 4 beta 1 or LFA-1. EMBO J. 11:405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown F.D., Rozelle A.L., Yin H.L., Balla T., Donaldson J.G. 2001. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154:1007–1017 10.1083/jcb.200103107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P., Norman J. 2008. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18:257–263 10.1016/j.tcb.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Spence H.J., Parsons M., White D.P., Clark K., Cheng K.W., Mills G.B., Humphries M.J., Messent A.J., Anderson K.I., et al. 2007. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell. 13:496–510 10.1016/j.devcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Chan M., Lindsay A.J., McCaffrey M.W., Boettiger D., Norman J.C. 2008. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183:143–155 10.1083/jcb.200804140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.I., Kong C., Su X., Stahl P.D. 2009. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J. Biol. Chem. 284:30328–30338 10.1074/jbc.M109.034546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coopman P.J., Thomas D.M., Gehlsen K.R., Mueller S.C. 1996. Integrin alpha 3 beta 1 participates in the phagocytosis of extracellular matrix molecules by human breast cancer cells. Mol. Biol. Cell. 7:1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C., Chavrier P. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7:347–358 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C., van Donselaar E., Hsu V.W., Yang C., Stahl P.D., Peters P.J. 1998. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J. Cell Biol. 140:603–616 10.1083/jcb.140.3.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G. 2003. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278:41573–41576 10.1074/jbc.R300026200 [DOI] [PubMed] [Google Scholar]
- Dunphy J.L., Moravec R., Ly K., Lasell T.K., Melancon P., Casanova J.E. 2006. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 16:315–320 10.1016/j.cub.2005.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Wolf K. 2003. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer. 3:362–374 10.1038/nrc1075 [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Onodera Y., Hashimoto A., Tanaka M., Hamaguchi M., Yamada A., Sabe H. 2004. Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. USA. 101:6647–6652 10.1073/pnas.0401753101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Hashimoto A., Yamada A., Onodera Y., Sabe H. 2005. Assays and properties of the ArfGAPs, AMAP1 and AMAP2, in Arf6 function. Methods Enzymol. 404:216–231 10.1016/S0076-6879(05)04021-8 [DOI] [PubMed] [Google Scholar]
- Hashimoto A., Hashimoto S., Ando R., Noda K., Ogawa E., Kotani H., Hirose M., Menju T., Morishige M., Manabe T., et al. 2011. GEP100-Arf6-AMAP1-cortactin pathway frequently used in cancer invasion is activated by VEGFR2 to promote angiogenesis. PLoS ONE. 6:e23359 10.1371/journal.pone.0023359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck L., Pontier S.M., Zuo D.M., Muller W.J. 2010. beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc. Natl. Acad. Sci. USA. 107:15559–15564 10.1073/pnas.1003034107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung A.H., Novick P.J. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Hynes N.E., Lane H.A. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 5:341–354 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- Ivaska J., Whelan R.D., Watson R., Parker P.J. 2002. PKC epsilon controls the traffic of beta1 integrins in motile cells. EMBO J. 21:3608–3619 10.1093/emboj/cdf371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C., Yamanouye N., Van Lint J., Laudenslager J., Vandenheede J.R., Faulkner D.J., Malhotra V. 1999. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell. 98:59–68 10.1016/S0092-8674(00)80606-6 [DOI] [PubMed] [Google Scholar]
- Kim M., Jang H.R., Kim J.H., Noh S.M., Song K.S., Cho J.S., Jeong H.Y., Norman J.C., Caswell P.T., Kang G.H., et al. 2008. Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis. 29:629–637 10.1093/carcin/bgm291 [DOI] [PubMed] [Google Scholar]
- Li J., Ballif B.A., Powelka A.M., Dai J., Gygi S.P., Hsu V.W. 2005. Phosphorylation of ACAP1 by Akt regulates the stimulation-dependent recycling of integrin beta1 to control cell migration. Dev. Cell. 9:663–673 10.1016/j.devcel.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Liljedahl M., Maeda Y., Colanzi A., Ayala I., Van Lint J., Malhotra V. 2001. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell. 104:409–420 10.1016/S0092-8674(01)00228-8 [DOI] [PubMed] [Google Scholar]
- Mayer B.J., Hirai H., Sakai R. 1995. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 5:296–305 10.1016/S0960-9822(95)00060-1 [DOI] [PubMed] [Google Scholar]
- Medeiros R.B., Dickey D.M., Chung H., Quale A.C., Nagarajan L.R., Billadeau D.D., Shimizu Y. 2005. Protein kinase D1 and the beta 1 integrin cytoplasmic domain control beta 1 integrin function via regulation of Rap1 activation. Immunity. 23:213–226 10.1016/j.immuni.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Menju T., Hashimoto S., Hashimoto A., Otsuka Y., Handa H., Ogawa E., Toda Y., Wada H., Date H., Sabe H. 2011. Engagement of overexpressed Her2 with GEP100 induces autonomous invasive activities and provides a biomarker for metastases of lung adenocarcinoma. PLoS ONE. 6:e25301 10.1371/journal.pone.0025301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailovic T., Marx M., Auer A., Van Lint J., Schmid M., Weber C., Seufferlein T. 2004. Protein kinase D2 mediates activation of nuclear factor kappaB by Bcr-Abl in Bcr-Abl+ human myeloid leukemia cells. Cancer Res. 64:8939–8944 10.1158/0008-5472.CAN-04-0981 [DOI] [PubMed] [Google Scholar]
- Molnar J., Hoekstra S., Ku C.S., Van Alten P. 1987. Evidence for the recycling nature of the fibronectin receptor of macrophages. J. Cell. Physiol. 131:374–383 10.1002/jcp.1041310309 [DOI] [PubMed] [Google Scholar]
- Morini M., Mottolese M., Ferrari N., Ghiorzo F., Buglioni S., Mortarini R., Noonan D.M., Natali P.G., Albini A. 2000. The alpha 3 beta 1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int. J. Cancer. 87:336–342 [DOI] [PubMed] [Google Scholar]
- Morishige M., Hashimoto S., Ogawa E., Toda Y., Kotani H., Hirose M., Wei S., Hashimoto A., Yamada A., Yano H., et al. 2008. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 10:85–92 10.1038/ncb1672 [DOI] [PubMed] [Google Scholar]
- Nam J.M., Onodera Y., Mazaki Y., Miyoshi H., Hashimoto S., Sabe H. 2007. CIN85, a Cbl-interacting protein, is a component of AMAP1-mediated breast cancer invasion machinery. EMBO J. 26:647–656 10.1038/sj.emboj.7601534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T., Shima D., Squire A., Bastiaens P.I., Gschmeissner S., Humphries M.J., Parker P.J. 1999. PKCalpha regulates beta1 integrin-dependent cell motility through association and control of integrin traffic. EMBO J. 18:3909–3923 10.1093/emboj/18.14.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Hashimoto S., Hashimoto A., Morishige M., Mazaki Y., Yamada A., Ogawa E., Adachi M., Sakurai T., Manabe T., et al. 2005. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 24:963–973 10.1038/sj.emboj.7600588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powelka A.M., Sun J., Li J., Gao M., Shaw L.M., Sonnenberg A., Hsu V.W. 2004. Stimulation-dependent recycling of integrin beta1 regulated by ARF6 and Rab11. Traffic. 5:20–36 10.1111/j.1600-0854.2004.00150.x [DOI] [PubMed] [Google Scholar]
- Prestle J., Pfizenmaier K., Brenner J., Johannes F.J. 1996. Protein kinase C mu is located at the Golgi compartment. J. Cell Biol. 134:1401–1410 10.1083/jcb.134.6.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.T., Tiganis T., Agarwal A., Djakiew D., Thompson E.W. 1999. Epidermal growth factor promotes MDA-MB-231 breast cancer cell migration through a phosphatidylinositol 3′-kinase and phospholipase C-dependent mechanism. Cancer Res. 59:5475–5478 [PubMed] [Google Scholar]
- Radhakrishna H., Donaldson J.G. 1997. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139:49–61 10.1083/jcb.139.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H., Klausner R.D., Donaldson J.G. 1996. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J. Cell Biol. 134:935–947 10.1083/jcb.134.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., Barry S., Woods A., van der Sluijs P., Norman J. 2001. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11:1392–1402 10.1016/S0960-9822(01)00442-0 [DOI] [PubMed] [Google Scholar]
- Sabe H. 2003. Requirement for Arf6 in cell adhesion, migration, and cancer cell invasion. J. Biochem. 134:485–489 10.1093/jb/mvg181 [DOI] [PubMed] [Google Scholar]
- Satoh T., Endo M., Nakafuku M., Nakamura S., Kaziro Y. 1990. Platelet-derived growth factor stimulates formation of active p21ras.GTP complex in Swiss mouse 3T3 cells. Proc. Natl. Acad. Sci. USA. 87:5993–5997 10.1073/pnas.87.15.5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya A., Sata M., Takeda K., Pacheco-Rodriguez G., Ferrans V.J., Moss J., Vaughan M. 2001. ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. USA. 98:2413–2418 10.1073/pnas.051634798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Sturany S., Van Lint J., Muller F., Wilda M., Hameister H., Hocker M., Brey A., Gern U., Vandenheede J., Gress T., et al. 2001. Molecular cloning and characterization of the human protein kinase D2. A novel member of the protein kinase D family of serine threonine kinases. J. Biol. Chem. 276:3310–3318 10.1074/jbc.M008719200 [DOI] [PubMed] [Google Scholar]
- Van Lint J., Rykx A., Maeda Y., Vantus T., Sturany S., Malhotra V., Vandenheede J.R., Seufferlein T. 2002. Protein kinase D: an intracellular traffic regulator on the move. Trends Cell Biol. 12:193–200 10.1016/S0962-8924(02)02262-6 [DOI] [PubMed] [Google Scholar]
- White D.E., Kurpios N.A., Zuo D., Hassell J.A., Blaess S., Mueller U., Muller W.J. 2004. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 6:159–170 10.1016/j.ccr.2004.06.025 [DOI] [PubMed] [Google Scholar]
- Woods A.J., White D.P., Caswell P.T., Norman J.C. 2004. PKD1/PKCmu promotes alphavbeta3 integrin recycling and delivery to nascent focal adhesions. EMBO J. 23:2531–2543 10.1038/sj.emboj.7600267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman C., Ayala M.I., Wright J.R., Bard F., Bossard C., Ang A., Maeda Y., Seufferlein T., Mellman I., Nelson W.J., Malhotra V. 2004. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat. Cell Biol. 6:106–112 10.1038/ncb1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.O., Shin S., Mercurio A.M. 2005. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer Res. 65:2761–2769 10.1158/0008-5472.CAN-04-4122 [DOI] [PubMed] [Google Scholar]
- Yu X., Wang F., Liu H., Adams G., Aikhionbare F., Liu D., Cao X., Fan L., Hu G., Chen Y., et al. 2011. ACAP4 protein cooperates with Grb2 protein to orchestrate epidermal growth factor-stimulated integrin β1 recycling in cell migration. J. Biol. Chem. 286:43735–43747 10.1074/jbc.M111.278770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.