Abstract
The cytoplasmic domains of EGF-like ligands, including EGF cytoplasmic domain (EGFcyt), have important biological functions. Using specific constructs and peptides of human EGF cytoplasmic domain, we demonstrate that EGFcyt facilitates lysosomal and proteasomal protein degradation, and this coincided with growth inhibition of human thyroid and glioma carcinoma cells. EGFcyt and exon 22–23-encoded peptide (EGF22.23) enhanced procathepsin B (procathB) expression and procathB-mediated lysosomal degradation of EGFR/ErbB1 as determined by inhibitors for procathB and the lysosomal ATPase inhibitor BafA1. Presence of mbEGFctF, EGFcyt, EGF22.23, and exon 23-encoded peptides suppressed the expression of the deubiqitinating enzyme ubiquitin C-terminal hydrolase-L1 (UCH-L1). This coincided with hyperubiquitination of total cellular proteins and ErbB1/2 and reduced proteasome activity. Upon small interfering RNA-mediated silencing of endogenously expressed UCH-L1, a similar hyperubiquitinylation phenotype, reduced ErbB1/2 content, and attenuated growth was observed. The exon 23-encoded peptide region of EGFcyt was important for these biologic actions. Structural homology modeling of human EGFcyt showed that this molecular region formed an exposed surface loop. Peptides derived from this EGFcyt loop structure may aid in the design of novel peptide therapeutics aimed at inhibiting growth of cancer cells.
Introduction
Members of the family of epidermal growth factor (EGF)-like ligands are synthesized as membrane-anchored proforms and are enzymatically cleaved to release extracellular and intracellular domains. The interaction of the extracellular part of EGF-like ligands with the membrane-bound EGF receptors, ErbB1–4, is well established and regulates normal growth, differentiation, and tumorigenesis [1]. The important roles of the cytoplasmic domain (cyt) of EGF-like ligands are emerging. The cyt domains of transforming growth factor α (TGFα-cyt) and amphiregulin (ARcyt) are important for cell polarity, intracellular vesicular trafficking [2–5], and basolateral sorting [6–8]. Soluble neuregulin-1 cyt acts as a nuclear transcriptional suppressor for several regulators of apoptosis [9,10], and its interaction with cytosolic LIM-kinase 1 is implicated in visual-spatial cognition [9,11]. Heparin binding-epidermal growth factor (HB-EGF) cyt can affect cell survival [12] and induces S-phase entry by interacting with the cyclin A transcriptional repressor promyelocytic leukemia zinc finger protein and its heterodimerization partner B-cell lymphoma 6 [13–15]. Nuclear localization of HB-EGFcyt is linked to aggressive transitional cell carcinoma [16]. Membrane-anchored betacellulin (BTC) undergoes intramembrane processing by presenilin 1 and/or presenilin 2-dependent gamma secretase to generate a free BTCcyt fragment that becomes palmitoylated as a prerequisite for gamma secretase-dependent processing and nuclear membrane localization of BTCcyt [17]. Impaired basolateral sorting of pro-EGF and inadequate renal EGFR activation caused by the P1070L mutation in human proEGFcyt results in rare autosomal recessive renal hypomagnesemia syndrome [18]. Transgenic (tg) mice overexpressing human proEGFcyt showed stunted growth [19]. In human thyroid carcinoma cells, EGFcyt alters the acetylation state of the microtubules and inhibits tumor cell invasion into elastin matrix by a reduced expression of (pro)cathepsin-L and impaired SNAP25-mediated process of vesicle-membrane fusion [20,21].
Ligand-dependent ErbB receptor degradation involves the binding of exogenous EGF to the EGFR (ErbB1), which triggers membrane-anchored surface receptor phosphorylation and initiates EGFR internalization/degradation along the endolysosomal pathway [22,23]. Lysosomal hydrolases of the cathepsin family proteolytically process EGFR [24], with procathepsin B (procathB) playing a major role in EGFR degradation [25,26]. ErbB degradation also involves the ubiquitin-proteasome system (UPS). UPS malfunction can increase the levels of activated ErbB1/EGFR and ErbB2/Neu antigen, and this may contribute to carcinogenesis [27,28]. Ubiquitination, the conjugation of a highly conserved 76-amino-acid ubiquitin (Ub) molecule to a substrate, is an energy-dependent multistep process [29]. Monoubiquitinin signal will target ErbB receptors for endosomal degradation, whereas a polyubiquitination tag is believed to mark the receptors for proteasomal degradation [30,31]. Before proteasomal degradation, deubiquitination enzymes (DUBs), including ubiquitin C-terminal hydrolase UCH-L1 and Ub-specific processing proteases (UBPs), cleave and recycle free Ub from its ubiquitinated substrate [32]. UCH-L1 maintains intracellular levels of free ubiquitin UCH-L1, and this preserves normal proteasome function [33]. UCH-L1 is expressed in mature neurons and testicular stem cells and is present during early embryogenesis [34]. Overexpression of UCH-L1 is found in numerous cancers and is correlated with advanced stages of disease and poor prognosis for patient survival [35].
In the present study, we demonstrate two novel roles of EGFcyt as an inducer of lysosomal procathB expression and a silencer of UCH-L1 expression in human thyroid and glioma cancer cells. Both events resulted in a decrease in cellular ErbB1/EGFR and ErbB2/Neu receptor levels and reduced cancer cell growth. This function was critically dependent on the presence of the exon 23-encoded peptide. Molecular modeling showed that this exon 23-encoded domain formed a loop structure in human EGFcyt.
Materials and Methods
Cell Culture
Human follicular thyroid carcinoma cell line FTC-133 and the human glioma cell line LN-18 were cultured in HAM-F12 medium (Fisher, Ottawa, Ontario, Canada) and 10% fetal calf serum (Sigma, Oakville, Ontario, Canada). Stable transfectants of the follicular thyroid carcinoma cell line FTC-133 harboring pcDNA4/HisMaxC empty plasmid control or pcDNA4/HisMaxC containing constructs encoding for the complete human EGF cytoplasmic domain (exons 22–24; EGFcyt), EGFcyt encoding for exon 22 and 23 (EGF22.23), a natural splice version of EGFcyt lacking exon 23 (EGFdel23) [20], and the human EGF, juxtamembrane, transmembrane, and cytoplasmic domain with C-terminal FLAG epitope (mbEGFctF; gift from Dr S. Wiley) [36] (Figure 1A) were described previously [19–21]. Transient transfectants of the human glioma cell line LN-18 were generated by using effectin (Qiagen, Toronto, Ontario, Canada) and using 0.4 µg of EGF constructs and empty vector (mock control). Protein lysates were harvested 48 hours after transient transfection for Western blot analysis.
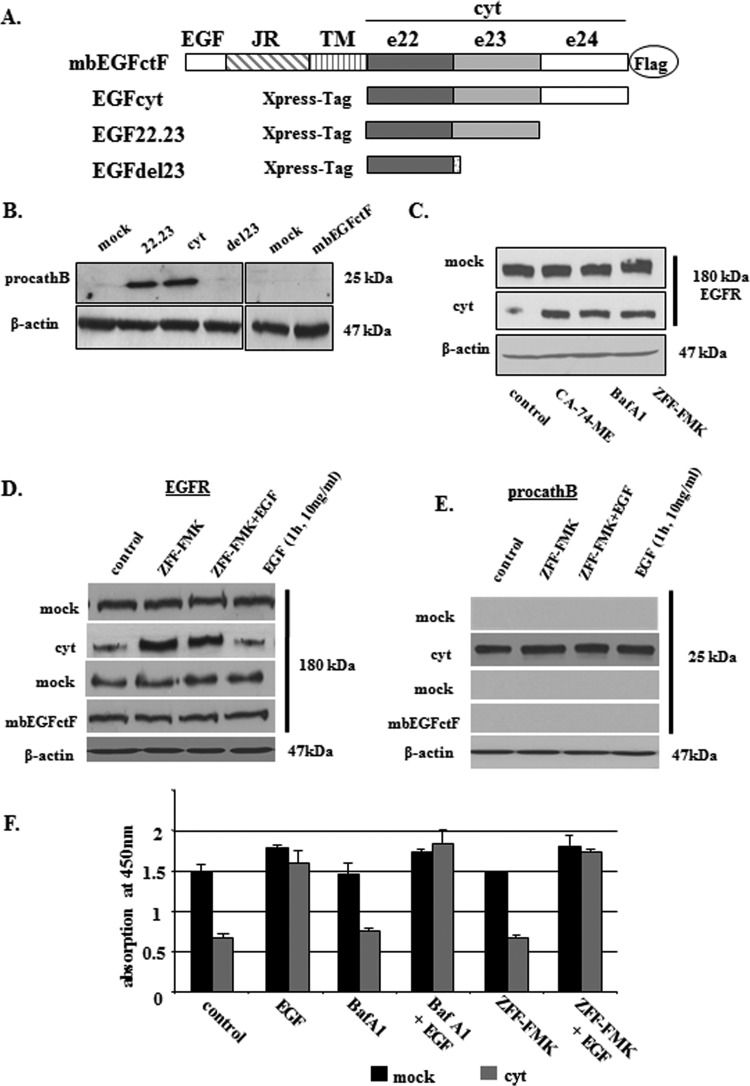
Figure 1.
(A) Schematic representation of human EGF constructs used in this study: cyt cytoplasmic domain; e22–24, exons 22 to 24; JR, juxtamembrane region; TM, transmembrane domain. (B) Western blot analysis revealed an exclusive up-regulation of procathB in stable human thyroid carcinoma transfectants expressing EGFcyt and EGF22.23. Deletion of the exon 23-encoded peptide region in EGFdel23 and membrane-bound mbEGFctF failed to show an induction of procathB expression. (C) Treatment with specific inhibitors for procathB/L (ZFF-FMK, 10 µM), cathB (CA-74-ME, 10 µM), and lysosomal ATPases (Baf-A1, 1 µM) resulted in enhanced EGFR protein levels in the presence of EGFcyt and EGF22.23 (not shown) but failed to alter procathB and EGFR protein levels in mock. (D) Western blot detection revealed an up-regulation of EGFR in the presence of ZFF-FMK (10 µM) in EGFcyt but not in mbEGFctF and mock transfectants. Addition of exogenous EGF did not oppose the ZFF-FMK effect nor did EGF alter cellular EGFR protein levels. (E) Exclusive expression of procathB protein in EGFcyt thyroid transfectants. Addition of exogenous EGF did not oppose the ZFF-FMK effect nor did EGF alter procathB protein levels. (F) BrdU proliferation assays revealed increased proliferation of proEGFcyt and mock cells on EGF treatment (10 ng/ml). The inhibitors ZFF-FMK (10 µM) and Baf-A1 (1 µM) did not alter cell growth nor did they interfere with the EGF-induced proliferation.
RNA Isolations and Reverse Transcription-Polymerase Chain Reaction
Total RNA of all transfectants was isolated with TRIzol reagent (Life Technologies, Burlington, Ontario, Canada), and RNA amounts were determined by spectrophotometry at 260 and 280 nm. Total RNA (1 µg) was used for first-strand complementary DNA (cDNA) synthesis using the SuperScript reverse transcriptase kit and 500 ng/ml of oligo d(T) primer (both Life Technologies). For reverse transcription-polymerase chain reaction (RT-PCR), 1 µl of cDNA and 25 pM of specific forward and reverse primer were used (Table 1). The PCR cycle profile consisted of an initial denaturation at 95°C for 15 minutes followed by 30 cycles of denaturation at 95°C for 1 minute, annealing at 60°C for 1 minute, and elongation at 72°C for 2 minutes. A negative control devoid of cDNA template was included. Single-PCR products were separated in a 1% agarose gel in TAE buffer (EMD, San Diego, CA), visualized by ethidium bromide staining, and sequenced to confirm identity.
Table 1.
Primer Sequences Used for the Amplification of Target Genes Listed.
| Target | Primer | Primer Sequence (from 5′ to 3〲) |
| UCHL-1 | Forward | CCC CGA GAT GCT GAA CAA AGT G GGG |
| Reverse | CTG CCT GTA TGG CCT CA | |
| UCHL-3 | Forward | GCT GGA GGC CAA TCC CGA GG |
| Reverse | GGT ATC TGG CTC GTT CTT CAG G | |
| EGFR | Forward | TGT GAG GTG GTC CTT GGG AAT TTG G |
| Reverse | TGC TGA CTA TGT CCC GCC ACT GGA | |
| EGFcyt | Forward | ACTCAGAAGCTGCTATCGAAAAACCC |
| Reverse | CTCACTGAGTCAGCTCCATTTGGTG | |
| ErbB2 | Forward | GAA CCC CCA GCT CTG CTA CCA |
| Reverse | CGT AGA AAG GTA GTT GTA GGG AC | |
| GAPDH | Forward | CATCACCATCTTCTAGGACCG |
| Reverse | TGACCTTGCCCACAGCCTTG |
Immunohistochemistry
Five-micrometer sections of paraformaldehyde-fixed and paraffin-embedded human thyroid tissues were treated with 3% hydrogen peroxidase to block endogenous peroxidase activity. Nonspecific binding were blocked with 3% bovine serum albumin and 10% goat normal serum (Sigma). The sections were incubated with a rabbit poly-clonal antiserum against UCH-L1 (1:100; Enzo Biochem, New York, NY) overnight at 4°C. Sections were washed 3 x 10 in Tris (pH 7.4) and 0.1% Tween 20 buffer and incubated with a secondary goat anti-rabbit antibody (1:200; New England Bio [NEB], Ipswich, MA) for 1 hour at room temperature. Staining was developed using the DAB kit (Pierce, Nepean, Ontario, Canada).
RNA Silencing
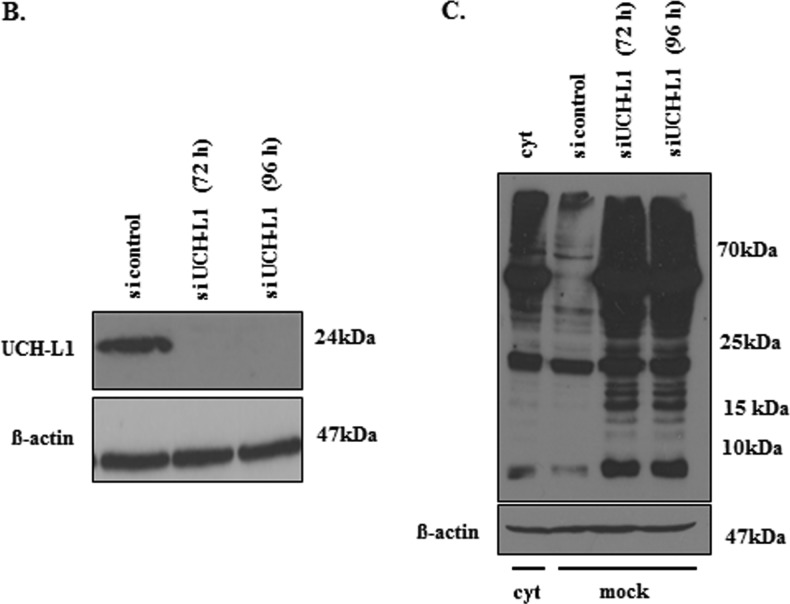
Mock and EGF23del transfectants at 60% confluence were transfected with 50 nM siRNA-UCHL-1 (AM51331, s105082, ID7345) or the nonsilencing scrambled sequence (5′AATTCTCCGAACGTGTCACGT3′) as a control (both Ambion, Austin, TX) using Lipofectamine 2000 (Life Technologies). After 48 and 96 hours, UCH-L1 gene expression and UCH-L1 protein levels were assessed by RT-PCR and Western blot analysis, respectively.
BrdU Proliferation Assay
Stable and transient transfectants were seeded at 103 cells/well into 96-well plates and cultured overnight in serum-free medium. Cells were preincubated with exogenous EGF (10 ng/ml) for 8 hours and with specific inhibitors. BrdU assays were performed according to the manufacturer's protocol (Roche, Mississauga, Ontario, Canada). Reactions were stopped using 1 M H2SO4, and the absorbance was measured in an ELISA reader at 450 nm.
Inhibitors
For BrdU assays and Western blots, cells were incubated with proteasome inhibitors MG132 (10 µM) or lactacystine (1 µM) for 2 hours and with cath-B inhibitor ZFF-FMK (10 µM), cath-B/-L inhibitor CA-74-ME (10 µM), and the inhibitor of lysosomal ATPases BafA1 (1 µM) for 3 hours (all Calbiochem, San Diego, CA) and ErBb1 inhibitor AG1478 (10 µM) for 1 hour (Sigma-Aldrich, Oakville, Ontario, Canada). Batimastat (Sigma-Aldrich) was used as described earlier [36]. Cellular proteins were extracted with 1x Laemmli buffer (4% SDS, Tris-HCl pH 7.6, mercaptoethanol, bromophenol blue).
MTT Cell Viability Assay
Cells were seeded at 103 cells/well into 96-well plates and cultured overnight in serum-free medium. Cells were incubated with specific inhibitors to determine their cytotoxicity effect on cells. MTT assays were performed according to the manufacturer's protocol (Life Technologies).
Transient Transfection with EGFcyt Peptides
Parental FTC-133 cells were transfected with N-terminally fluorescein isothiocyanate (FITC)-labeled 10-to 12-mer synthetic peptides (EZBioLab, Carmel, IN) derived from the exon 22- and exon 23-encoding peptide sequences of human EGFcyt using the Chariot kit according to manufacturer's instructions (Ambion; Table 2). Transfection efficiency was consistently more than 70% as determined by counting FITC-positive cells in a Z1 upright fluorescent microscope (Zeiss, Jena, Germany).
Table 2.
Peptide Sequences Used in This Study.
| EGFcyt Coding Region of Peptides | Peptide No. | Peptide Sequence (N′- to C′-terminus) |
| Exon 22 | P1 | TQKLLSKNPKNP |
| P2 | KNPYEESSRDVR | |
| P3 | DVRSRRPADTED | |
| P4 | TEDGMSSCPQP | |
| Exon 23 | P1 | PQPWFVVIKE |
| P2 | VIKEHQDLKN | |
| P3 | DLKNGGQPVA | |
| P4 | QPVAGEDGQAAD | |
| Scrambled peptide | P5 | QPVIHDGVAAE |
Peptides (10–12 mer) were designed to cover the complete exons 22 and 23 and have a three amino acid overlap at the N-terminus with each consecutive C-terminal peptide. All peptides were FITC conjugated at the N-terminus to monitor cellular uptake.
Subcellular Fractionation and Western Blot Analysis
Cells were seeded in 25-cm2 flasks for 3 days under normal serum conditions. At 80% confluence, cell protein lysates were taken using a nuclear, cytoplasmic, and membrane extraction kit according to the company protocol (Pierce). For Western blot, transfectants were seeded at 8.5 x 104 cells/6 wells for 24 hours in serum-free medium with and without treatment of MG132 and lactacystine (both at 1 µM). For Western blot analysis, 20-µg/ml proteins were transferred onto a Hybond nitrocellulose membrane (Thermo Scientific, Nepean, Ontario, Canada) and nonspecific binding sites were saturated for 1 hour at room temperature in blocking buffer containing Tris-buffered saline/0.01% Tween 20 plus 5% skimmed milk. Membranes were incubated overnight at 4°C with rabbit polyclonal antibodies to UCHL-1 (1:1000; Enzo Biochem), histone 1 (H1), ErbB1, ErbB1Tyr1045, and ErB2 (all 1:1000; NEB), and a monoclonal antibody (mAb) to ubiquitin (1:2000; Enzo Biochem), respectively. All antibodies were diluted in blocking buffer. A mouse mAb against β-actin (1:20,000; Sigma) was used to determine equal protein loading per lane. On incubation with a horseradish peroxidase-conjugated goat anti-rabbit (1:2000; NEB) or goat antimouse secondary antibody (1:20,000; Sigma) for 2 hours at room temperature, specific immunoreactive bands were detected with an ECL kit (Thermo Scientific, Nepean, Ontario, Canada).
Immunoprecipitation
FTC-133-EGFcyt, FTC-133-mbEGFctF, and corresponding mock transfectants (1 x 106 cells) were harvested at 80% confluence, lysed in 400 µl of lysis buffer (50 mM HEPES [pH 7.4], 1% Triton X-100, 100 mM NaCl, 10 mM NaF, 1 mM EDTA, 1 mM sodium vanadate) for 1 hour on ice and centrifuged at 13,000g for 15 minutes at 4°C. Anti-EGFR antibody (10 µg/ml; Santa Cruz, Santa Cruz, CA) was added in lysis buffer and incubated overnight at 4°C. Immunocomplexes were purified by protein A/G agarose beads (30 µl/200 µl; Sigma) with gentle rocking for 4 hours at 4°C, pelleted for 60 seconds, and washed 3x in 500 µl of ice-cold lysis buffer. Pellets were resuspended twice in 100 µl of 3x SDS sample buffer (1.25 M Tris, pH 6.8, 10% SDS, 20% glycerol, 10% mercapthoethanol, 2% bromophenol blue), heated to 90°C for 5 minutes followed by Western blot detection of immunoprecipitated EGFR (1:1000, NEB). Membranes were stripped with buffer containing 0.2 M glycine and 0.05 Tween 20 for 15 minutes at room temperature, washed, blocked with 5% milk, and probed for c-Cbl (1:1000; Santa Cruz) and ubiquitin (Ub; 1:2000) (Cedarlane, Burlington, Ontario, Canada).
Proteasome Activity Assay
Transfectants expressing cytosolic EGFcyt, mbEGFctF, and mock cells were seeded in 96-well plates at 1 x 103 cells/well. Cells were pretreated with 10 µM of MG132 for 1 hour and soluble EGF for 8 hours (10 ng/ml). Proteasomal activity assays were performed according to the manufacturer's protocol (Promega, Madison, WI).
Homology Modeling of the EGFcyt Domain
The homology modeling approach was used to generate 3D structures of EGFcyt composed of 149 amino acids [37]. The Modeller Web-based server was used in model building. On the basis of sequence identity values ranging between 35% and 38%, the structure of the receptor binding protein P2 of PRD1 (PDB code: 1n7v) was used as one of the templates. The strain energy of the model was generated separately in the GROMOS force field using the 53A6 parameter set optimized for molecular dynamic simulations. After building the 3D model of human EGFcyt, all atomic positions were locked, and required hydrogen atoms were added to the backbone structure of the molecule. Molecular dynamics simulation was performed at 1 x 106 for 50 picoseconds for further optimization in GROMACS (version 3.3). This involved a brief, steepest-descent run that used a maximum step protocol of 1.5 Å, and a maximum tolerance of 2000 kJ/mol per nanometer followed by a more extensive conjugate gradient minimization with tolerances of 200 kJ/mol per nanometer. After building and refinement of the 3D EGFcyt model, the Protein Structure & Model Assessment Tools were used to verify the quality of the model. This tool is capable of verifying a number of aspects of model qualities such as (i) local model quality estimation (anolea atomic mean force potential, empirical force field, composite scoring function for model quality estimation), (ii) global model quality estimation (all atom distance-dependent statistical potential), (iii) stereochemistry check (protein structure verification, stereochemical quality check; minimum resolution, 2.5 Å), and (iv) structural features (secondary structure, geometrical features, and solvent exposure assignment, analysis of protein structure motifs). On the basis of these assessment results, model quality was evaluated according to the Ramachandran plot and the amino acid residues in the allowed, disallowed region and overall G factor.
Statistical Analysis
All experiments were repeated at least three times for two different clones. For multiple experiment comparisons, analysis of variance table and Tukey test were used with P < .05 considered significant.
Results
EGFcyt Affects Cathepsin-B-Mediated EGFR Degradation in Cancer Cells
Cathepsin-B (cathB) plays an important role in the lysosomal degradation of EGFR [26]. Here we demonstrate that of all EGF constructs tested (Figure 1A), soluble EGFcyt and EGF22.23 up-regulated procathB in thyroid cancer (Figure 1B) and glioma cells (Figure W1A). This effect was not observed with membrane-anchored mbEGFctF, the EGFdel23 splice variant with a deletion of exon 23, or mock cells (Figures 1B and W1A), suggesting cellular compartment-specific EGFcyt effects and a functional role for the EGFcyt exon 23 peptide region. Both immunoreactive cathB and CD63 were colocalized in mock and EGFcyt clones, indicating lysosomal localization of procathB (data not shown). The presence of EGFcyt coincided with diminished EGFR and ErbB2 expression in stably transfected thyroid cancer cells (Figures 1C and 6, A and B) and transiently transfected LN-18 glioma cells (Figure W1B). To determine an involvement of lysosomal procathB in this EGFR down-regulation, EGFcyt thyroid transfectants and mock were incubated with specific inhibitors for cathB (ZFF-FMK, 10 µM), cathL/B (CA-74-ME, 10 µM), or the inhibitor of lysosomal ATPases BafA1 (1 µM). Treatment with these three inhibitors resulted in a marked up-regulation of EGFR in the EGFcyt transfectants but not in mock (Figure 1C). ZFF-FMK also caused the up-regulation of EGFR in EGFcyt glioma transfectants (Figure W1C). However, in mbEGFctF and mock transfectants cellular levels of EGFR (Figures 1D and W1C) and procathB (Figures 1E and W1D) remained unchanged. The EGFR present in the thyroid (Figure W4, B–D) and EGFcyt glioma (Figures W1E and W3) transfectants was bioactive in response to exogenous EGF and caused a maximal proliferative response similar to mock, whereas BafA1 and ZFF-FMK alone had no effect on proliferation (Figures 1F and W1E).
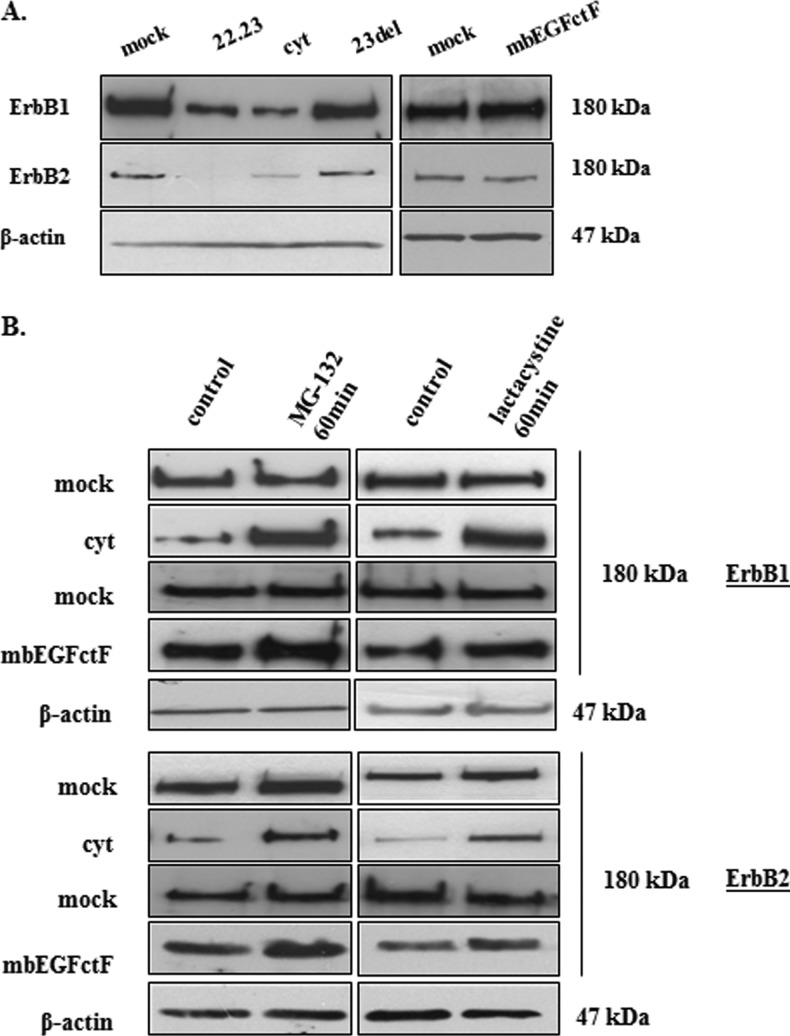
Figure 6.
(A) Although RT-PCR did not show a difference in ErbB1 and ErbB2 gene expression (data not shown), Western blot analysis revealed a decrease of ErbB1 and ErbB2 protein in the presence of EGFcyt and EGF22.23. No change in ErbB status was detected in the presence of mbEGFctF, EGFdel23, and mock. (B) Treatment with the proteasome inhibitors MG132 and lactacystine increased the amount of ErbB1 and ErbB2 protein in EGFcyt and mbEGFctF transfectants as shown in representative Western blots. Proteasome inhibitors had no effect on ErbB1/2 protein levels in EGFdel23 (data not shown) and mock transfectants. Equal protein loading was assessed by detecting β-actin.
EGFcyt Affects Proteasome Activity in Cancer Cells
We determined the ability of structurally distinct EGFcyt domains to affect the proliferation of human thyroid and brain cancer cells. Cancer cells expressing soluble EGFcyt and EGF22.23 showed significantly reduced BrdU incorporation when compared with mock and/or EGFdel23 (Figure 2A) and mbEGFctF (Figures 2B and W2A) transfectants. This reduction in cancer cell growth was not the result of increased cell death as determined by cytotoxicity assays (data not shown). EGF-induced EGFR activation antagonized the growth inhibitory effect of EGFcyt in thyroid cancer (Figures 2, A and C, and W4, A and B) and glioma (Figure W3) cells. Treatment of mock, EGFdel23, and mbEGFctF clones with soluble EGF caused a small increase in cell proliferation (Figures 2, A–D, W3, and W4, A and B). We determined if cleavage of the extracellular EGF part of mbEGFctF may account for the increased proliferation of mbEGFctF clones compared with EGFcyt transfectants. BrdU proliferation assays in the presence of the matrix metalloproteinase (MMP) inhibitor batimastat failed to alter the proliferation of mbEGFctF in thyroid transfectants, indicating that the increased proliferation observed in mbEGFctF clones was not a result of liberation of EGF from the mbEGFctF protein with subsequent EGFR activation (Figure W4A). However, batimastat was able to decrease the proliferation of LN-18, suggesting EGFR activation as a result of proteolytic release of endogenous EGF-like ligand(s) in these glioma cells. We detected the expression of endogenous proEGF by PCR in LN-18 cells (data not shown). The decrease in proliferation with batimastat was significant in EGFcyt and mbEGFctF LN-18 transfectant but not in the mock controls (Figure W3).
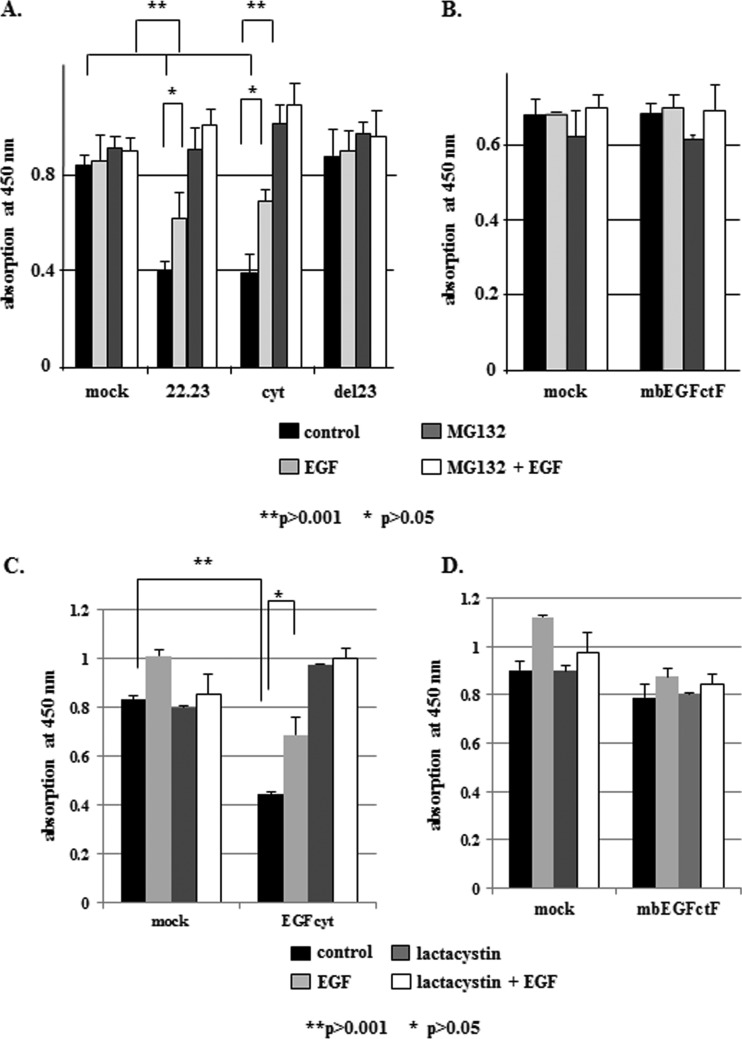
Figure 2.
(A, C) In thyroid cancer cells, EGFcyt and EGF22.23 caused a 50% decrease in proliferation when compared to EGFdel23 and mock. Treatment with exogenous EGF (10 ng/ml), MG132 (10 µM), and lactacystine (1 µM) either completely or partially antagonized the growth inhibitory action of EGFcyt and EGF22.23, respectively. Dual treatment of cells with exogenous EGF plus MG132 or lactacystine (1 µM) showed that the proteasome inhibitor did not antagonize the growth-promoting action of EGF. (B and D) MbEGFctF had no growth-inhibitory effect and treatments with EGF (10 ng/ml) MG132 (10 µM) and lactacystine (1 µM) failed to affect cell proliferation in mbEGFctF and mock cells.
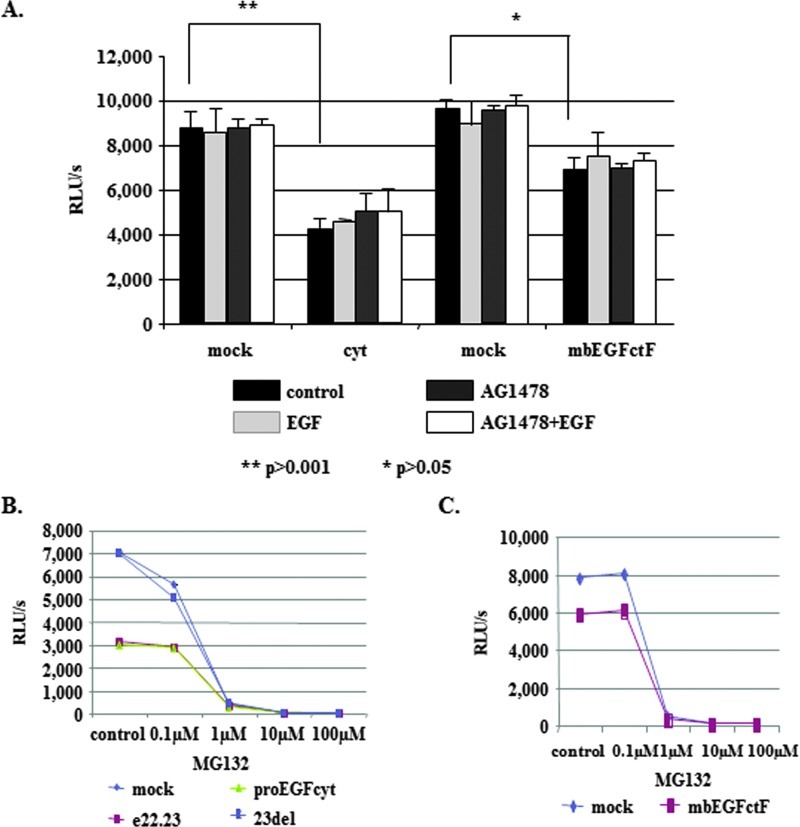
The proteasome inhibitors MG132 and lactacystine increased EGFR levels in thyroid cancer cells (Figure 6B) and glioma (Figure W2B) and enhance growth rates of EGFcyt and EGF22.23 transfectants (Figure 2, A and C). We detected a significant reduction in proteasomal activity in the presence of EGFcyt and EGF22.23 (Figure 3, A and B) and, to a lesser extent, in mbEGFctF (Figure 3, A and C). Proteasomal activity assays in the presence of increasing concentrations of MG132 confirmed the inhibitory effect of EGFcyt, EGF22.23 (Figure 3B) and mbEGFctF (Figure 3C) and indicated similar inhibition kinetics. Almost complete proteasomal inhibition was observed at 1 µM MG132 in all transfectants (Figure 3, B and C). Treatment with soluble EGF and/or the EGFR inhibitor AG1478 failed to increase proteasomal activity in EGFcyt clones (Figure 3A). Cytotoxicity assays done in the presence of MG132, lactacystine, and EGF confirmed that the reduction of proteasomal activity in the EGFcyt clones was not the result of increased cell death (data not shown).
Figure 3.
(A and B) Proteasome activity was significantly decreased in the presence of EGFcyt and, to a lesser extent, in mbEGFctF clones when compared to the corresponding mock. EGF-mediated EGFR activation and the inhibition thereof by EGFR inhibitor AG1478 had no effect on proteasomal activity in these cells. Despite attenuated proteasome activity in the presence of EGFcyt and EGF22.23, proteasome activity in all clones investigated was inhibited at 1 µM of MG132. (A and C) The same results were obtained with mbEGFctF, suggesting a similar mechanism by which EGFcyt, EGF22.23, and mbEGFctF can decrease proteasome activity. These results also emphasized the importance of the exon 23-encoded peptide region in this process because EGFdel23 and mock failed to affect proteasome activity.
EGFcyt Is a Novel Regulator of UCH-L1 in Cancer Cells
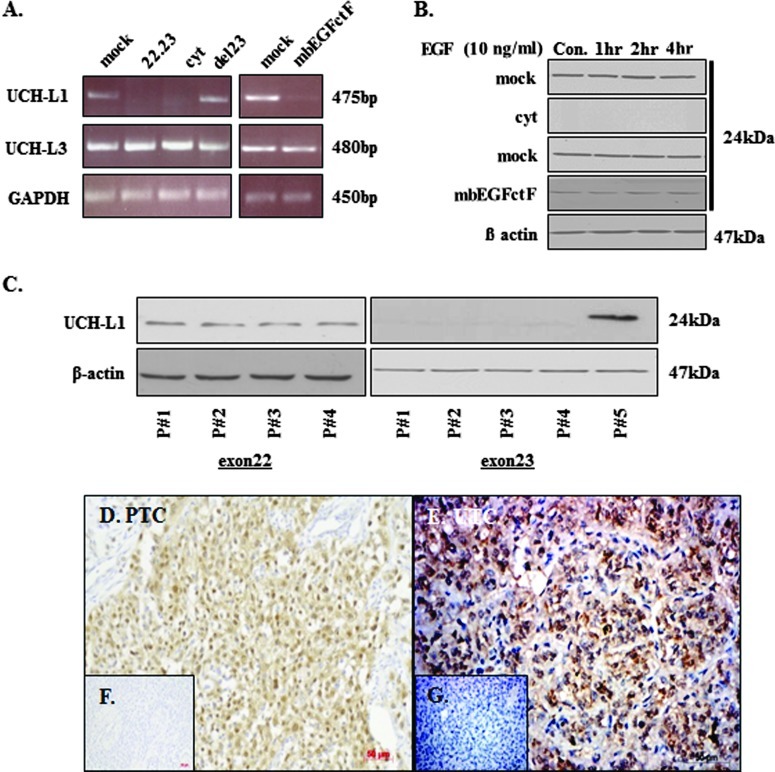
Microarray analysis identified and subsequent RT-PCR and Western blot data identified ubiquitin C-terminal hydrolase (UCH-L1) as a target gene in thyroid cancer cells. Thyroid cancer transfectants expressing EGFcyt and EGF22.23 showed the silencing of UCH-L1 at the mRNA (Figure 4A) and protein levels (Figure 4B). In the presence of mbEGFctF, UCH-L1 gene expression was attenuated at the mRNA (Figure 4A) and protein levels (Figure 4B). Treatment with soluble EGF for up to 4 hours did not change UCH-L1 protein levels in the EGFcyt and mbEGFctF clones (Figure 4B). LN-18 glioma cells showed strong UCH-L1 expression, but transient transfection with EGFcyt or mbEGFctF caused markedly reduced UCHL-1 protein levels (Figure W2C). UCH-L1 down-regulation by EGFcyt, EGF22.23, and mbEGfctF was specific for UCH-L1 and not observed for UCHL3 in glioma (data not shown) and thyroid cancer cells (Figure 4A). UCH-L1 is a member of the DUB protein family and plays an important role in the processing of Ub as part of protein degradation. To identify particular peptide regions in the EGFcyt domain responsible for this down-regulation of UCH-L1, we used FITC-labeled 10- to 12-mer peptides that covered the entire exon 22 and exon 23 region of human EGFcyt (Table 2). Thyroid cells were transfected with the different FITC-peptide conjugates, and transfection efficiency was determined in each case to be at 60% to 70% (counted fraction of FITC-stained to unstained cells). All peptides derived from the exon 23 peptide region caused a complete loss of UCH-L1 protein, whereas all exon 22-encoded peptides and the scrambled control peptide failed to affect UCH-L1 expression (Figure 4C). Thus, the exon 23-encoded peptide region of the human EGF cytoplasmic domain, as a membrane-bound or soluble form, was identified to have an essential role in suppressing UCH-L1 expression in cancer cells.
Figure 4.
(A) UCH-L1 gene activity and (B) UCH-L1 protein were exclusively downregulated in the presence of EGFcyt, EGF22.23, and mbEGFctF. (C) Specific 10-to 12-mer peptides covering the complete EGFcyt peptide region encoded by exons 22 and 23 were used to determine the specificity of exon-derived peptides for the silencing of UCH-L1 by EGFcyt. Exclusively peptides encoded by the exon 23-derived peptide sequences of EGFcyt, but not scrambled peptides or peptides derived from pEGFcyt exon 22, were capable of downregulating UCH-L1 expression at the gene and protein levels. Representative immunodetection of UCH-L1 in (D) papillary and (E) undifferentiated human thyroid cancer tissues. (F and G) No specific immunostaining was detected in normal human thyroid tissues (data not shown) and when the primary antiserum was replaced by a species-specific nonimmune serum at the same concentration. Magnification, x20.
The expression of UCH-L1 in human thyroid tissues has not been reported. We performed immunohistochemistry and demonstrated, for the first time, the presence of immunoreactive UCH-L1 in human papillary (Figure 4D) and undifferentiated (Figure 4E) thyroid carcinoma tissues. Normal human thyroid tissue (not shown) and control sections with primary antiserum replaced by nonimmune serum were devoid of UCH-L1 (Figure 4, F and G). Our results suggest the exclusive expression of UCH-L1 in thyroid cancer tissues.
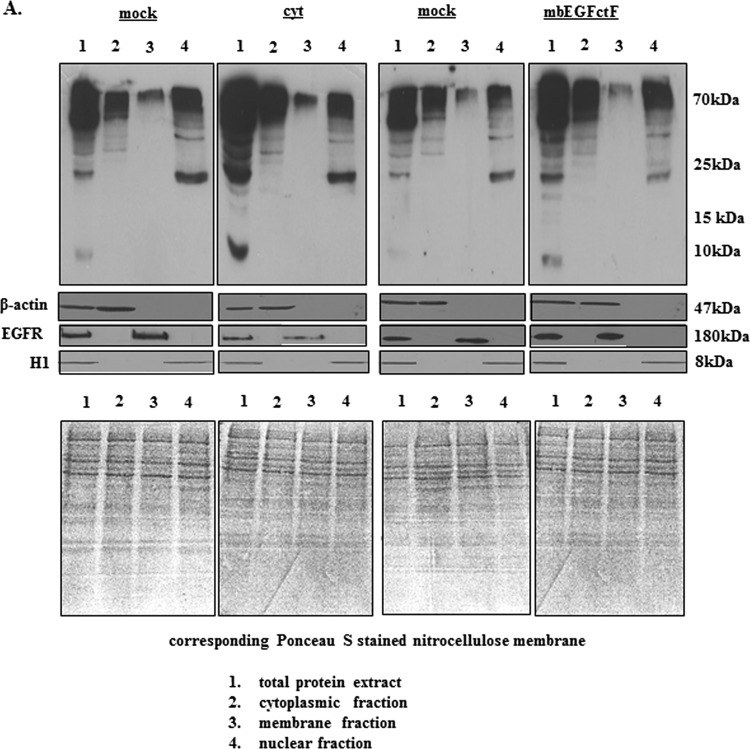
EGFcyt Decreases UCH-L1, Causes Hyperubiquitination of Cellular Proteins, and Reduces ErbB1/2 Receptor Levels
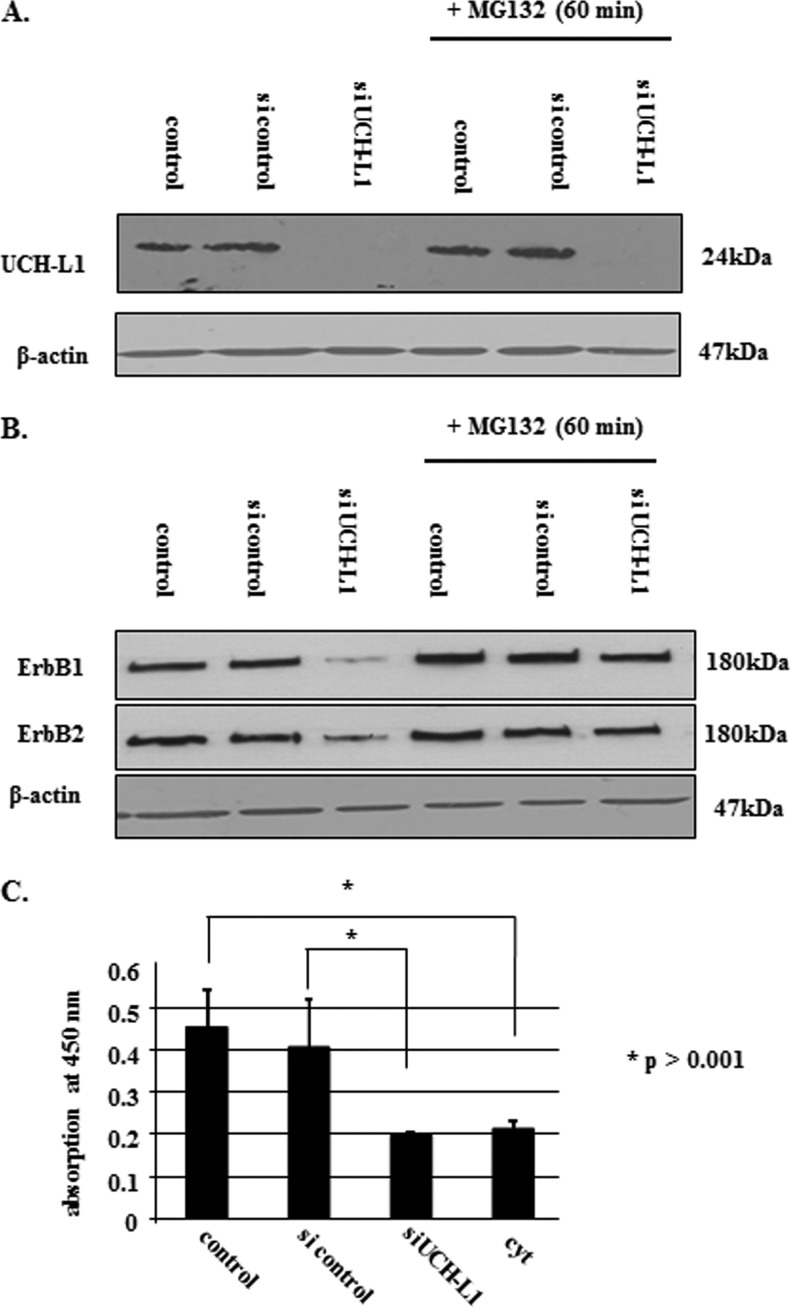
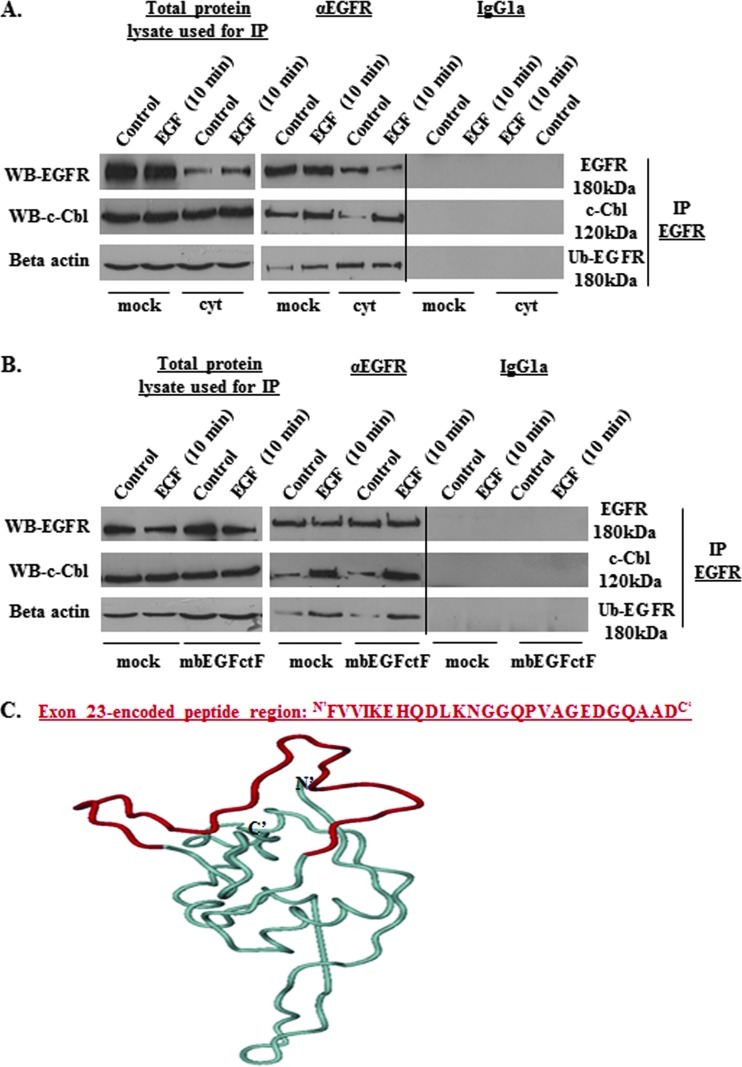
Cells with impaired UCH-L1-mediated DUB function are known to show high levels of protein ubiquitination [38]. Western blot analysis of total, cytoplasmic, nuclear, and membrane-bound protein fractions showed increased levels of ubiquitinated cellular proteins in the presence of either EGFcyt or mbEGFctF (Figure 5A). RT-PCR and Western blot analysis confirmed the successful specific knockdown of UCH-L1 for more than 96 hours in UCH-L1-positive EGFdel23 and mock clones (Figure 5B). Silencing of UCH-L1 resulted in significantly elevated levels of ubiquitinated proteins at 72 and 96 hours in mock transfectants (Figure 5C). Next, we addressed if a reduced level of UCH-L1 can affect the level of ErbB receptors, particularly ErbB1/EGFR and ErbB2/Neu antigen, and whether this may contribute to the attenuated cell growth in the presence of EGFcyt. We failed to detect changes in ErbB1 gene expression in any of the transfectants investigated (data not shown) but observed a significant decrease in ErbB1 and ErbB2 protein content exclusively in the presence of EGFcyt (and EGF22.23) in thyroid cancer cells (Figure 6, A and B) and glioma transfectants (Figure W1B). No change in ErbB1/2 protein content was detectable in mbEGFctF, EGFdel23, or mock cells (Figures 6, A and B, and W1B). Treatment with proteasome inhibitors MG132 and lactacystine resulted in increased ErbB1/2 protein levels exclusively in proEGFcyt and mbEGFctF transfectants (Figures 6B and W2B). Upon si-UCH-L1 treatment, we detected decreased levels of UCH-L1 mRNA (data not shown), UCH-L1 protein (Figure 7, A and B) and ErbB1/2 protein (Figure 7B). Treatment with MG132 failed to affect UCH-L1 levels in siUCH-L1-treated cells (Figure 7A) but abolished the siUCH-L1-induced decrease in ErbB1/2 (Figure 7B), resulting in protein levels similar to untreated and si-control-treated mock (Figure 7, A and B). The siUCH-L1-treated mock cells with a decrease in ErbB1/2 levels showed significantly reduced cell proliferation (Figure 7C). We performed immunoprecipitation (IP) assays with a specific antibody to EGFR on mock and UCH-L1-negative EGFcyt thyroid transfectants plus/minus EGF treatment to determine whether the presence of EGFcyt coincided with the increased level of ubiquitinated EGFR. Irrespective of EGF treatment, the IP revealed attenuated total EGFR protein levels in EGFcyt clones compared with mock (Figure 8A). Ubiquitinated EGFR was clearly observed in untreated EGFcyt transfectants but only weakly detectable in mock (Figure 8A). In mock, but not in EGFcyt transfectants, EGF treatment resulted in an increased presence of ubiquitinated EGFR (Figure 8A). IP results for mbEGFcyt and mock transfectants show similar EGFR levels, and in both cases, we observed an increase in ubiquitinated EGFR on EGF treatment (Figure 8B). When the anti-EGFR was replaced by a nonimmune IgG1a control serum, no immunoprecipitated proteins were detected (Figure 8, A and B). In all transfectants investigated, EGF treatment caused a marked increase in the phosphorylation of EGFR tyrosine residue 1045 (EGFR-Tyr1045) (Figure W4C), which was inhibited in the presence of the EGFR inhibitor AG1478 (Figure W4D). We performed co-IP experiments for c-Cbl, known to ubiquitinate [22,39,40] EGFR-Tyr1045 [41] and assist in the sorting process of the EGFR [42]. EGF treatment resulted in significantly increased c-Cbl coimmunoprecipitation with EGFR in mock, EGFcyt (Figure 8A, and mbEGFctF clones (Figure 8B).
Figure 5.
(A) Representative Western blots showing the level of ubiquitinated proteins in EGFcyt and mock cells. In the presence of EGFcyt and mbEGFctF, the level of ubiquitinated total protein and protein fractions derived from subcellular compartments was increased. This enhanced protein ubiquitination was not observed with EGFdel23 (not shown) and mock. Ponceau S-stained blots served as loading control. EGFR served as marker for the membrane fraction, β-actin for the total and cytoplasmic fraction and histone 1 (H1) as marker for the nuclear fraction. (B) Transfection with a specific siUCH-L1 (50 nM) construct for 72 and 96 hours successfully silenced UCH-L1 expression in UCH-L1+ mock cells. (C) Western blot analysis showed that silencing of UCH-L1 protein coincided with increased levels of total ubiquitinated (Ub-) proteins in siUCH-L1 treated mock compared with UCH-L1+ mock and UCH-L1- EGFcyt cells treated with scrambled siRNA.
Figure 7.
(A) UCH-L1 siRNA treatment (50 nM) successfully silenced endogenous UCH-L1 expression in mock. MG132 (10 µM) treatment did not influence silencing of UCH-L1 in mock cells. (B) On UCH-L1 knockdown, Western blot analysis showed reduced ErbB1 and ErbB2 protein levels. However, in mock treated simultaneously with MG132 (10 µM) and siUCH-L1 (50 nM), this receptor down-regulation was abolished, suggesting that proteasome activity was essential for the degradation of ubiquitinated ErbB1/2. β-Actin was used to assess equal protein loading. (C) Silencing of UCH-L1 gene activity in mock resulted in an approximately 50% reduction in proliferation, which was similar to the growth rates determined for UCH-L1-negative EGFcyt transfectants.
Figure 8.
(A) Western blot detection of total fractions of EGFR and c-Cbl plus/minus treatment with EGF before IP; β-actin was used to assess equal protein loading. IP experiments and subsequent Western blot analysis confirmed the attenuated EGFR levels in the presence of EGFcyt. Unstimulated EGFcyt cells revealed enhanced levels of ubiquitinated EGFR similar to mock treated with EGF. In the presence of EGFcyt, coimmunoprecipitation of c-Cbl with EGFR revealed reduced c-Cbl-EGFR interaction compared with mock, which increased markedly on EGF treatment. An IgG1a isotype control antibody was used as a negative control in the IP experiments. (B) Western blot detection of total fractions of EGFR and c-Cbl plus/minus treatment with EGF before IP; β-actin was used to assess equal protein loading. IP results for EGFR, c-Cbl and ubiquitinated EGFR in mbEGFctF and mock cells plus/minus EGF treatment showed comparable results. Treatment with exogenous EGF caused phosphorylation of EGFR at residue Tyr1045 (see Figure W4, C and D) and enhanced the interaction between c-Cbl and activated EGFR in all transfectants studied. An IgG1a isotype control antibody was used as a negative control in the IP experiments. (C) A structural model of human EGFcyt was generated based on the structure of the receptor binding protein P2 of PRD1 (PDB code: 1n7v) used as template. This molecule has up to 38% amino acids identity with human EGFcyt. The exon 23-encoded peptide region marked in red was found to form an exposed loop.
Rendered Structural Modeling Reveals an Exposed Exon 23-Encoded Peptide Loop in EGFcyt
The molecular structure of human EGFcyt has not been reported. We used homology modeling based on the receptor binding protein P2 of PRD1 (PDB code: 1n7v), a modeling template displaying up to 38% amino acid identity with human EGFcyt. The rendered model of the cytoplasmic domain of human EGF revealed the exon 23-encoded peptide region we identified as a biologically important molecular module of EGFcyt to form an exposed loop structure potentially capable of participating in protein interactions involving EGFcyt (Figure 8C).
Discussion
In the present study, we identified soluble EGFcyt as a novel factor affecting lysosomal (pro)cathB-mediated degradation and proteasomal activity in cancer cells. We had previously demonstrated the presence of soluble human EGFcyt in the cytoplasm of cancer cells [19,20]. The increased procathB expression and down-regulation in EGFR protein were dependent on the exon 23-encoded peptide region present in EGFcyt and EGF22.23, confirming our previous findings of the important role of this peptide region as an essential molecular determinant for the in vitro and in vivo biologic activity of the EGF cytoplasmic domain [19–21]. CathB has been recognized as an important and specific degradation mechanism for EGFR in the endolysosomal compartment [26]. EGF ligand-mediated activation results in monoubiquitination at multiple sites within the tyrosine activation domain of EGFR [30,43], which serves as a signal for receptor internalization [30,44,45] and regulates sorting of endosomal EGFR to lysosomes [46,47]. Continuous EGFR ubiquitination by the E3 ubiquitin ligase c-Cbl is required for lysosomal targeting and degradation of the EGFR [42,48]. The specific inhibition of cathB and the elevation of the acidicpHinthe endosomal-lysosomal compartment by the vacuolar-type ATPase (V-ATPase) inhibitor bafilomycin A1 (BafA1), known to disrupt the vesicular proton gradient and raise the pH of acidic vesicles [49], effectively blocked EGFR degradation in the presence of EGFcyt in cancer cells.
The presence of soluble and membrane-anchored EGFcyt and EGF22.23, all containing the exon 23-encoded peptide region, was also associated with reduced proteasome activity which could be further suppressed with increasing concentrations of MG132, a non-selective proteasome inhibitor known to also inhibit procathB [50]. To discriminate between these two inhibitory effects, we replaced MG132 with the selective proteasome inhibitor lactacystine [51] but obtained similar results. We identified the deubiquitin esterase UCH-L1, a major cellular ubiquitin hydrolase [33], as a novel transcriptional target that was specifically downregulated or completely silenced in the presence of mbEGFctF and EGFcyt, respectively. Intriguingly, the silencing of UCH-L1 was also achieved using small peptides designed according to the exon 23-encoded peptide sequence of EGFcyt. UCH-L1 is involved in diverse aspects of tumor cell adhesion and motility [52,53], but its role in tumor cell growth is still unclear and may depend on the specific cell context [54–57]. Uch-L1 transgenic mice recently provided conclusive evidence for a role of UCH-L1 as an oncogene. These mice displayed a high incidence of sporadic tumors, primarily consisting of lymphomas and lung adenoma [52]. In humans, prostate cancer is among numerous cancers shown to express UCH-L1 [58]. Here we provide evidence for the presence of UCH-L1 in human thyroid carcinoma. UCH-L1 was absent in normal thyroid tissues, suggesting that UCH-L1 may qualify as a novel marker for thyroid cancer cells.
Proper control of cellular ubiquitin levels is crucial for normal cell functions and requires the balanced actions of ubiquitin ligases and DUB enzymes [47,59–61]. Malfunction of the ubiquitin pathway due to UCH-L1 depletion in the presence of proEGFcyt, proEGF22.23, and exon23 peptides or UCH-L1 decrease in mbEGFctF clones coincided with increased ubiquitination of total, cytoplasmic, membrane, and nuclear protein fractions, including EGFR. This likely triggered a depletion of the cellular pool of free ubiquitin that caused the decrease in proteasome activity observed in thyroid cancer cells [33,62,63]. Furthermore, the UCH-L1negative, procathBhigh EGFcyt phenotype resulted in lysosomal degradation of ErbB1/2 receptors as determined by our cathB/lysosomal inhibitor studies and significantly reduced cell growth in our cancer cell models. Importantly and contrary to human fibroblasts from patients with eight different lysosomal storage diseases, the attenuated UCH-L1 expression in our cancer cell models was not associated with increased cell death [62].
EGFR-activating EGF-like ligands can antagonize the function of EGFcyt [21]. We used the general MMP inhibitor batimastat to account for the possibility of a proteolytic release of (a) the bioactive extracellular EGF part of the membrane-anchored mbEGFctF that may stimulate EGFR (Figure 1A) [36] and/or (b) the MMP-mediated cleavage of endogenous membrane-bound EGF-like ligands. Such effects would explain differences observed between the EGFcyt and mbEGFctF transfectants because a liberated EGF/endogenous EGF-like ligand is expected to counteract the actions of the EGFcyt domain. Although batimastat had no effect in thyroid transfectants, it caused a small but significant decrease in proliferation with the glioma EGFcyt and mbEGFctF clones. The fact that we detected endogenous EGF expression in LN-18 may explain that both EGFcyt and mbEGFctF clones showed similar responses to batimastat and made proteolytic release of the EGF domain of the mbEGFctF construct unlikely. In addition, the batimastat experiments clearly showed that the release of endogenous EGF-like ligand had no significant effect on the responses measured, suggesting that the cellular effects observed for EGFcyt and mbEGFctF in thyroid and glioma cancer cells were genuine and, in the case of LN-18, only marginally affected by endogenously induced EGFR activity. Further support for a lack of enzymatic processing of mbEGFctF or its involvement in juxtacrine activation of EGFR on neighboring cells [36] came from the finding that mbEGFctF and mock clones showed similar levels of c-Cbl-EGFR interaction. The E3 ubiquitin ligase c-Cbl is known to ubiquitinate EGFR [22,39,40] and assist in the sorting process of this tyrosine kinase receptor [42]. As expected, the proto-oncogene and RING finger member c-Cbl coimmunoprecipitated with ubiquitinylated EGFR [42,64] and EGF-mediated EGFR phosphorylation caused the enhanced c-Cbl interactions with activated EGFR, suggesting that neither membrane-anchored nor soluble EGFcyt interfered with the ligandinduced c-Cbl-EGFR protein interaction.
This study demonstrated for the first time that EGFcyt can down-regulate ErbB2/Neu. The ErbB2 receptor is commonly considered an endocytosis-impaired receptor and ligand-activated EGFR-ErbB2 dimers largely recycle back to the cell membrane for reactivation [65–67]. Despite its known association with the two E3 ubiquitin ligases CHIP and c-Cbl [67–70], the impaired ability of ErbB2 to undergo ubiquitination has been suggested as a potential cause [71]. Recently, the deubiquitinating activity of POH1, a component of the 19S proteasomal lid, was shown to have a critical role in the regulation of ubiquitinated ErbB2 levels [72]. We identified a novel role of UCH-L1 in facilitating the EGFcyt-mediated down-regulation of ErbB2, implicating UCH-L1 as an important novel DUB in EGFcyt-mediated regulation of both ErbB1 and ErbB2 [73].
Our data provide further evidence for a potential tumor suppressor function of the small EGFcyt peptide region encoded by exon 23 in cancer cells [19–21], and this involves UCH-L1. Our structural homology modeling predicted this exon 23-encoded peptide region to be an exposed loop structure accessible for protein-protein interactions. The combination of existing ErbB inhibitors and the UCH-L1 gene silencing effect of specific exon 23-encoded peptides may be a novel attractive therapeutic strategy for the treatment of patients with aggressive ErbB1/2-dependent cancers.
Supplementary Material
Acknowledgments
The authors thank Christine Froehlich and Katrin Hammje for their skillful technical assistance and Steven Wiley, Pacific Northwest National Laboratory, USA, for providing the mbEGFctF construct.
Footnotes
C.H.V. and T.K. extend their thanks to the Mildred Schell Cancer Research Foundation (Deutsche Krebshilfe) for generous financial support. T.K. thanks the Natural Science and Engineering Council of Canada, Health Science Centre Foundation, and Manitoba Institute of Child Health for financial support.
This article refers to supplementary materials, which are designated by Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Massague J, Pandiella A. Membrane-anchored growth factors. Annu Rev Biochem. 1993;62:515–541. doi: 10.1146/annurev.bi.62.070193.002503. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Larrea J, Merlos-Suarez A, Urena JM, Baselga J, Arribas J. A role for a PDZ protein in the early secretory pathway for the targeting of proTGF-α to the cell surface. Mol Cell. 1999;3:423–433. doi: 10.1016/s1097-2765(00)80470-0. [DOI] [PubMed] [Google Scholar]
- 3.Kuo A, Zhong C, Lane WS, Derynck R. Transmembrane transforming growth factor-α tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franklin JL, Yoshiura K, Dempsey PJ, Bogatcheva G, Jeyakumar L, Meise KS, Pearsall RS, Threadgill D, Coffey RJ. Identification of MAGI-3 as a transforming growth factor-α tail binding protein. Exp Cell Res. 2005;303:457–470. doi: 10.1016/j.yexcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Franklin JL, Graves-Deal R, Jerome WG, Cao Z, Coffey RJ. Myristoylated Naked2 escorts transforming growth factor α to the basolateral plasma membrane of polarized epithelial cells. Proc Natl Acad Sci USA. 2004;101:5571–5576. doi: 10.1073/pnas.0401294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dempsey PJ, Coffey RJ. Basolateral targeting and efficient consumption of transforming growth factor-α when expressed in Madin-Darby canine kidney cells. J Biol Chem. 1994;269:16878–16889. [PubMed] [Google Scholar]
- 7.Dempsey PJ, Meise KS, Coffey RJ. Basolateral sorting of transforming growth factor-α precursor in polarized epithelial cells: characterization of cytoplasmic domain determinants. Exp Cell Res. 2003;285:159–174. doi: 10.1016/s0014-4827(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 8.Brown CL, Coffey RJ, Dempsey PJ. The proamphiregulin cytoplasmic domain is required for basolateral sorting, but is not essential for constitutive or stimulus-induced processing in polarized Madin-Darby canine kidney cells. J Biol Chem. 2001;276:29538–29549. doi: 10.1074/jbc.M102114200. [DOI] [PubMed] [Google Scholar]
- 9.Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao J, Lin H, Ouyang Y, Lei D, Osman A, Kim TW, Mei L, Dai P, Ohlemiller KK, Ambron RT. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat Neurosci. 2004;7:1250–1258. doi: 10.1038/nn1342. [DOI] [PubMed] [Google Scholar]
- 11.Wang JY, Frenzel KE, Wen D, Falls DL. Transmembrane neuregulins interact with LIM kinase 1, a cytoplasmic protein kinase implicated in development of visuospatial cognition. J Biol Chem. 1998;273:20525–20534. doi: 10.1074/jbc.273.32.20525. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Hutchinson L, Gaston SM, Raab G, Freeman MR. BAG-1 is a novel cytoplasmic binding partner of the membrane form of heparin-binding EGF-like growth factor: a unique role for proHB-EGF in cell survival regulation. J Biol Chem. 2001;276:30127–30132. doi: 10.1074/jbc.M010237200. [DOI] [PubMed] [Google Scholar]
- 13.Hieda M, Isokane M, Koizumi M, Higashi C, Tachibana T, Shudou M, Taguchi T, Hieda Y, Higashiyama S. Membrane-anchored growth factor, HB-EGF, on the cell surface targeted to the inner nuclear membrane. J Cell Biol. 2008;180:763–769. doi: 10.1083/jcb.200710022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinugasa Y, Hieda M, Hori M, Higashiyama S. The carboxylterminal fragment of pro-HB-EGF reverses Bcl6-mediated gene repression. J Biol Chem. 2007;282:14797–14806. doi: 10.1074/jbc.M611036200. [DOI] [PubMed] [Google Scholar]
- 15.Nanba D, Higashiyama S. Dual intracellular signaling by proteolytic cleavage of membrane-anchored heparin-binding EGF-like growth factor. Cytokine Growth Factor Rev. 2004;15:13–19. doi: 10.1016/j.cytogfr.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Adam RM, Danciu T, McLellan DL, Borer JG, Lin J, Zurakowski D, Weinstein MH, Rajjayabun PH, Mellon JK, Freeman MR. A nuclear form of the heparin-binding epidermal growth factor-like growth factor precursor is a feature of aggressive transitional cell carcinoma. Cancer Res. 2003;63:484–490. [PubMed] [Google Scholar]
- 17.Stoeck A, Shang L, Dempsey PJ. Sequential and gamma-secretase-dependent processing of the betacellulin precursor generates a palmitoylated intracellular-domain fragment that inhibits cell growth. J Cell Sci. 2010;123:2319–2331. doi: 10.1242/jcs.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groenestege WM, Thebault S, van der Wijst J, van den Berg D, Janssen R, Tejpar S, van den Heuvel LP, van Cutsem E, Hoenderop JG, Knoers NV, et al. Impaired basolateral sorting of pro-EGF causes isolated recessive renal hypomagnesemia. J Clin Invest. 2007;117:2260–2267. doi: 10.1172/JCI31680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klonisch T, Glogowska A, Gratao AA, Grzech M, Nistor A, Torchia M, Weber E, de Angelis MH, Rathkolb B, Cuong HV, et al. The C-terminal cytoplasmic domain of human proEGF is a negative modulator of body and organ weights in transgenic mice. FEBS Lett. 2009;583:1349–1357. doi: 10.1016/j.febslet.2009.03.051. [DOI] [PubMed] [Google Scholar]
- 20.Pyka J, Glogowska A, Dralle H, Hoang-Vu C, Klonisch T. Cytoplasmic domain of proEGF affects distribution and post-translational modification of microtubuli and increases microtubule-associated proteins 1b and 2 production in human thyroid carcinoma cells. Cancer Res. 2005;65:1343–1351. doi: 10.1158/0008-5472.CAN-04-2030. [DOI] [PubMed] [Google Scholar]
- 21.Glogowska A, Pyka J, Kehlen A, Los M, Perumal P, Weber E, Cheng SY, Hoang-Vu C, Klonisch T. The cytoplasmic domain of proEGF negatively regulates motility and elastinolytic activity in thyroid carcinoma cells. Neoplasia. 2008;10:1120–1130. doi: 10.1593/neo.08580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterman H, Katz M, Rubin C, Shtiegman K, Lavi S, Elson A, Jovin T, Yarden Y. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. EMBO J. 2002;21:303–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannessen LE, Pedersen NM, Pedersen KW, Madshus IH, Stang E. Activation of the epidermal growth factor (EGF) receptor induces formation of EGF receptor- and Grb2-containing clathrin-coated pits. Mol Cell Biol. 2006;26:389–401. doi: 10.1128/MCB.26.2.389-401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 25.Mort JS, Buttle DJ. Cathepsin B. Int J Biochem Cell Biol. 1997;29:715–720. doi: 10.1016/s1357-2725(96)00152-5. [DOI] [PubMed] [Google Scholar]
- 26.Authier F, Metioui M, Bell AW, Mort JS. Negative regulation of epidermal growth factor signaling by selective proteolytic mechanisms in the endosome mediated by cathepsin B. J Biol Chem. 1999;274:33723–33731. doi: 10.1074/jbc.274.47.33723. [DOI] [PubMed] [Google Scholar]
- 27.Ohta T, Fukuda M. Ubiquitin and breast cancer. Oncogene. 2004;23:2079–2088. doi: 10.1038/sj.onc.1207371. [DOI] [PubMed] [Google Scholar]
- 28.Baulida J, Kraus MH, Alimandi M, Di Fiore PP, Carpenter G. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 30.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 31.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 34.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 35.Hibi K, Westra WH, Borges M, Goodman S, Sidransky D, Jen J. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am J Pathol. 1999;155:711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong J, Opresko LK, Chrisler W, Orr G, Quesenberry RD, Lauffenburger DA, Wiley HS. The membrane-anchoring domain of epidermal growth factor receptor ligands dictates their ability to operate in juxtacrine mode. Mol Biol Cell. 2005;16:2984–2998. doi: 10.1091/mbc.E04-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell GI, Fong NM, Stempien MM, Wormsted MA, Caput D, Ku LL, Urdea MS, Rall LB, Sanchez-Pescador R. Human epidermal growth factor precursor: cDNA sequence, expression in vitro and gene organization. Nucleic Acids Res. 1986;14:8427–8446. doi: 10.1093/nar/14.21.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu X, Bryington M, Fisher AB, Liang X, Zhang X, Cui D, Datta I, Zuckerman KS. Ubiquitin/proteasome-dependent degradation of D-type cyclins is linked to tumor necrosis factor-induced cell cycle arrest. J Biol Chem. 2002;277:16528–16537. doi: 10.1074/jbc.M109929200. [DOI] [PubMed] [Google Scholar]
- 39.Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 40.Rubin C, Gur G, Yarden Y. Negative regulation of receptor tyrosine kinases: unexpected links to c-Cbl and receptor ubiquitylation. Cell Res. 2005;15:66–71. doi: 10.1038/sj.cr.7290268. [DOI] [PubMed] [Google Scholar]
- 41.Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, Raiborg C, Stenmark H, Madshus IH. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol Biol Cell. 2004;15:3591–3604. doi: 10.1091/mbc.E04-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ettenberg SA, Magnifico A, Cuello M, Nau MM, Rubinstein YR, Yarden Y, Weissman AM, Lipkowitz S. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J Biol Chem. 2001;276:27677–27684. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- 43.Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 44.Shih SC, Sloper-Mould KE, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 47.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 48.Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 49.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 50.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- 51.Imajoh-Ohmi S, Kawaguchi T, Sugiyama S, Tanaka K, Omura S, Kikuchi H. Lactacystin, a specific inhibitor of the proteasome, induces apoptosis in human monoblast U937 cells. Biochem Biophys Res Commun. 1995;217:1070–1077. doi: 10.1006/bbrc.1995.2878. [DOI] [PubMed] [Google Scholar]
- 52.Hussain S, Foreman O, Perkins SL, Witzig TE, Miles RR, van Deursen J, Galardy PJ. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. 2010;24:1641–1655. doi: 10.1038/leu.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 54.Mizukami H, Shirahata A, Goto T, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, Hibi K. PGP9.5 methylation as a marker for metastatic colorectal cancer. Anticancer Res. 2008;28:2697–2700. [PubMed] [Google Scholar]
- 55.Bittencourt Rosas SL, Caballero OL, Dong SM, da Costa Carvalho Mda G, Sidransky D, Jen J. Methylation status in the promoter region of the human PGP9.5 gene in cancer and normal tissues. Cancer Lett. 2001;170:73–79. doi: 10.1016/s0304-3835(01)00449-9. [DOI] [PubMed] [Google Scholar]
- 56.Mandelker DL, Yamashita K, Tokumaru Y, Mimori K, Howard DL, Tanaka Y, Carvalho AL, Jiang WW, Park HL, Kim MS, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–4968. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 57.Yamashita K, Park HL, Kim MS, Osada M, Tokumaru Y, Inoue H, Mori M, Sidransky D. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res. 2006;66:3921–3927. doi: 10.1158/0008-5472.CAN-05-1511. [DOI] [PubMed] [Google Scholar]
- 58.Jang MJ, Baek SH, Kim JH. UCH-L1 promotes cancer metastasis in prostate cancer cells through EMT induction. Cancer Lett. 2011;302:128–135. doi: 10.1016/j.canlet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 59.Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 60.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimura Y, Tanaka K. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem. 2010;147:793–798. doi: 10.1093/jb/mvq044. [DOI] [PubMed] [Google Scholar]
- 62.Bifsha P, Landry K, Ashmarina L, Durand S, Seyrantepe V, Trudel S, Quiniou C, Chemtob S, Xu Y, Gravel RA, et al. Altered gene expression in cells from patients with lysosomal storage disorders suggests impairment of the ubiquitin pathway. Cell Death Differ. 2007;14:511–523. doi: 10.1038/sj.cdd.4402013. [DOI] [PubMed] [Google Scholar]
- 63.Komada M. Controlling receptor downregulation by ubiquitination and deubiquitination. Curr Drug Discov Technol. 2008;5:78–84. doi: 10.2174/157016308783769469. [DOI] [PubMed] [Google Scholar]
- 64.de Melker AA, van der Horst G, Borst J. c-Cbl directs EGF receptors into an endocytic pathway that involves the ubiquitin-interacting motif of Eps15. J Cell Sci. 2004;117:5001–5012. doi: 10.1242/jcs.01354. [DOI] [PubMed] [Google Scholar]
- 65.Austin CD, De Maziere AM, Pisacane PI, van Dijk SM, Eigenbrot C, Sliwkowski MX, Klumperman J, Scheller RH. Endocytosis and sorting of ErbB2 and the site of action of cancer therapeutics trastuzumab and geldanamycin. Mol Biol Cell. 2004;15:5268–5282. doi: 10.1091/mbc.E04-07-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abella JV, Park M. Breakdown of endocytosis in the oncogenic activation of receptor tyrosine kinases. Am J Physiol Endocrinol Metab. 2009;296:E973–E984. doi: 10.1152/ajpendo.90857.2008. [DOI] [PubMed] [Google Scholar]
- 67.Roepstorff K, Grovdal L, Grandal M, Lerdrup M, van Deurs B. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer. Histochem Cell Biol. 2008;129:563–578. doi: 10.1007/s00418-008-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou P, Fernandes N, Dodge IL, Reddi AL, Rao N, Safran H, DiPetrillo TA, Wazer DE, Band V, Band H. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 69.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 70.Raja SM, Clubb RJ, Bhattacharyya M, Dimri M, Cheng H, Pan W, Ortega-Cava C, Lakku-Reddi A, Naramura M, Band V, et al. A combination of trastuzumab and 17-AAG induces enhanced ubiquitinylation and lysosomal pathway-dependent ErbB2 degradation and cytotoxicity in ErbB2-overexpressing breast cancer cells. Cancer Biol Ther. 2008;7:1630–1640. doi: 10.4161/cbt.7.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu H, Buus R, Clague MJ, Urbe S. Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS One. 2009;4:e5544. doi: 10.1371/journal.pone.0005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niendorf S, Oksche A, Kisser A, Lohler J, Prinz M, Schorle H, Feller S, Lewitzky M, Horak I, Knobeloch KP. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27:5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.