Loss of both meiosis-specific kleisins in mice reveals conserved functions of the meiotic cohesin complexes in axial element formation during spermatogenesis.
Abstract
Cohesin is a conserved multisubunit protein complex that participates in chromosome segregation, DNA damage repair, chromatin regulation, and synaptonemal complex (SC) formation. Yeast, but not mice, depleted of the cohesin subunit Rec8 are defective in the formation of the axial elements (AEs) of the SC, suggesting that, in mammals, this function is not conserved. In this paper, we show that spermatocytes from mice lacking the two meiosis-specific cohesin subunits RAD21L and REC8 were unable to initiate RAD51- but not DMC1-mediated double-strand break repair, were not able to assemble their AEs, and arrested as early as the leptotene stage of prophase I, demonstrating that cohesin plays an essential role in AE assembly that is conserved from yeast to mammals.
Introduction
In mammalian spermatogenesis, a subset of spermatogonia undergoes a terminal round of DNA replication and then enters meiosis. Meiosis is a specialized process in which two successive rounds of chromosome segregation and cell divisions occur without intervening DNA replication. This reduces the number of each chromosome from four copies in the meiocyte to one copy in haploid gametes. At the initiation of the prophase I, a proteinaceous structure called the axial element (AE) begins to form along replicated sister chromatids. Subsequently, the AEs of homologues become juxtaposed by transverse element proteins (e.g., SYCP1, beginning in zygonema), and the paired axes joined by transverse elements form the tripartite synaptonemal complexes (SCs) that connect all homologues at pachynema (Yang and Wang, 2009). This dynamic protein complex provides the structural framework in which homologous chromosomes undergo close juxtaposition and repair of double-strand breaks (DSBs) by recombination. Subsequently, but before the first meiotic division, homologues desynapse but retain stable connections formed by resolution of certain recombination events as crossovers (visible as chiasmata). In most organisms, chiasmata are required to enable proper orientation of homologues on the meiosis I spindle before the first meiotic division. Sister chromatid cohesion distal to crossovers maintains chiasmata at their initial positions until anaphase I.
During meiosis, sister chromatid cohesion is lost in two consecutive steps. Loss of chromosome arm cohesion in anaphase I releases the linkage between homologues, allowing them to segregate to opposite poles (Page and Hawley, 2003). However, maintenance of centromeric cohesion by the action of Shugoshin-like–2 in mammals ensures the generation of tension by the proper attachment of sister chromatids to the meiosis II spindle, enabling their proper segregation to opposite poles (Llano et al., 2008; Gutiérrez-Caballero et al., 2012).
During the mitotic cell cycle, sister chromatid cohesion is mediated by the multisubunit cohesin complex between S phase and anaphase (Gruber et al., 2003; Unal et al., 2007; Haering et al., 2008; Zhang et al., 2008). Structurally, the mitotic cohesin complex comprises four core proteins: SMC1α, SMC3, RAD21, and a HEAT repeat domain protein (STAG1 and STAG2; Watanabe, 2005; Hirano, 2006). SMC1α and SMC3 are members of the structural maintenance of a chromosome family of ATPases, which heterodimerize in an antiparallel orientation. The α-kleisin subunit RAD21 closes the ring cohesin complex and is the substrate of the protease separase (Uhlmann et al., 2000). There are meiosis-specific mammalian paralogues of RAD21, SMC1α, and STAG1-2, namely, REC8 and RAD21L, SMC1β, and STAG3, respectively (Parisi et al., 1999; Prieto et al., 2001; Gruber et al., 2003; Gutiérrez-Caballero et al., 2011), which lead to a variety of meiosis-specific cohesin complexes (Ishiguro et al., 2011; Gutiérrez-Caballero et al., 2011). Interestingly, yeast Rec8 is necessary for stepwise release of sister chromatid cohesion in meiosis, and this role is widely conserved across eukaryotes (Klein et al., 1999; Golubovskaya et al., 2006; Severson et al., 2009). Furthermore, the analysis of yeast mutants of Rec8 revealed additional functions in AE assembly, pairing of homologues, synapsis, and recombination (Klein et al., 1999; Watanabe and Nurse, 1999).
In mammals, cohesins colocalize and interact with the structural AE components SYCP3 and SYCP2 (Suja and Barbero, 2009). However, there is disagreement as to whether the mammalian cohesin complex is an integral part of the AE itself (Eijpe et al., 2003) or constitutes a different structure of the chromosomal core (Pelttari et al., 2001). Mouse spermatocytes lacking SMC1β, REC8, or RAD21L are able to assemble AEs but undergo meiotic arrest at the zygotene or early pachytene stages, with partially synapsed chromosomes (Bannister et al., 2004; Revenkova et al., 2004; Herrán et al., 2011). REC8 is also dispensable for AE and SC assembly in many higher eukaryotes, suggesting that cohesin may not be universally required for AE assembly (Bhatt et al., 1999; Bannister et al., 2004). However, Caenorhabditis elegans depleted of the three meiosis-specific kleisins (Rec8, COH-3, and COH-4) are unable to form AEs, similar to yeast bearing rec8Δ or smc3Δ alleles (Klein et al., 1999; Severson et al., 2009). These results suggest that this AE assembly function might be obscured in other higher eukaryotes, such as mammals, owing to the involvement of multiple kleisins (Gutiérrez-Caballero et al., 2011; Ishiguro et al., 2011; Lee and Hirano, 2011).
To better understand the role that cohesins play in AE and SC assembly in mammals, we performed a genetic depletion of the two meiosis-specific kleisins REC8 and RAD21L in mice (Bannister et al., 2004; Herrán et al., 2011). Our results reveal that either of these kleisins is each sufficient for association of the AE proteins SYCP3 and SYCP2 with chromosomes and that AE formation fails only in mice lacking both kleisins. This failure to assemble AEs leads to accumulation of cells with leptotene-like morphology, which is, to the best of our knowledge, the earliest arrest of mouse spermatogenesis ever reported. This evidence indicates that meiotic cohesin complexes are essential structural components of the AE from yeast to mammals. In addition, we show that meiotic cohesins function downstream of the SPO11-mediated DSB formation and upstream of RAD51- but not DMC1-mediated DSB repair.
Results and discussion
Mice lacking RAD21L and REC8 develop normally but are infertile
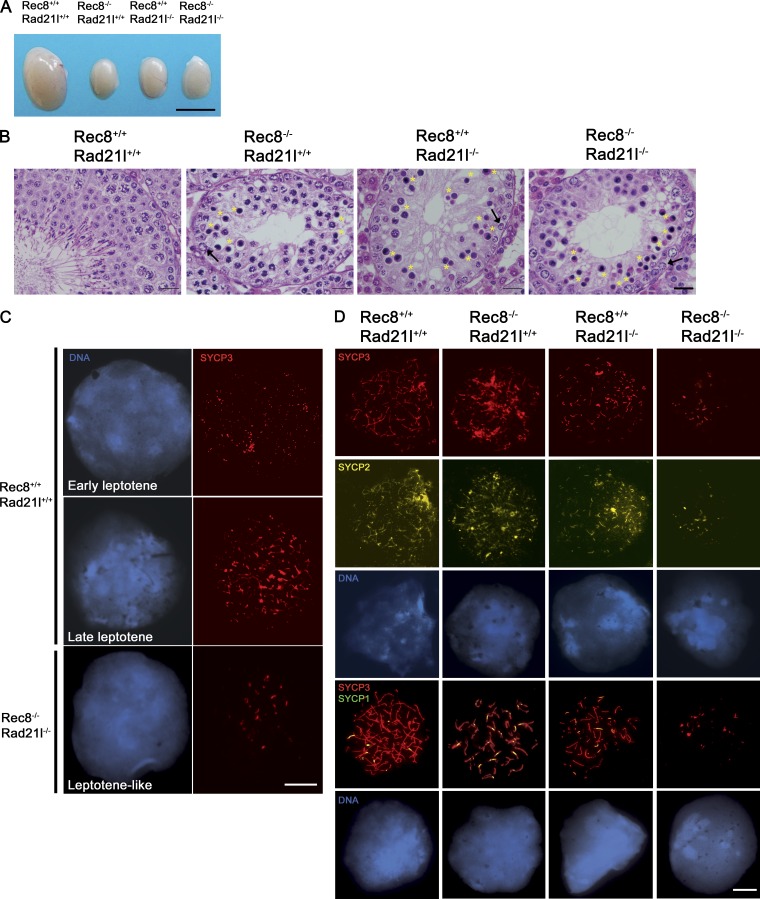
Mice lacking either of two meiosis-specific cohesin subunits, REC8 or RAD21L, show similar defects in meiosis. To test for possible overlap in the functions of these two kleisin proteins, we generated kleisin double-knockout (dKO) mice. Rad21l−/− Rec8−/−, henceforth dKO-kls animals, were obtained from crosses of double heterozygotes (Rec8+/− and Rad21l+/−) in the expected Mendelian ratios and analyzed. dKO-kls mice developed normally and displayed no overt defects besides infertility. As expected from the phenotypes of the single Rec8 and Rad21l mutants, all the dKO-kls mice (n = 11) were infertile (unpublished data; Bannister et al., 2004; Herrán et al., 2011). The testes from dKO-kls mice were reduced in size, weighing 30 ± 3 mg at 6–8 wk of age compared with 103 ± 8 mg in wild-type males (Fig. 1 A and Table S1). Furthermore, histological examination of testes from 6–8-wk-old dKO-kls males revealed seminiferous tubules that were always devoid of postmeiotic cell types, despite the presence of spermatogonia, and Sertoli and Leydig cells (Fig. 1 B and not depicted). Similar defects were also observed in the Rec8-deficient and Rad21L-deficient mice (Fig. 1, A and B; Bannister et al., 2004; Herrán et al., 2011).
Figure 1.
Absence of RAD21L and REC8 arrest mouse spermatogenesis in early prophase I. (A) dKO-kls mice show a 70% reduction in testes size compared with wild type. Similar reductions are observed in Rad21l−/− and Rec8−/− males. (B) Mutation of both Rad21l and Rec8 elicits an arrest of spermatogenesis at stage IV characterized by intermediate spermatogonia (arrows) in a representative section of a seminiferous tubule. Massive apoptosis of spermatocytes (condensed nuclei indicated by asterisks) and absence of mature spermatozoa/spermatids are observed in the dKO-kls tubules. A similar arrest is observed in seminiferous tubules from singly mutant Rad21l and Rec8 mice. (C) Immunolabeling for SYCP3 in spermatocytes from a wild-type mouse at early and late leptonema and spermatocytes arrested at a leptotene-like stage from a dKO-kls mouse. (D) dKO-kls spermatocytes arrested at the leptotene-like stage show absence of chromosomal synapsis. Double immunolabeling for SYCP3 and SYCP2 or SYCP1 shows SYCP3/SYCP2 aggregates without synapsis as indicated by the lack of SYCP1 labeling in double mutant spermatocytes. Spermatocytes from Rad21l−/− (zygotene-like arrest), Rec8−/− (zygotene-like arrest), and wild-type (zygotene stage) mice show AEs and synapsed LEs with stretches of SYCP1. Bars: (A) 5 mm; (B) 25 µm; (C and D) 100 µm.
Meiotic kleisins are essential for the formation of AEs in spermatocytes
We staged and examined spermatocyte spreads by immunolocalization of SYCP3. In the absence of RAD21L and REC8, AE assembly and synapsis between homologues were disrupted very early (Fig. 1, C and D). dKO-kls spermatocytes arrested at a leptotene-like stage (100% of cells) were characterized by the punctate aggregates of SYCP3. In contrast, thin threads were observed in late leptotene spermatocytes from wild-type and single mutant mice (Fig. 1, C and D). SYCP2, another axial protein, colocalized with SYCP3 in these aggregates (Fig. 1 D). Antibodies against SYCP1 were used to determine whether transverse components of the SC assembled in dKO-kls mutant spermatocytes. The aggregates of AE proteins did not show reactivity with SYCP1 antibodies, indicating an absence of transverse filament assembly (n = 120; Fig. 1 D). In agreement with previous studies, spermatocytes of Rad21l and Rec8 single mutant mice arrested at a zygotene-like stage, displaying several fragmented AEs and some partially synapsed lateral elements (LEs) that never progressed to the expected 19 fully synapsed autosomal bivalent chromosomes observed in wild-type pachynema (unpublished data; Bannister et al., 2004; Herrán et al., 2011).
Tubule section in wild-type mice can be categorized in stages running from I to XII according to the spectrum of germ cell types that are present (Russell et al., 1990). Following these criteria, dKO-kls, Rad21l, and Rec8 mutant mice appeared to be arrested at stage IV of the seminiferous epithelium cycle (Fig. 1 B; Herrán et al., 2011). In wild-type tubules, stage IV typically corresponds to midpachynema. At this stage, mutant spermatocytes that fail to complete recombination and/or chromosome synapsis will usually undergo apoptosis (de Rooij and de Boer, 2003). As examples, MSH5-, DMC1-, or SPO11-deficient spermatocytes arrest at stage IV, but the terminal stage was described as zygonema, late zygonema, and midpachynema, respectively, based on the progression of synapsis (Yoshida et al., 1998; de Vries et al., 1999; Baudat et al., 2000). Interestingly, dKO-kls spermatocytes arrest at a stage best described as leptonema because they lack AEs yet remain viable up to stage IV. The absence of later stages indicates that these spermatocytes then undergo apoptosis, likely caused by activation of meiotic checkpoints. Thus, disruption of the meiotic cohesin complexes leads to absence of AEs, which elicits spermatocyte death after an extended leptotene-like stage.
DSBs are formed but not repaired in dKO-kls male mice
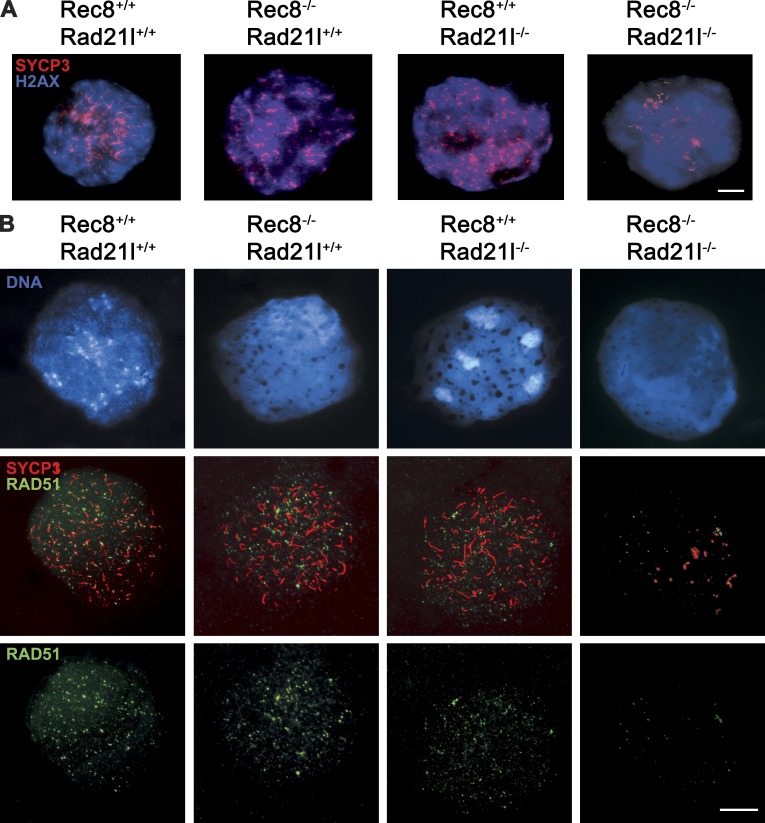
We next sought to determine how recombination mechanisms are affected by the absence of RAD21L and REC8. Programmed DSBs that initiate meiotic recombination are normally generated by the nuclease SPO11 at the early leptotene stage (Keeney, 2001). We analyzed break formation in mutant spermatocytes using antibodies against the γ-H2AX, a histone variant that is phosphorylated during early prophase I in response to SPO11-induced DSBs in an ATM-dependent manner (Mahadevaiah et al., 2001). As seen in Fig. 2 A, 100% of leptotene-like arrested spermatocytes from dKO-kls mice showed a positive staining that was similar to that observed in the wild-type mice (87 ± 30 vs. 92 ± 56; Table S1). This suggests that the formation of programmed DSBs is not markedly altered in dKO-kls mice. However, the presence of γ-H2AX staining in all the arrested spermatocytes from dKO-kls animals indicates that breaks are not repaired efficiently.
Figure 2.
Meiotic cohesins are dispensable for DSB formation but not for normal loading of RAD51. (A) Double immunolabeling of SYCP3 and γ-H2AX. In wild-type spermatocytes, γ-H2AX labels the chromatin from leptonema to pachynema when the signal disappears from autosomal chromosomes and remains only on sex body chromatin (not depicted). In the single and dKO-kls mutants, γ-H2AX also labels leptotene chromatin. (B) Double immunolabeling of SYCP3 and RAD51. In wild-type spermatocytes and spermatocytes of single Rad21l or Rec8 mutants, RAD51 localizes to AEs/LEs at leptonema. In Rec8−/− Rad21l−/− spermatocytes, there is an 8.5-fold reduction in the number of RAD51 foci. Bars, 100 µm.
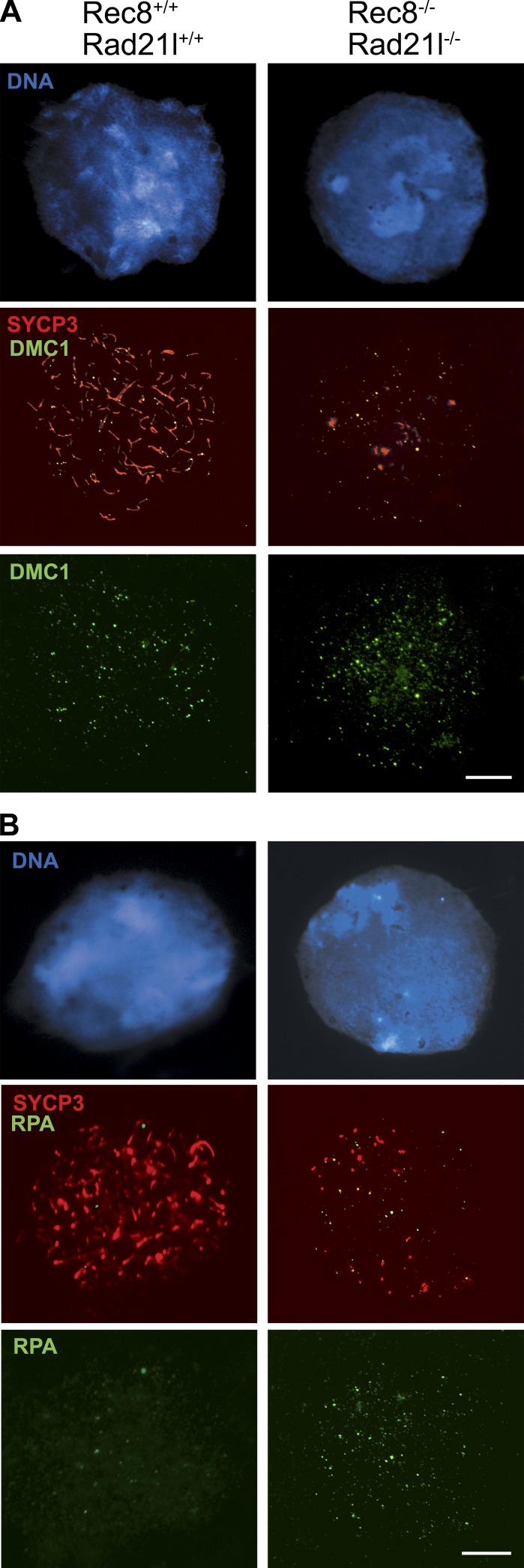
We further analyzed the recombination process to address why DSBs are not repaired in the double mutant spermatocytes. After DSBs are induced, the broken ends are resected, and the strand invasion enzymes RAD51 and DMC1 are recruited to the resulting single-strand DNA overhangs to promote homologue pairing and DNA repair (Symington and Gautier, 2011). In wild-type leptotene spermatocytes, RAD51 and DMC1 assemble on the AEs/LEs of bivalents and disappear toward pachynema, with the exception of the unsynapsed portions of the sex chromosome AEs (Tarsounas et al., 1999). As shown in Fig. 2 B, wild-type spermatocytes showed 70–150 RAD51 foci (111 ± 32). In contrast, the dKO-kls contained only a few RAD51 foci (14 ± 5, P < 0.05; Table S1). Next, we monitored the loading of DMC1, a meiosis-specific paralogue of RAD51. In wild-type spermatocytes, both recombinases colocalize extensively such that most recombination-associated foci contain both proteins (Tarsounas et al., 1999). Intriguingly, immunofluorescence with a DMC1-specific antibody revealed no detectable difference in numbers of DMC1 foci between wild type and dKO-kls in leptonema (52 ± 20 vs. 54 ± 19; Table S1 and Fig. 3 A). The presence of an equivalent number of DMC1 foci in double mutant leptotene spermatocytes implies that the γ-H2AX labeling in the dKO-kls spermatocytes is very likely generated by the meiotic SPO11 nuclease.
Figure 3.
Normal loading of DMC1 and RPA accumulation at leptonema-arrested dKO-kls spermatocytes. Double immunolabeling of SYCP3 and DMC1 or RPA. (A) In wild-type and dKO-kls spermatocytes, DMC1 protein localizes to AEs/LEs at leptonema. (B) In dKO-kls spermatocytes, there is a threefold increase in the numbers of RPA foci at the leptotene-like arrest in comparison with wild-type spermatocytes. Bars, 100 µm.
Next, we determined the distribution of the replication protein A (RPA) in dKO-kls spermatocytes. RPA is a single-stranded DNA-binding protein that enhances the formation of RAD51 and DMC1 filaments in vitro. RPA is first detected in few foci at leptonema. The initial binding of RPA to single-strand DNA at the resected ends of DSBs is supposed to be too transient to be cytologically detected by immunofluorescence because RPA is rapidly displaced by RAD51/DMC1 (Yang et al., 2008). Subsequently, after RAD51/DMC1 loading, abundant RPA foci are detected in the synapsed regions of the LEs at zygonema (Krogh and Symington, 2004; Moens et al., 2007). Intriguingly, despite failure to proceed beyond leptonema, RPA foci were increased threefold in the dKO-kls spermatocytes relative to wild type (55 ± 19 vs. 18 ± 10, P < 0.05; Table S1 and Fig. 3 B), which is very likely caused by the sharp reduction in the loading of RAD51 (Fig. 2 B; Roig et al., 2010). TRIP13 is the mammalian orthologue of the yeast Pch2 (pachytene checkpoint 2) gene, and its deletion leads to a block of spermatogenesis and oogenesis because of defects in DSB repair (Li and Schimenti, 2007; Roig et al., 2010). Interestingly, Trip13 mutant spermatocytes accumulate RPA foci in leptonema and also show a sharply reduced number of RAD51 foci and an accumulation of DMC1 foci at leptonema (Roig et al., 2010). Thus, the RAD51 and DMC1 homologous recombination pathways, although very similar biochemically (Kagawa and Kurumizaka, 2010), can be genetically dissected. Collectively, our results indicate that meiotic cohesins are dispensable for the loading of DMC1 but are essential for normal loading of RAD51 (Xu et al., 2003; Sharan et al., 2004; Yang et al., 2008; Roig et al., 2010).
Cohesin complexes in mutant spermatocytes
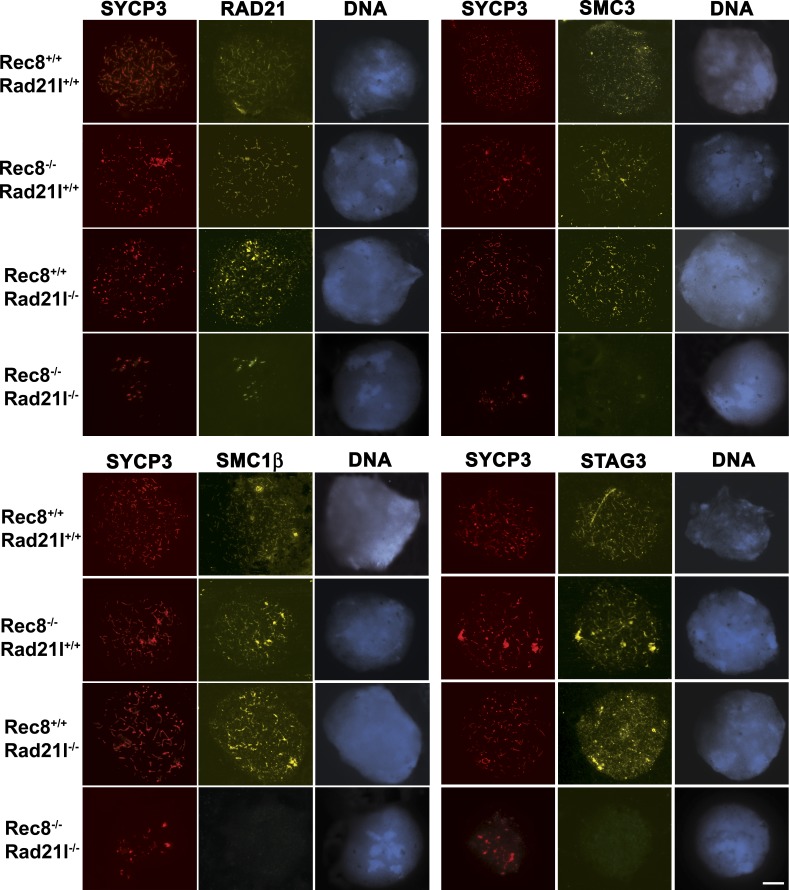
In the absence of SYCP3, the cohesin complex persists at a residual AE-like chromosomal core in mouse meiocytes (Pelttari et al., 2001; Kolas et al., 2004; Kouznetsova et al., 2005). We evaluated whether deficiency of both RAD21L and REC8 compromised the loading of other cohesin subunits by immunostaining for the cohesin subunits STAG3, SMC3, SMC1β, and RAD21 (Fig. 4). Surprisingly, we observed robust staining only for RAD21, which colocalized with SYCP3 labeling, and much fainter staining of SMC3 (Fig. 4). We did not detect either STAG3 or SMC1β (Fig. 4), suggesting that these cohesin subunits are complexed in vivo with REC8 and RAD21L kleisins. Collectively, these data suggest that the two α-kleisins, RAD21L and REC8, are not epistatic and sustain most, if not all, meiosis-specific cohesin complexes involved in AE formation and SC assembly. Our results are consistent with the observation that RAD21 is present on chromosome axes from leptonema to diplonema in mouse spermatocytes (this paper; Herrán et al., 2011) but contrast with one study in which RAD21 was first detected in the pachytene stage (Ishiguro et al., 2011). Because we observed RAD21 immunolabeling in the Rad21l−/−, Rec8−/−, and dKo-kls spermatocytes (Fig. 4), cross-reactivity of our antibody with RAD21L and/or REC8 is not likely the reason for these contrasting results, as previously claimed (Ishiguro et al., 2011). This early localization of RAD21 would suggest a direct role of this kleisin in establishing cohesion in premeiotic S phase.
Figure 4.
Cohesin complexes in the absence of RAD21L and REC8. Double immunofluorescence of SYCP3 and either RAD21, SMC3, SMC1β, or STAG3. In wild-type leptotene spermatocytes, the cohesins RAD21, SMC3, SMC1β, and STAG3 colocalize with SYCP3 along the AEs of the chromosomes. In Rad21l and Rec8 single mutant leptotene spermatocytes, RAD21, SMC3, SMC1β, and STAG3 colocalize also with SYCP3 along the AEs/LEs of the chromosomes. In spermatocytes from dKO-kls, arrested at leptonema, SMC1β and STAG3 are not detected by immunofluorescence, whereas immunolabeling for RAD21 and SMC3 renders robust and very faint fluorescence signals, respectively. Bar, 100 µm.
Sister chromatid cohesion in the absence of meiotic kleisins
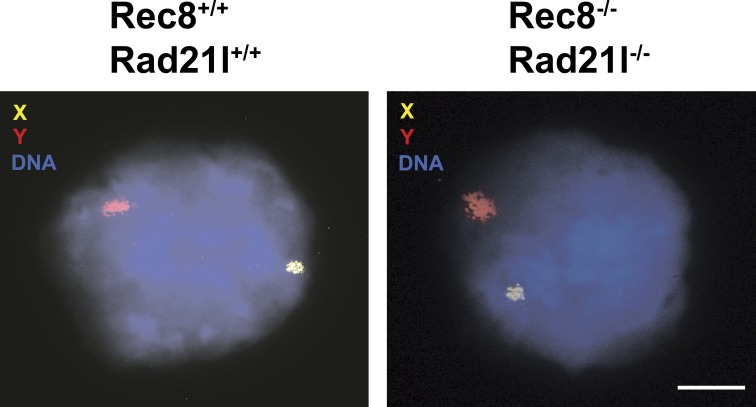
In addition to their role in AE formation and DNA looping (Revenkova and Jessberger, 2006; Novak et al., 2008), cohesin complexes must tether the two sister chromatids from S phase until the onset of anaphase, when separase is activated, cleaves the α-kleisin RAD21 subunit, and leads to opening of the cohesin ring (Hauf et al., 2001). Thus, a deficiency for two out of the three mammalian kleisins could partially affect chromosomal cohesion and/or replication of the premeiocyte/meiocyte. To test this possibility, we performed FACS analysis of whole cells from seminiferous tubules, which showed the existence of tetraploid compartment in dKO-kls testes (Fig. S1). Next, we performed FISH analysis (one probe from the X and one probe from the Y chromosomes) in dKO-kls spermatocytes (Fig. 5). The results revealed a single signal for each chromosome probe (corresponding to two closed and tethered sister chromatids). Collectively, these results suggest that sister chromatid cohesion is properly established in the preceding S phase through the somatic α-kleisin RAD21 subunit complexed with the SMC3 and SMC1α or SMC1β subunits. This result implies that meiosis-specific cohesin complexes are dispensable for cohesion, by itself, and are specifically required for SC structure.
Figure 5.
dKO-kls spermatocytes are proficient for sister chromatid cohesion. Hybridization of two DNA probes to the X and Y chromosomes renders a single dot signal for each probe in both wild-type and dKO-kls spermatocytes. Clear FISH signals for both probes can be observed in 63% of the 90 leptonema-like cells analyzed. Bar, 100 µm.
To our knowledge, deletion of both REC8 and RAD21L leads to the earliest arrest of mouse meiotic prophase reported to date, including the mutations for the individual cohesins REC8, SMC1β, and RAD21L and the structural additional components SYCP3, SYCP1, and SYCP2 (Yuan et al., 2000; Bannister et al., 2004; Revenkova et al., 2004; de Vries et al., 2005; Yang et al., 2006; Herrán et al., 2011). Interestingly, it was reported very recently that SYCP1 and SYCP3 double deficiency does not abolish the loading of the meiotic cohesin complexes nor the presynaptic pairing and early homologous recombination in mouse meiocytes (Kouznetsova et al., 2011). Thus, the meiotic cohesin complexes are required to initiate AE/LE assembly in mammals (and not vice versa), which is a prerequisite for SC formation. Our findings provide evidence for a crucial role for meiotic cohesins (REC8- and RAD21L-containing cohesin complexes) in the very early assembly of the AEs of the mammalian SC, revealing that this process is conserved from yeast to mammals.
Materials and methods
Immunocytology and antibodies
Testes were detunicated and processed for spreading using the “dry-down” technique (Peters et al., 1997). The slides were rinsed three times for 5 min in PBS and incubated overnight at room temperature with primary antibodies diluted in PBS. The primary antibodies used for immunofluorescence were rabbit αSMC3 serum K987 (1:20), rabbit αSMC1β serum K974 (1:20; Prieto et al., 2004), rabbit αSTAG3 serum K403 (1:20; Prieto et al., 2001), rabbit αRAD21 IgG K854 (1:5; Prieto et al., 2002), rabbit αSYCP2 serum K1035 (1:20), mouse αSYCP3 IgG sc-74569 (1:100), rabbit αRAD51 sc-8349 (1:30), and PC130 (1:5; EMD; provided by J. Page, Universidad Autonoma de Madrid, Madrid, Spain), rabbit αDMC1 sc-22768 (1:20; Santa Cruz Biotechnology, Inc.), rabbit αSYCP1 IgG ab15090 (1:200; Abcam), rabbit anti–γ-H2AX (ser139) IgG (#07-164; 1:200; Millipore), and rabbit αRPA IgG (1:300; provided by E. Marcon, Toronto University, Toronto, Canada). The secondary antibodies used were TRITC α-mouse 115–095-146/α-rabbit 111–025-144 and FITC α-mouse 115–095-146/α-rabbit 111–095-045 (Jackson ImmunoResearch Laboratories, Inc.; all 1:100). Slides were visualized at room temperature using a microscope (Axioplan 2; Carl Zeiss) with 63× objectives with an aperture of 1.4 (Carl Zeiss). Images were taken with a digital camera (ORCA-ER; Hamamatsu Photonics) and processed with OPENLAB 4.0.3 (PerkinElmer) and Photoshop (Adobe).
Mice
The mutation at the Rad21l−/− locus is a null allele generated by gene targeting using an insertional targeting vector (Herrán et al., 2011). Mice were genotyped by genomic Southern blot analysis of tail genomic DNA digested with SpeI and hybridized with a 5′ external probe. The probe (1 kb) was generated by PCR using the primers 5′-ACTAGTCTAAATAAAGGTCTT-3′ and 5′-GATTTAAGCATGAATGAAGTAAC-3′. The mutation at the Rec8 locus is a null allele (premature stop codon at exon 6) isolated in a forward genetic screen for mouse infertility (Bannister et al., 2004). Mice were genotyped by direct sequencing of the PCR-amplified exon 6 from genomic DNA using the primers 5′-CCTTTACATCCCTGTTCTC-3′ and 5′-ACAGGAACACCAACTAACTC-3′. dKO-kls mice were obtained by genetic crossing of double heterozygote mice (Rec8+/− Rad21l+/−) and compared with single mutant and wild-type littermates. All animal experiments were reviewed and approved by the Consejo Superior de Investigaciones Científicas (Spanish Research Council) and the Universidad de Salamanca National Committee on Bioethics.
FISH analysis
The mouse Y-specific probes were obtained by PCR using the following three set of primers: 1S, 5′-TAGGATGGTAAGCCCAATGC-3′; 1AS, 5′-TTGGTTGGTTAATTGTTTGGG-3′; 2S, 5′-CATGCCCCTTGGACTGAC-3′; 2AS, 5′-CTTTTTTTTTCCCCCTCTGG-3′; 3S, 5′-TCCTCTTGCAGAGAAGGGAC-3′; and 3AS, 5′-CCTCCGCTCCAATCCTATCA-3′ (Navin et al., 1996). The X-specific DNA probe is a pericentromeric DNA fragment cloned in a plasmid (Disteche et al., 1985). Both probes were labeled by Nick translation with Dig-11–deoxy-UTP and Bio-16–deoxy-UTP and hybridized to spermatocyte spreads following standard procedures. In brief, slides were pretreated with pepsin (0.005% in 0.01 N HCl for 5 min at 37°C), dehydrated, and RNase treated (0.1 mg/ml in PBS for 1 h at 37°C). After denaturation in 70% formamide during 3 min at 75°C, slides were dehydrated in ice-cold ethanol and hybridized overnight at 37°C to a denatured DNA probe. After washing (50% formamide and 2× SSC at 42°C), biotin- and digoxigenin-labeled probes were immunodetected using streptavidin-Cy3/goat biotinylated antistreptavidin/streptavidin-Cy3 (Jackson ImmunoResearch Laboratories, Inc.) and mouse antidigoxigenin-FITC/goat anti–mouse/rabbit anti–mouse-FITC (Roche), respectively (Pendás et al., 1994).
FACS analysis
Wild-type, Rad21l−/−, and dKO-kls testicular cell preparation and their DNA content measurement were performed as previously described (Malkov et al., 1998). In brief, testes were dissected in separation medium (4 mM l-glutamine, 1.5 mM sodium pyruvate, 10% fetal calf serum, and 75 mg/ml ampicillin in DME containing nonessential amino acids). After decapsulation, the tubules were then treated with collagenase for 5 min at 37°C. The sedimented seminiferous cords were washed in separation medium and treated with 2.5 µg/ml trypsin and 1 U/ml DNase I for 2 min at 37°C. Using a pair of scalpels, the tubules were disintegrated, and the resulting tissue suspension was passed through a 50-µm nylon mesh, washed twice by centrifugation, and counted. Cells were brought to a concentration of 2 × 106 cells/ml in separation medium, diluted 1:1 with propidium iodide solution (10 mM Tris, pH 8, 1 mM NaCl, 0.1% Nonidet P-40, 0.7 mg/ml RNase A, and 0.05 mg/ml propidium iodide), and freshly analyzed by a cell-sorting instrument (FACSort; BD).
Online supplemental material
Fig. S1 shows that the absence of RAD21L and REC8 does not impair DNA replication and does not provoke loss of chromatid cohesion in dKO-kls spermatocytes. Table S1 shows the number of early recombination-associated foci in spermatocytes and the weight of testis from wild-type and dKO-kls mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201100/DC1.
Supplementary Material
Acknowledgments
We wish to express our sincere thanks to E. Marcon and J. Page for providing antibodies (RPA and RAD51, respectively), I. Ramos-Fernández and Teresa Hernandez for excellent technical assistance, and C. López-Otín for his support. We specially appreciate the comments from an anonymous reviewer.
This work was supported by grant SAF2011-25252 and Junta de Castilla y León. C. Gutiérrez-Caballero and Y. Herrán are supported by Fondo de Investigación Sanitaria and Formación de Personal Investigador fellowships, respectively. E. Llano is the recipient of a Ramón y Cajal Research contract.
Footnotes
Abbreviations used in this paper:
- AE
- axial element
- dKO
- double knockout
- DSB
- double-strand break
- LE
- lateral element
- RPA
- replication protein A
- SC
- synaptonemal complex
References
- Bannister L.A., Reinholdt L.G., Munroe R.J., Schimenti J.C. 2004. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 40:184–194 10.1002/gene.20085 [DOI] [PubMed] [Google Scholar]
- Baudat F., Manova K., Yuen J.P., Jasin M., Keeney S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 6:989–998 10.1016/S1097-2765(00)00098-8 [DOI] [PubMed] [Google Scholar]
- Bhatt A.M., Lister C., Page T., Fransz P., Findlay K., Jones G.H., Dickinson H.G., Dean C. 1999. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19:463–472 10.1046/j.1365-313X.1999.00548.x [DOI] [PubMed] [Google Scholar]
- de Rooij D.G., de Boer P. 2003. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet. Genome Res. 103:267–276 10.1159/000076812 [DOI] [PubMed] [Google Scholar]
- de Vries F.A., de Boer E., van den Bosch M., Baarends W.M., Ooms M., Yuan L., Liu J.G., van Zeeland A.A., Heyting C., Pastink A. 2005. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19:1376–1389 10.1101/gad.329705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S.S., Baart E.B., Dekker M., Siezen A., de Rooij D.G., de Boer P., te Riele H. 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13:523–531 10.1101/gad.13.5.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C.M., Tantravahi U., Gandy S., Eisenhard M., Adler D., Kunkel L.M. 1985. Isolation and characterization of two repetitive DNA fragments located near the centromere of the mouse X chromosome. Cytogenet. Cell Genet. 39:262–268 10.1159/000132155 [DOI] [PubMed] [Google Scholar]
- Eijpe M., Offenberg H., Jessberger R., Revenkova E., Heyting C. 2003. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J. Cell Biol. 160:657–670 10.1083/jcb.200212080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya I.N., Hamant O., Timofejeva L., Wang C.J., Braun D., Meeley R., Cande W.Z. 2006. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119:3306–3315 10.1242/jcs.03054 [DOI] [PubMed] [Google Scholar]
- Gruber S., Haering C.H., Nasmyth K. 2003. Chromosomal cohesin forms a ring. Cell. 112:765–777 10.1016/S0092-8674(03)00162-4 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Caballero C., Herrán Y., Sánchez-Martín M., Suja J.A., Barbero J.L., Llano E., Pendás A.M. 2011. Identification and molecular characterization of the mammalian α-kleisin RAD21L. Cell Cycle. 10:1477–1487 10.4161/cc.10.9.15515 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Caballero C., Cebollero L.R., Pendás A.M. 2012. Shugoshins: from protectors of cohesion to versatile adaptors at the centromere. Trends Genet. 10.1016/j.tig.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Haering C.H., Farcas A.M., Arumugam P., Metson J., Nasmyth K. 2008. The cohesin ring concatenates sister DNA molecules. Nature. 454:297–301 10.1038/nature07098 [DOI] [PubMed] [Google Scholar]
- Hauf S., Waizenegger I.C., Peters J.M. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 293:1320–1323 10.1126/science.1061376 [DOI] [PubMed] [Google Scholar]
- Herrán Y., Gutiérrez-Caballero C., Sánchez-Martín M., Hernández T., Viera A., Barbero J.L., de Álava E., de Rooij D.G., Suja J.Á., Llano E., Pendás A.M. 2011. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 30:3091–3105 10.1038/emboj.2011.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7:311–322 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- Ishiguro K., Kim J., Fujiyama-Nakamura S., Kato S., Watanabe Y. 2011. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 12:267–275 10.1038/embor.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W., Kurumizaka H. 2010. From meiosis to postmeiotic events: uncovering the molecular roles of the meiosis-specific recombinase Dmc1. FEBS J. 277:590–598 10.1111/j.1742-4658.2009.07503.x [DOI] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1–53 10.1016/S0070-2153(01)52008-6 [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 98:91–103 10.1016/S0092-8674(00)80609-1 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Yuan L., Hoog C., Heng H.H., Marcon E., Moens P.B. 2004. Male mouse meiotic chromosome cores deficient in structural proteins SYCP3 and SYCP2 align by homology but fail to synapse and have possible impaired specificity of chromatin loop attachment. Cytogenet. Genome Res. 105:182–188 10.1159/000078188 [DOI] [PubMed] [Google Scholar]
- Kouznetsova A., Novak I., Jessberger R., Höög C. 2005. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J. Cell Sci. 118:2271–2278 10.1242/jcs.02362 [DOI] [PubMed] [Google Scholar]
- Kouznetsova A., Benavente R., Pastink A., Höög C. 2011. Meiosis in mice without a synaptonemal complex. PLoS ONE. 6:e28255 10.1371/journal.pone.0028255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233–271 10.1146/annurev.genet.38.072902.091500 [DOI] [PubMed] [Google Scholar]
- Lee J., Hirano T. 2011. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J. Cell Biol. 192:263–276 10.1083/jcb.201008005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.C., Schimenti J.C. 2007. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 3:e130 10.1371/journal.pgen.0030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano E., Gómez R., Gutiérrez-Caballero C., Herrán Y., Sánchez-Martín M., Vázquez-Quiñones L., Hernández T., de Alava E., Cuadrado A., Barbero J.L., et al. 2008. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 22:2400–2413 10.1101/gad.475308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner J.M., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27:271–276 10.1038/85830 [DOI] [PubMed] [Google Scholar]
- Malkov M., Fisher Y., Don J. 1998. Developmental schedule of the postnatal rat testis determined by flow cytometry. Biol. Reprod. 59:84–92 10.1095/biolreprod59.1.84 [DOI] [PubMed] [Google Scholar]
- Moens P.B., Marcon E., Shore J.S., Kochakpour N., Spyropoulos B. 2007. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 120:1017–1027 10.1242/jcs.03394 [DOI] [PubMed] [Google Scholar]
- Navin A., Prekeris R., Lisitsyn N.A., Sonti M.M., Grieco D.A., Narayanswami S., Lander E.S., Simpson E.M. 1996. Mouse Y-specific repeats isolated by whole chromosome representational difference analysis. Genomics. 36:349–353 10.1006/geno.1996.0473 [DOI] [PubMed] [Google Scholar]
- Novak I., Wang H., Revenkova E., Jessberger R., Scherthan H., Höög C. 2008. Cohesin Smc1β determines meiotic chromatin axis loop organization. J. Cell Biol. 180:83–90 10.1083/jcb.200706136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. 2003. Chromosome choreography: the meiotic ballet. Science. 301:785–789 10.1126/science.1086605 [DOI] [PubMed] [Google Scholar]
- Parisi S., McKay M.J., Molnar M., Thompson M.A., van der Spek P.J., van Drunen-Schoenmaker E., Kanaar R., Lehmann E., Hoeijmakers J.H.J., Kohli J. 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19:3515–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelttari J., Hoja M.R., Yuan L., Liu J.G., Brundell E., Moens P., Santucci-Darmanin S., Jessberger R., Barbero J.L., Heyting C., Höög C. 2001. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 21:5667–5677 10.1128/MCB.21.16.5667-5677.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendás A.M., Moran P., Freije J.P., Garcia-Vazquez E. 1994. Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet. Cell Genet. 67:31–36 10.1159/000133792 [DOI] [PubMed] [Google Scholar]
- Peters A.H., Plug A.W., van Vugt M.J., de Boer P. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66–68 10.1023/A:1018445520117 [DOI] [PubMed] [Google Scholar]
- Prieto I., Suja J.A., Pezzi N., Kremer L., Martínez-A C., Rufas J.S., Barbero J.L. 2001. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat. Cell Biol. 3:761–766 10.1038/35087082 [DOI] [PubMed] [Google Scholar]
- Prieto I., Pezzi N., Buesa J.M., Kremer L., Barthelemy I., Carreiro C., Roncal F., Martinez A., Gomez L., Fernandez R., et al. 2002. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 3:543–550 10.1093/embo-reports/kvf108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto I., Tease C., Pezzi N., Buesa J.M., Ortega S., Kremer L., Martínez A., Martínez-A C., Hultén M.A., Barbero J.L. 2004. Cohesin component dynamics during meiotic prophase I in mammalian oocytes. Chromosome Res. 12:197–213 10.1023/B:CHRO.0000021945.83198.0e [DOI] [PubMed] [Google Scholar]
- Revenkova E., Jessberger R. 2006. Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma. 115:235–240 10.1007/s00412-006-0060-x [DOI] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Hodges C.A., Hunt P.A., Liebe B., Scherthan H., Jessberger R. 2004. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6:555–562 10.1038/ncb1135 [DOI] [PubMed] [Google Scholar]
- Roig I., Dowdle J.A., Toth A., de Rooij D.G., Jasin M., Keeney S. 2010. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 6:e1001062 10.1371/journal.pgen.1001062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L.D., Ren H.P., Sinha Hikim I., Schulze W., Sinha Hikim A.P. 1990. A comparative study in twelve mammalian species of volume densities, volumes, and numerical densities of selected testis components, emphasizing those related to the Sertoli cell. Am. J. Anat. 188:21–30 10.1002/aja.1001880104 [DOI] [PubMed] [Google Scholar]
- Severson A.F., Ling L., van Zuylen V., Meyer B.J. 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23:1763–1778 10.1101/gad.1808809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S.K., Pyle A., Coppola V., Babus J., Swaminathan S., Benedict J., Swing D., Martin B.K., Tessarollo L., Evans J.P., et al. 2004. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 131:131–142 10.1242/dev.00888 [DOI] [PubMed] [Google Scholar]
- Suja J.A., Barbero J.L. 2009. Cohesin complexes and sister chromatid cohesion in mammalian meiosis. Genome Dyn. 5:94–116 10.1159/000166622 [DOI] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- Tarsounas M., Morita T., Pearlman R.E., Moens P.B. 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147:207–220 10.1083/jcb.147.2.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 103:375–386 10.1016/S0092-8674(00)00130-6 [DOI] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J.M., Koshland D. 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science. 317:245–248 10.1126/science.1140637 [DOI] [PubMed] [Google Scholar]
- Watanabe Y. 2005. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 21:405–412 10.1016/j.tig.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 400:461–464 10.1038/22774 [DOI] [PubMed] [Google Scholar]
- Xu X., Aprelikova O., Moens P., Deng C.X., Furth P.A. 2003. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 130:2001–2012 10.1242/dev.00410 [DOI] [PubMed] [Google Scholar]
- Yang F., Wang P.J. 2009. The mammalian synaptonemal complex: a scaffold and beyond. Genome Dyn. 5:69–80 10.1159/000166620 [DOI] [PubMed] [Google Scholar]
- Yang F., De La Fuente R., Leu N.A., Baumann C., McLaughlin K.J., Wang P.J. 2006. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 173:497–507 10.1083/jcb.200603063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Eckardt S., Leu N.A., McLaughlin K.J., Wang P.J. 2008. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J. Cell Biol. 180:673–679 10.1083/jcb.200709057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T. 1998. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell. 1:707–718 10.1016/S1097-2765(00)80070-2 [DOI] [PubMed] [Google Scholar]
- Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Höög C. 2000. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 5:73–83 10.1016/S1097-2765(00)80404-9 [DOI] [PubMed] [Google Scholar]
- Zhang N., Kuznetsov S.G., Sharan S.K., Li K., Rao P.H., Pati D. 2008. A handcuff model for the cohesin complex. J. Cell Biol. 183:1019–1031 10.1083/jcb.200801157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.