Abstract
Deregulation of the mammalian target of rapamycin (mTOR) signaling pathway has been found in a variety of human cancers. However, the exact molecular mechanism how the mTOR signaling pathway is regulated remains largely elusive. Recently, DEPTOR was identified as an endogenous mTOR inhibitor that could suppress mTOR activity in vivo. More importantly, accumulated evidence has implicated that DEPTOR plays a pivotal role in the development and progression of human malignances, which could in part be mediated through its inhibitory role toward mTOR. Furthermore, three independent laboratories including our own have demonstrated that the stability of DEPTOR is controlled by the SCFβ-TrCP E3 ubiquitin ligase and deregulated DEPTOR destruction might contribute to hyperactivation of mTOR in pathologic conditions including cancer. This review discusses the recent literature regarding the function of DEPTOR involved in cell growth, apoptosis, autophagy, epithelial-mesenchymal transition, and drug resistance, all of which are associated with the pathogenesis of human cancers. Moreover, we also summarize that targeting DEPTOR may be a novel strategy for achieving better anticancer treatments.
Introduction
Although many scientists and physicians have made significant efforts to fight cancers for the past several decades, cancer is still one of the most common causes of deaths in the world [1]. For example, statistical evidence shows that there is one in four deaths due to cancer in the United States. Moreover, it has an estimated 1,638,910 new cancer cases and 577,190 cancer-causing deaths that are projected to occur in 2012 [1]. Unfortunately, the 5-year relative survival rate for all human cancers diagnosed is 66% at present [1], suggesting that cancer is still a major health problem in the United States, just falling short of cardiovascular/heart disease. The causes responsible for cancer development and progression have been intensively investigated during the past 50 years; however, the molecular mechanisms of carcinogenesis are still poorly understood.
Toward the major strides to better understand cancer-causing mechanisms, in the recent years, many studies have demonstrated that multiple signaling pathways are involved in tumorigenesis such as phosphatidylinositol-3 kinase (PI3K)/Akt, Hedgehog, mammalian target of rapamycin (mTOR), nuclear factor κB, epidermal growth factor receptor, platelet-derived growth factor, extracellular signal-regulated kinase, fibroblast growth factor, mitogen-activated protein kinase, insulin-like growth factor, Notch, Ras, and the WNT/β-catenin signaling pathway [2–13]. Among these signaling pathways, the mTOR pathway received increased attention because mounting evidence has demonstrated that targeting the mTOR pathway could be a promising anticancer therapeutic option [14]. More importantly, many mTOR inhibitors that have been and are undergoing clinical trials to fight against various types of human cancers have been developed [15]. However, the outputs of these agents that have already underwent clinical trials are not favorable due in part to the complicated regulatory mechanisms composed of multiple feedback loops for the mTOR signaling pathway. Therefore, a better understanding of how mTOR activity is regulated by the interwoven network of signaling circuits in normal physiology as well as in various pathologic conditions will provide insights into better clinical intervention with the mTOR inhibitors. To that end, the group led by Dr David Sabatini recently found an endogenous mTOR inhibitor, DEPTOR, which is overexpressed in multiple myeloma cells and required for their survival [16]. More recently, we and other two groups independently reported that the stability of DEPTOR is governed by SCFβ-TrCP (β-transducin repeat-containing protein), an E3 ubiquitin ligase [17–19]. Accumulated evidence has also demonstrated that DEPTOR plays pivotal roles in tumorigenesis. Therefore, in the following sections, we will summarize what is currently known regarding DEPTOR in the tumorigenesis and how these newly generated knowledge in DEPTOR regulation would be helpful for designing novel therapeutic strategies through targeting DEPTOR toward achieving a better treatment outcome for a subset of human cancer patients.
mTOR Signaling Pathway
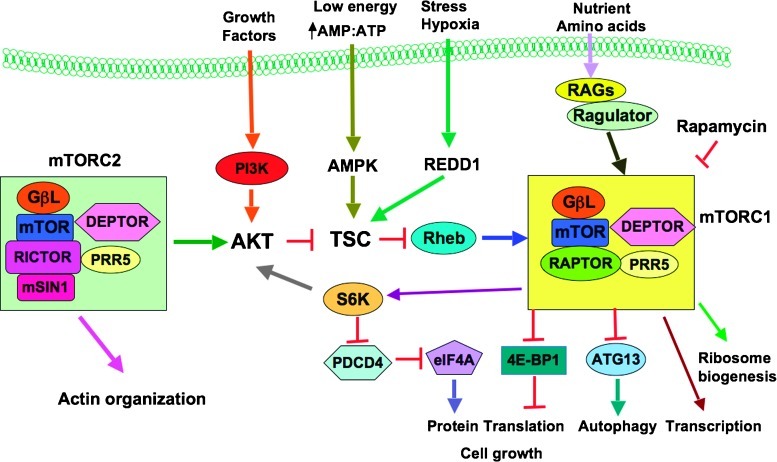
mTOR belongs to the PI3K-related protein kinase subfamily that plays a pivotal role in the regulation of various cellular processes including cell growth, apoptosis, and cell metabolism [20]. As illustrated in Figure 1, mTOR consists of two distinct subcomplexes, termed mTOR complex 1 (mTORC1) and 2 (mTORC2), which have slight differences in the subunit compositions, and thus physiological functions [20]. Specifically, mTORC1 consists of five components, mTOR, Raptor (regulatory-associated protein of mTOR), mLST8/GβL (mammalian lethal with Sec13 protein 8/G protein β-subunit-like protein), PRAS40 (proline-rich Akt substrate of 40 kDa), and DEPTOR (DEP-domain-containing mTOR-interacting protein) [20]. mTORC2 is composed of six components, including mTOR, Rictor (rapamycin-insensitive companion of mTOR), mLST8/GβL, DEPTOR, PROTOR (protein observed with Rictor 1)/PRR5 (proline-rich protein 5), and mitogen-activated protein kinase-associated protein 1 (also known as mSIN1) [20]. Both biochemical and genetic studies have established that mTOR is an important sensor of various metabolic and nutrient stresses, receiving and interpreting environmental inputs and transducing them to the downstream signaling pathways to control cellular metabolism, cellular growth, and survival [21].
Figure 1.
Schematic illustration of the mTOR signaling pathway. AMPK indicates AMP-activated kinase; ATG13, autophagy-related protein 13; DEPTOR, DEP-domain-containing mTOR-interacting protein; 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; HIF-1α, hypoxia-inducible factor 1α; mLST8/GβL, mammalian lethal with Sec13 protein 8/G protein β-subunit-like protein; mSin1, stress-activated protein kinase interacting protein; mTOR, mammalian target of rapamycin; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; PDCD4, programmed cell death 4; PI3K, phosphatidylinositol 3-kinase; PRAS40, proline-rich Akt substrate of 40 kDa; PRR5, proline-rich protein 5; PROTOR, protein observed with Rictor-1; Raptor, regulatory-associated protein of mTOR; REDD1, regulated in development and DNA damage response 1; Rheb, Ras homolog enriched in brain; Rictor, rapamycin-insensitive companion of mTOR; S6K, ribosomal p70 S6 kinase; TSC, tuberous sclerosis complex.
It is known that mTORC1 activates protein synthesis, nutrient transport, lipid synthesis, cell growth, and other processes in response to several stresses including growth factors, low energy, and nutrients. In addition, functioning as a nutrient and energy sensor, mTORC1 negatively regulates autophagy, a process that is necessary for cell survival under the starvation condition [14]. More specifically, under unfavorable nutrient conditions (low-energy state) including serum or glucose starvation, mTORC1 kinase activity is kept low, and cellular metabolism and growth are therefore restrained at basal levels. When cells are shifted to a more favorable environment (high-energy state), mTORC1 signaling is activated to stimulate cell metabolism and cell growth [14,22]. mTORC1 exerts its regulatory functions partly through phosphorylating downstream targets such as two well-characterized target proteins S6K (p70 S6 ribosomal kinase) and 4E-BP1 (the eukaryotic translation initiation factor 4E-binding protein 1) [14]. Nevertheless, mTORC2 functions as an Akt-and SGK1-activating kinase, which phosphorylates the hydrophobic motif of Akt at Ser473 and SGK1 (serum/glucocorticoid regulated kinase 1) at Ser422, leading to full activation of the Akt and SGK1 kinase [14]. In addition, mTORC2 mediates the actin cytoskeletal organization, thereby playing a critical role in tumor cell motility, invasiveness and metastasis [23]. However, given the critical role of mTOR kinases in cell fate control, it remains largely unknown how the activation of mTOR kinase activity is regulated in vivo.
DEPTOR as an mTOR Inhibitor
It is noteworthy that the exact functions as well as the regulatory mechanisms of each component in mTORC1 and mTORC2 currently are not fully understood. Recently, Peterson et al. [16] made a groundbreaking contribution by demonstrating a critical function of DEPTOR in mTOR pathway. They found that DEPTOR is part of mTORC1 and mTORC2 and interacts specifically with mTOR [16]. Moreover, DEPTOR depletion activates both mTORC1 and mTORC2 signaling and enhances the in vitro kinase activities of both complexes, suggesting that DEPTOR is an inhibitor of mTORC1 and mTORC2 kinase activities [16]. Furthermore, both mTORC1 and mTORC2 could negatively regulate the expression of DEPTOR at both transcriptional and posttranslational levels [16]. More importantly, mTOR-dependent phosphorylation of DEPTOR promotes its release from mTOR, thereby releasing its inhibitory role. Consistent with this notion, overexpression of DEPTOR inhibits mTORC1 signaling. However, elevated expression of DEPTOR unexpectedly activates the Akt signaling, in part due to DEPTOR-mediated suppression of the S6K-dependent negative feedback loop toward the IRS1-PI3K-Akt signaling axis [16]. Taken together, these findings suggest that DEPTOR functions as an endogenous inhibitor of mTOR.
Regulation of DEPTOR by the E3 Ubiquitin Ligase SCFβ-TrCP
The 26S proteasomal destruction pathway has been previously implicated in the regulation of the mTOR signaling pathway [24]. It has been found that cells displayed a significant decrease in mTORC1 activity when treated with the proteasomal inhibitor, MG132 [24]. Although Raptor was demonstrated to associate with Cullin-4 [24], it remains largely unknown how the ubiquitin destruction pathway participates in the regulation of the mTOR signaling pathway. Recently, our studies provided strong experimental evidence that SCFβ-TrCP-dependent destruction of the mTOR inhibitor, DEPTOR, governs mTOR activity and, subsequently, cellular response to environmental stresses. At the same time, two other independent groups also identified that the stability of DEPTOR is regulated by the β-TrCP-containing E3 ubiquitin ligase [17–19]. Here, we will briefly discuss how β-TrCP can influence mTOR signaling by promoting the ubiquitination and subsequent destruction of its endogenous inhibitor, DEPTOR.
SCFβ-TrCP as the Putative E3 Ligase Responsible for DEPTOR Destruction
β-TrCP is one of the F-box family of proteins, which forms a multi-component SCF (Skp1-Cullin-1-F-box) type of E3 ubiquitin ligase complex. Recent studies have identified multiple specific substrates of β-TrCP including Cdc25a [25,26], β-catenin [27,28], caspase 3 [29], Emi1 [30,31], Mdm2 [32], IκB [33], PDCD4 (programmed cell death 4) [34], Snail [35], Claspin [36], REST (RE1-silencing transcription factor) [37,38], BimEL (Bcl-2 interacting mediator of cell death extra long) [39], and Wee1 [40]. However, until now, there is no characterized role for β-TrCP in either cellular metabolism or mTOR signaling regulation. To that end, we recently found that DEPTOR interacts with β-TrCP and that SCFβ-TrCP controls DEPTOR abundance, suggesting that SCFβ-TrCP could be a candidate E3 ubiquitin ligase for DEPTOR [18]. Importantly, the research group led by Dr Michele Pagano also found that serum stimulation led to a significant increase in the binding of DEPTOR to endogenous β-TrCP, indicating that DEPTOR is a serum-dependent ubiquitin substrate of β-TrCP [17]. Moreover, silencing endogenous β-TrCP increased the DEPTOR half-life on serum stimulation, indicating that β-TrCP indeed controls the DEPTOR stability, a critical negative regulator of mTOR activation [17]. Consistent with this notion, the results reported independently from Dr Yi Sun's laboratory also showed that DEPTOR binds to β-TrCP and SCF complex through which its stability is negatively regulated by β-TrCP [19]. Furthermore, β-TrCP shortens the DEPTOR protein half-life and promotes DEPTOR ubiquitination [19]. Taken together, three groups independently identified SCFβ-TrCP as an E3 ligase that governs DEPTOR degradation.
DEPTOR Degron Is Controlled by Phosphorylation
It is known that there are two required conditions for β-TrCP to promote the degradation of its substrates: 1) most substrates typically containing the canonical DSGxxS phospho-degron and 2) proper phosphorylation of the substrate by one or a combination of various kinasesisrequiredfor recognitionby β-TrCP [41]. Unexpectedly, DEPTOR does not contain a canonical DpSGxxpS degron that could be recognized by SCFβ-TrCP; instead, it contains a derivative pS/TpSGxxpS phospho-degron variant [18]. More importantly, Duan et al. [17] found that phosphorylation of all three serine residues within the putative DEPTOR phospho-degron (Ser286, Ser287, and Ser291) is required for the interaction between DEPTOR and β-TrCP. Consistently, Zhao et al. [19] reported that phosphorylation of Ser293 and Ser299 may function to prime the phosphorylation of Ser286, Ser287, and Ser291. Similarly, using a combination of biochemical assays coupled with mass spectrometry analysis, we also identified that phosphorylation of multiple sites (S286, S287 S265, S286, S293, T295, and S299) within DEPTOR is necessary for its interaction with β-TrCP [18].
Casein Kinase I Participates in the Regulation of DEPTOR Stability
Because phosphorylation of the substrates within their phospho-degrons is a prerequisite for proper destruction mediated by the SCFβ-TrCP E3 ubiquitin ligase complex, we were therefore interested in further defining the upstream kinases responsible for phosphorylating DEPTOR, which triggers its interaction with β-TrCP. Our dedicated efforts with various biochemical analyses revealed a possible role for casein kinase I (CKI) in the regulation of DEPTOR stability. CKI, one of the first identified kinases, phosphorylates many downstream targets to participate in a wide range of cellular processes including cell cycle progression, circadian cycle, and apoptosis [42,43]. Interestingly, many well-characterized β-TrCP substrates including β-catenin [44], Period 1 [45] and Mdm2 [32] are CKI substrates as well, and phosphorylation by CKI is required for their efficient recognition and ubiquitination by β-TrCP. Unlike other kinases such as Akt and mTOR [46], the activity of CKI is considered relatively constant in vivo. However, previous studies have demonstrated that, like many other kinases such as glycogen synthase kinase 3 or CKII, CKI requires a priming phosphorylation event at the -3 position for efficient phosphorylation of the substrate [42].
Therefore, we reasoned that given the constant activity of CKI, the priming kinase might determine the timing of the CKI-mediated phosphorylation and subsequent destruction of the substrate. In this regard, our work further showed that, although ectopic expression of CKI greatly enhanced DEPTOR interaction with β-TrCP, this effect was significantly reduced on mTOR inhibition [18]. Furthermore, using mass spectrometry analysis, we found that mTOR could phosphorylate DEPTOR at pS265, pS286, pS293, pT295, and pS299 [18]. This result suggests that mTOR might be the sought-after priming kinase, which phosphorylates DEPTOR to trigger subsequent CKI-mediated phosphorylation of DEPTOR at the pS286 or pS287 sites within the identified phospho-degron. In keeping with this idea, the study from Pagano et al. found that mTOR phosphorylates Ser293 and Ser299 to promote degron phosphorylation by CKIα [17]. Interestingly, Dr Sun's group demonstrated that DEPTOR is phosphorylated by S6K1 and RSK1 (ribosomal S6 kinase 1) on the three identified serine residues (Ser286, Ser287, and Ser291) on serum stimulation. Specifically, S6K1 could act as a prime kinase to facilitate DEPTOR phosphorylation by RSK1, resulting in β-TrCP binding and subsequent DEPTOR ubiquitination and degradation [19]. Because both RSK1 and S6K are the mTOR downstream signaling pathways that could be activated by serum stimulation, without a doubt, we recognize that more thorough studies are required to investigate the critical role of mTOR/S6K/RSK1 in a phosphorylation-dependent degradation of DEPTOR.
Regulation of DEPTOR in Specific Conditions
Our studies revealed that DEPTOR protein levels are increased under unfavorable conditions, contributing to the suppression of mTOR kinase activity. More importantly, we found that DEPTOR is rapidly degraded in an mTOR-dependent manner when cells are under favorable conditions to allow for the timely activation of the mTOR signaling pathway. Therefore, our studies provide experimental evidence to reveal the dual inhibitory nature between DEPTOR and mTOR, which allows the formation of an intricate regulatory circuit to govern the timely response of mTOR kinase activity to environmental stresses. As a result, in response to unfavorable conditions, reduced mTOR activity leads to impaired DEPTOR destruction and further reduction in mTOR signaling. However, in response to growth-stimulatory signals, induction of mTOR kinase activity leads to rapid destruction of DEPTOR and further facilitating the activation of mTOR signaling. Thus, our study expands the current understanding of the physiological function of β-TrCP by pinpointing its critical role in regulating mTOR activity, by which β-TrCP may participate in governing cellular metabolism and nutrient sensing.
Role of DEPTOR in Human Cancers
The function of DEPTOR in tumorigenesis is still controversial. Because DEPTOR binds and inhibits mTOR, whose activity is mostly hyperactivated in many human tumors, DEPTOR is considered as a tumor suppressor protein. Indeed, down-regulation of DEPTOR has been found in many types of human cancers [17,47]. However, DEPTOR has been found to be overexpressed in many other tumor types including breast cancer, prostate cancer, chronic myeloid leukemia, and lung cancer [17,48–51]. For example, DEPTOR is overexpressed in multiple myeloma cells, and this overexpression is necessary for Akt activation and cell survival [16]. Recently, Pei et al. [52] reported that DEPTOR is significantly increased in thyroid carcinoma cells and tissues compared with normal cells and adjacent normal tissues. Moreover, DEPTOR expression is associated with lymph node status and distant metastasis [52]. Furthermore, patients diagnosed with thyroid carcinoma exhibiting overexpression of DEPTOR are susceptible to earlier recurrence and poorer survival, suggesting that DEPTOR expression is an independent prognostic marker for thyroid carcinoma [52]. In support of this notion, DEPTOR expression was found to correlate with poor survival in patients with multiple myeloma [53]. Taken together, the potential role of DEPTOR as an oncogene or a tumor suppressor may be cell context or tissue specific. In the following paragraphs, we will briefly discuss the potential roles of DEPTOR in cell growth, apoptosis, autophagy, epithelial-mesenchymal transition (EMT), and drug resistance.
Role of DEPTOR in Cell Growth
Accumulated evidence has suggested that DEPTOR plays a critical role in tumor cell growth. Peterson et al. [16] found that suppression of DEPTOR in multiple myeloma cells abolished cell proliferation, overexpression of DEPTOR was necessary for maintaining cell survival, whereas up-regulation of DEPTOR contributes to Akt activation, in part by suppressing the ability of S6K to inactivate the IRS1/PI3K/Akt signaling axis. Moreover, DEPTOR depletion increases cell size, and rapamycin treatment reversed this phenotype [16]. These results suggest that inhibiting DEPTOR expression or disrupting the DEPTOR-mTOR interaction could be a promising strategy for better treatment of multiple myeloma. Recently, Kazi et al. [54] found that DEPTOR knockdown also increased the myoblast size and the rate of proliferation. In addition, DEPTOR knockdown increased the percent of cells in the S-phase, coincident with an increased pRb phosphorylation [54]. However, the study from Dr Pagano's group showed that T98G cells expressing nondegradable mutant DEPTOR displayed reduced cell size, arguing that up-regulation of DEPTOR levels may be a viable therapeutic option for the treatment of human cancers with low DEPTOR levels. Interestingly, Zhao et al. [19] showed that DEPTOR accumulation inhibited cell death when cells were grown at high-paclitaxel conditions, whereas DEPTOR knockdown abrogated this cell growth inhibition. Furthermore, DEPTOR plays a survival-promoting role under high-paclitaxel conditions in part through activation of Akt and inhibition of paclitaxel-induced PARP cleavage [19]. Although Akt has been indicated as a major signaling node through which overexpressed DEPTOR may facilitate tumorigenesis, suggesting that further in-depth investigation is warranted to fully elucidate the role of DEPTOR in cell growth. We are also curious to further understand whether there are mTOR-independent functions of DEPTOR that contribute to this observed promoting role of DEPTOR to cell growth and cell survival.
Role of DEPTOR in Apoptosis
DEPTOR has been reported to be involved in cell apoptosis [16]. Peterson et al. [16] demonstrated that down-regulation of DEPTOR in HeLa cells protects cells from apoptosis due to increased poly ADP ribose polymerase (PARP) and cleaved caspase 3. Specifically, reductions in the serum concentration of the medium caused a decrease in Akt phosphorylation and a decrease in the cleaved forms of caspase 3 and PARP in HeLa cells, whereas DEPTOR-deficient HeLa cells were resistant to caspase 3 and PARP cleavage [16]. Moreover, they showed that DEPTOR suppression protects HeLa cells from apoptosis through inhibition of SGK1 activity [16]. Furthermore, inhibition of DEPTOR also prevented caspase 3 cleavage in serum-deprived HT-29 cells [16], suggesting that DEPTOR plays a role in apoptosis in other cancer cell line. However, it remains unclear whether this critical role of DEPTOR in cellular apoptosis is largely owing to its inhibitory role of mTOR kinase activity, and thus, further studies are required to explore whether there is mTOR-independent function of DEPTOR in governing cellular apoptotic pathways.
Role of DEPTOR in Autophagy
Recently, multiple groups have reported that DEPTOR is involved in autophagy pathway. It is known that autophagy is a catabolic process involving the massive degradation of cellular organelles and aggregate-prone proteins in lysosomes [55]. Autophagy is a tightly regulated process that allows the cells to maintain a balance between the synthesis, degradation, and subsequent recycling of intracellular constituents [55]. Therefore, autophagy is considered as an alternative source of energy to provide nutrients and energy in response to certain stresses such as serum starvation [55]. It has been well accepted that autophagy has a dual role as a key regulator of multiple aspects in human cancer [56,57]. On the one hand, autophagy is a tumor suppressor mechanism, which is connected to its roles in the prevention of oxidative stress and genomic instability [57]. In this sense, autophagy can serve as a barrier to limit tumor initiation. However, after neoplastic lesions are established, tumor cells can exploit autophagy to survive in nutrient starvation and hypoxic condition for malignant progression and tumor maintenance. Tumor cells can also induce autophagy in response to chemotherapeutics, leading to limiting drug efficacy[57]. Therefore, the role of autophagy in cancer is rather complex, which could be dependent on tumor type, stage, and the genetic context [56].
Recently, we and other groups reported that DEPTOR induced the autophagy through suppression of the mTOR activity [18,19]. We found that accumulation of DEPTOR by depletion of β-TrCP induced LC3 autophagy marker. Moreover, we observed that overexpression of a nondegradable form of DEPTOR led to a marked increase of LC3-II and a decrease in p62 abundance, indicating that these cells have an increased autophagic flux [18]. Furthermore, the autophagic vesicle marker monodansylcadaverine is increased in these cells expressing nondegradable DEPTOR. In addition, using Cherry-GFP-LC3B fusion protein to measure autophagic flux, we observed an activation of autophagy in HeLa cells after the absence of glucose, whereas depletion of DEPTOR decreased autophagic flux on glucose deprivation [18]. Consistent with our results, Dr Sun's group also independently found that DEPTOR knockdown blocks autophagy induced by glucose deprivation, as evidenced by reduced levels of LC3-II [19]. In addition, DEPTOR knockdown suppresses autophagy as evidenced by p62 accumulation. More importantly, increased numbers of AO (acridine orange)-positive cells as an autophagy marker on glucose deprivation were inhibited by DEPTOR knockdown as well [19]. More recently, Liu et al. [58] found that transcriptional levels of DEPTOR were increased time-dependently within 18 hours and then decreased after nutrient depletion, suggesting that DEPTOR contributes to autophagy in this process. Taken together, DEPTOR plays a pivotal role in autophagy pathway in response to certain stresses such as energy depletion, in part through negatively regulating the activity of the mTOR signaling pathway, one of the well-characterized critical regulators of the autophagy process.
Role of DEPTOR in EMT
Interestingly, DEPTOR has also been reported to be involved in the EMT process [59]. EMT is a process that involves remarkable morphologic changes from an epithelial cobblestone phenotype to an elongated fibroblastic phenotype, including loss of cell-cell adhesion, actin cytoskeleton reorganization, loss of cell polarity, and increased cell motility and invasion [60]. Specifically, EMT has been characterized by down-regulation of E-cadherin and up-regulation of mesenchymal molecular markers such as vimentin, Snail, Slug, and fibronectin [60]. It is well accepted that EMT plays an important role in cancer invasion and metastasis [60,61]. Recently, Chen et al. [59] demonstrated that DEPTOR depletion could induce EMT in cancer cells by reducing the expression of epithelial marker E-cadherin and inducing the expression of mesenchymal marker N-cadherin. They also showed that DEPTOR depletion increased the Snail protein in the nucleus. Moreover, DEPTOR depletion also increased Akt phosphorylation, suggesting that the Akt pathway may be involved in DEPTOR depletion-mediated acquisition of EMT [59]. Furthermore, this group found that big mitogen-activated protein kinase 1 suppresses EMT and cancer metastasis through destabilization of Snail by the DEPTOR/PI3K/mTORC2/Akt/glycogen synthase kinase 3β signaling pathways [59]. Without a doubt, further studies are required to determine the molecular mechanisms underlying DEPTOR depletion-mediated induction of EMT, which will provide critical insights into whether targeting DEPTOR could become a useful therapeutic strategy to fight against metastatic cancers.
Potential Role of DEPTOR in Drug Resistance
Recently, we and others have shown that DEPTOR plays important roles in the regulation of drug sensitivity [18,19,62]. Drug resistance is the most common cause of tumor recurrence because it leads to chemotherapeutic failure and fails to eliminate all tumor cells [63]. To that end, increasing drug sensitivity may shed light on the development of novel strategy for achieving better treatment overcome of human cancers. Therefore, we will briefly discuss the potential function of DEPTOR in the regulation of drug resistance in the following paragraphs.
A relatively large body of literature has demonstrated that DEPTOR is associated with drug resistance in human malignancies [18,19,62]. For example, Dr Sun's group reported that under β-TrCP knockdown/DEPTOR-accumulated condition, cancer cells become more resistant to rapamycin, an mTOR inhibitor whose cell growth inhibition depends on mTOR activity [19]. Moreover, the resistant to rapamycin was rescued by knockdown of DEPTOR. This indicates that DEPTOR expression might dictate the cellular response to mTOR inhibitors, in part due to the inhibitory role of DEPTOR toward the mTOR signaling pathway.
More interestingly, we found that, in a panel of multiple myeloma cell lines, DEPTOR expression levels inversely correlate with their responsiveness to Velcade treatment [18]. Multiple myeloma cell lines bearing a low expression of DEPTOR are much more sensitive to Velcade than those with relatively high DEPTOR expression [18], which seems to correlate with the unstable nature of DEPTOR that accumulates on Velcade treatment to suppress mTOR kinase activity. This might be because DEPTOR is accumulated to higher levels on Velcade treatment, which suppresses the mTOR pathway to trigger cellular apoptosis. Further studies will be necessary to fully understand whether DEPTOR is critical for Velcade sensitivity. In the meantime, our work provides some rationale for designing targeted therapies for multiple myeloma patients with Velcade; however, further work is required to validate whether DEPTOR expression could be useful to predict Velcade responsiveness.
More recently, Boyd et al. [64] demonstrated that a high DEPTOR expression is associated with improved survival in patients treated with thalidomide, suggesting that the expression levels of DEPTOR are predictive of the response to thalidomide in myeloma. In addition, Foster et al. [62] reported that DEPTOR is highly expressed in paclitaxel-resistant ovarian cancer cell lines, suggesting that, in a stage of drug resistance, overexpression of DEPTOR could promote cancer progression through increased survival. Consistently, Dr Sun's group demonstrated that cancer cells become more resistant to paclitaxel after accumulation of DEPTOR [19]. To elucidate the mechanism of paclitaxel resistance on DEPTOR accumulation, Dr Sun's group used multiple methods such as small interfering RNA knockdown of Akt or RICTOR, inhibition of PI3K by LY294003, and inhibition of mTORC1/2 by Torin. Importantly, they found that blockage of mTORC1/2 and PI3K/Akt pathways largely abrogated paclitaxel resistance [19], suggesting that DEPTOR-mediated paclitaxel resistance is due to the activation of these two pathways. Although these recent studies have shown a close relationship between DEPTOR expression and drug resistance, more thorough studies are required to further understand the biologic significance of DEPTOR and the underlying molecular mechanisms in conferring drug resistance under DEPTOR-overexpressing conditions.
DEPTOR as a Potential Drug Target for Cancer Therapy
Given the importance of DEPTOR in tumor cell growth, apoptosis, autophagy, EMT, and drug resistance, DEPTOR has recently emerged as an attractive pharmacological target for the development of novel cancer therapy. In multiple myeloma cell lines with a high expression of DEPTOR, inhibition of DEPTOR by its inhibitor could be a therapeutic strategy for multiple myeloma. However, currently, no such DEPTOR inhibitor has been developed. Nevertheless, up-regulation of DEPTOR is needed for better treatment in human cancers with a low expression of DEPTOR to regulate mTOR activity. In this regard, Liu et al. [65,66] recently showed that resveratrol, a naturally occurring polyphenol, promotes the interaction between mTOR and DEPTOR, leading to inhibition of mTOR signaling. The key question is how resveratrol increases the association between mTOR and DEPTOR. The authors reasoned that resveratrol might directly interact with mTOR or DEPTOR, resulting in an enhanced interaction between these two proteins [66]. Another possibility is that resveratrol indirectly promotes the interaction between mTOR and DEPTOR through binding to an auxiliary protein [66]. Alternatively, resveratrol may activate other signaling pathway that causes a chemical modification of DEPTOR or mTOR, leading to enhanced interaction between these proteins [66]. Because it has no experimental evidence to tease out the exact molecular mechanism underlying the role of resveratrol in regulating the mTOR pathway, further investigation is required to formally test these possibilities. It is also interesting to further explore whether other naturally occurring compounds could deliver similar effects in regulating the DEPTOR/mTOR signaling axis, thereby presenting as promising agents to treat pathologic conditions with aberrant mTOR activation including diabetes and cancers.
Conclusion
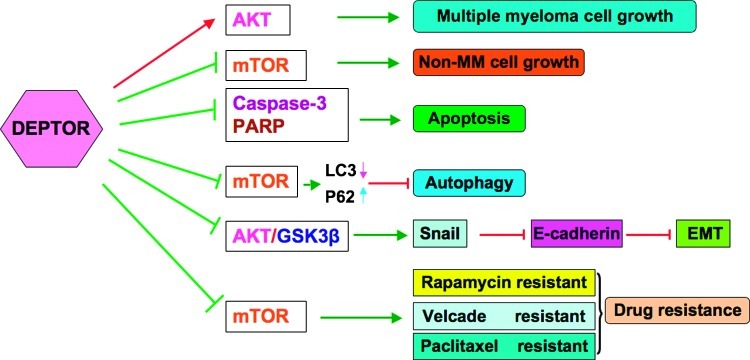
In conclusion, DEPTOR plays a critical role in the development and progression of human cancers by regulating many cellular processes such as cell growth, apoptosis, autophagy, EMT, and drug resistance (Figure 2). Moreover, DEPTOR has a dual role as tumor suppressor gene or oncogene dependent on specific tumor types. Therefore, development of inhibitors to target DEPTOR could be a novel strategy for the treatment of cancers with a high expression of DEPTOR, whereas up-regulation of DEPTOR may be a viable therapeutic option for cancers characterized by low DEPTOR levels and activation of mTOR. One alternative strategy may be to target several signaling pathways that control DEPTOR expression, such as CKI and β-TrCP. Furthermore, the natural compound resveratrol inhibits mTOR signaling by targeting DEPTOR. Because of nontoxic features, targeting DEPTOR by natural agents combined with conventional chemotherapeutics could be a promising and safer approach for achieving better treatment of human malignancies. However, because DEPTOR's role in cancer development seems to be tissue specific, further in-depth mechanistic studies are required to investigate the exact molecular mechanism how DEPTOR affects the mTOR pathway in different tissues using novel in vitro and animal models. Further testing is also required to test whether DEPTOR expression could serve as a new predictive biomarker of response to chemotherapeutics for the treatment of various types of human cancers.
Figure 2.
Illustrated presentation of the various potential functions of DEPTOR in different types of human cancers. GSK3β indicates glycogen synthase kinase 3β; MM, multiple myeloma.
Footnotes
This work was supported by grants from the National Institute of General Medicines, National Institutes of Health (NIH; GM089763 and GM094777 to W.W.).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Ng JM, Curran T. The Hedgehog's tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011;11:493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 4.Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 5.Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010;10:342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 6.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 7.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 8.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Ahmad A, Li Y, Kong D, Azmi AS, Banerjee S, Sarkar FH. Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim Biophys Acta. 2010;1806:122–130. doi: 10.1016/j.bbcan.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perkins ND. The diverse and complex roles of NF-κBsubunitsin cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 11.Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat Rev Cancer. 2010;10:618–629. doi: 10.1038/nrc2900. [DOI] [PubMed] [Google Scholar]
- 12.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 13.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 16.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, et al. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44:304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 21.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 22.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, Luo Y, Huang S. Updates of mTOR inhibitors. Anticancer Agents Med Chem. 2010;10:571–581. doi: 10.2174/187152010793498663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh P, Wu M, Zhang H, Sun H. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle. 2008;7:373–381. doi: 10.4161/cc.7.3.5267. [DOI] [PubMed] [Google Scholar]
- 25.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 26.Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFβ-TrCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latres E, Chiaur DS, Pagano M. The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 28.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβ-TrCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y, Lu H, Sun Y. SAG/ROC-SCF β-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042–1054. doi: 10.1593/neo.06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, Margottin-Goguet F, Jackson PK, Yamasaki L, Pagano M. Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 31.Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(βTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 32.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(β-TrCP) ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strack P, Caligiuri M, Pelletier M, Boisclair M, Theodoras A, Beer-Romero P, Glass S, Parsons T, Copeland RA, Auger KR, et al. SCF(β-TrCP) and phosphorylation dependent ubiquitination of IκBα catalyzed by Ubc3 and Ubc4. Oncogene. 2000;19:3529–3536. doi: 10.1038/sj.onc.1203647. [DOI] [PubMed] [Google Scholar]
- 34.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and βTrCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Lee SH, Kim HS, Kim NH, Piao S, Park SH, Jung YS, Yook JI, Park BJ, Ha NC. Role of CK1 in GSK3β-mediated phosphorylation and degradation of snail. Oncogene. 2010;29:3124–3133. doi: 10.1038/onc.2010.77. [DOI] [PubMed] [Google Scholar]
- 36.Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, Medema RH, Freire R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr Biol. 2006;16:1950–1955. doi: 10.1016/j.cub.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 37.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, et al. Control of chromosome stability by the β-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW. SCFβ-TrCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J, Pagano M. βTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knippschild U, Wolff S, Giamas G, Brockschmidt C, Wittau M, Wurl PU, Eismann T, Stoter M. The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development. Onkologie. 2005;28:508–514. doi: 10.1159/000087137. [DOI] [PubMed] [Google Scholar]
- 43.Price MA. CKI, there's more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- 44.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 45.Shirogane T, Jin J, Ang XL, Harper JW. SCFβ-TrCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 46.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gera J, Lichtenstein A. The mammalian target of rapamycin pathway as a therapeutic target in multiple myeloma. Leuk Lymphoma. 2011;52:1857–1866. doi: 10.3109/10428194.2011.580478. [DOI] [PubMed] [Google Scholar]
- 48.Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M, Naderi A, Roberts I, Barbosa-Morais NL, Garcia MJ. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- 49.Joos S, Granzow M, Holtgreve-Grez H, Siebert R, Harder L, Martin-Subero JI, Wolf J, Adamowicz M, Barth TF, Lichter P, et al. Hodgkin's lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int J Cancer. 2003;103:489–495. doi: 10.1002/ijc.10845. [DOI] [PubMed] [Google Scholar]
- 50.van Duin M, van Marion R, Vissers K, Watson JE, van Weerden WM, Schroder FH, Hop WC, van der Kwast TH, Collins C, van Dekken H. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer. 2005;44:438–449. doi: 10.1002/gcc.20259. [DOI] [PubMed] [Google Scholar]
- 51.van Duin M, van Marion R, Watson JE, Paris PL, Lapuk A, Brown N, Oseroff VV, Albertson DG, Pinkel D, de Jong P, et al. Construction and application of a full-coverage, high-resolution, human chromosome 8q genomic microarray for comparative genomic hybridization. Cytometry A. 2005;63:10–19. doi: 10.1002/cyto.a.20102. [DOI] [PubMed] [Google Scholar]
- 52.Pei L, Xie P, Zhou E, Yang Q, Luo Y, Tang Z. Overexpression of DEP domain containing mTOR-interacting protein correlates with poor prognosis in differentiated thyroid carcinoma. Mol Med Report. 2011;4:817–823. doi: 10.3892/mmr.2011.503. [DOI] [PubMed] [Google Scholar]
- 53.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Kazi AA, Hong-Brown L, Lang SM, Lang CH. Deptor knockdown enhances mTOR activity and protein synthesis in myocytes and ameliorates disuse muscle atrophy. Mol Med. 2011;17:925–936. doi: 10.2119/molmed.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eskelinen EL. The dual role of autophagy in cancer. Curr Opin Pharmacol. 2011;11:294–300. doi: 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Chen Y, Wen L, Cui G. Molecular mechanisms underlying the time-dependent autophagy and apoptosis induced by nutrient depletion in multiple myeloma: a pilot study. J Huazhong Univ Sci Technolog Med Sci. 2012;32:1–8. doi: 10.1007/s11596-012-0001-2. [DOI] [PubMed] [Google Scholar]
- 59.Chen R, Yang Q, Lee JD. BMK1 kinase suppresses epithelial-mesenchymal transition through the Akt/GSK3ss signaling pathway. Cancer Res. 2012;72:1579–1587. doi: 10.1158/0008-5472.CAN-11-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, Sarkar FH. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 2011;8:27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 62.Foster H, Coley HM, Goumenou A, Pados G, Harvey A, Karteris E. Differential expression of mTOR signalling components in drug resistance in ovarian cancer. Anticancer Res. 2010;30:3529–3534. [PubMed] [Google Scholar]
- 63.Wang Z, Li Y, Ahmad A, Azmi AS, Kong D, Banerjee S, Sarkar FH. Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13:109–118. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyd KD, Walker BA, Wardell CP, Ross FM, Gregory WM, Davies FE, Morgan GJ. High expression levels of the mammalian target of rapamycin inhibitor DEPTOR are predictive of response to thalidomide in myeloma. Leuk Lymphoma. 2010;51:2126–2129. doi: 10.3109/10428194.2010.509893. [DOI] [PubMed] [Google Scholar]
- 65.Liu M, Liu F. Resveratrol inhibits mTOR signaling by targeting DEPTOR. Commun Integr Biol. 2011;4:382–384. doi: 10.4161/cib.4.4.15309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu M, Wilk SA, Wang A, Zhou L, Wang RH, Ogawa W, Deng C, Dong LQ, Liu F. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. J BiolChem. 2010;285:36387–36394. doi: 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]