Abstract
Pancytopenia is a major cause of morbidity in acute myeloid leukemia (AML), yet its cause is unclear. Normal osteoblastic cells have been shown to support hematopoiesis. To define the effects of leukemia on osteoblastic cells, we used an immunocompetent murine model of AML. Leukemic mice had inhibition of osteoblastic cells, with decreased serum levels of the bone formation marker osteocalcin. Osteoprogenitor cells and endosteal-lining osteopontin+ cells were reduced, and osteocalcin mRNA in CD45− marrow cells was diminished. This resulted in severe loss of mineralized bone. Osteoclasts were only transiently increased without significant increases in bone resorption, and their inhibition only partially rescued leukemia-induced bone loss. In vitro data suggested that a leukemia-derived secreted factor inhibited osteoblastic cells. Because the chemokine CCL-3 was recently reported to inhibit osteoblastic function in myeloma, we tested its expression in our model and in AML patients. Consistent with its potential novel role in leukemic-dependent bone loss, CCL-3 mRNA was significantly increased in malignant marrow cells from leukemic mice and from samples from AML patients. Based on these results, we propose that therapeutic mitigation of leukemia-induced uncoupling of osteoblastic and osteoclastic cells may represent a novel approach to promote normal hematopoiesis in patients with myeloid neoplasms.
Introduction
Efficacy of treatment for AML, the most common adult acute leukemia, is limited and recurrence is common. Therefore, the identification of additional therapeutic targets is needed. One of the major causes of morbidity and mortality of acute leukemia is the disruption of normal hematopoiesis, causing neutropenia, anemia, and thrombocytopenia. Hematopoietic damage often occurs before overt systemic leukemia, which suggests that leukemic cells play an active role in the inhibition of normal hematopoiesis. The mechanisms by which AML inhibits normal hematopoiesis are poorly understood, and it is unclear whether this is a direct effect of the leukemic cells on the normal hematopoietic cells in the marrow or whether the microenvironment mediates leukemia-dependent hematopoietic damage.1
Cells of the mesenchymal/osteoblastic lineage play an essential role in the regulation of normal hematopoietic stem cells (HSCs).2–4 In addition to data suggesting that activation of osteoblastic cells expands HSCs and that osteoblastic injury results in myeloablation,2,4 specific disruptions of the osteoblastic compartment without genetic manipulation of the hematopoietic system result in a myeloproliferative disorder, demonstrating the important role osteoblasts play in hematopoietic stem and progenitor cell regulation.5 Osteoclasts as well as endothelial cells have also been shown to play a role in normal hematopoiesis and regulation of HSCs, particularly in their mobilization from the marrow.6–8
In xenograft models, human acute myeloid leukemia (AML) cells reside at the endosteal surface of bone,9,10 where they are found in close proximity to osteoblastic and osteoclastic cells; however, the interactions between leukemia and these microenvironmental cells have not been clearly defined. Moreover, xenograft models, although beginning to elucidate in vivo intercellular relationships, juxtapose hematopoietic and nonhematopoietic cells from different species, and may not recapitulate normal leukemia-microenvironment regulatory interactions. Nonetheless, a number of xenograft studies have suggested that leukemia disrupts molecular mechanisms used by normal HSCs to home to the endosteal niche, including CD4411,12 and the well-established interaction between CXCR4 and its ligand CXCL12.13–15
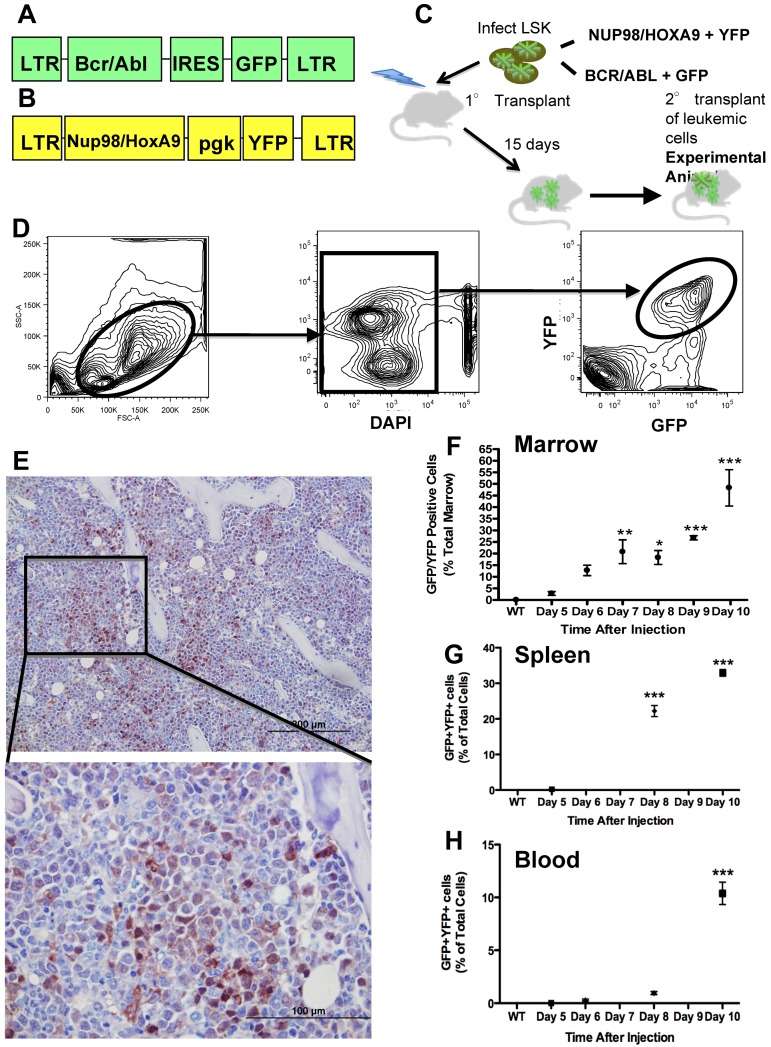
We hypothesized that leukemic cells alter osteoblastic and osteoclastic cell function resulting in measurable skeletal changes, which may impair support of normal hematopoiesis. To study interactions between leukemia and the marrow microenvironment, we used a well-characterized murine model of myelogenous leukemia.16,17 In this model, leukemia is initiated by immature hematopoietic cells that have been engineered to coexpress the BCR/ABL and Nup98/HoxA9 fusion products (Figure 1A-C), both of which have been documented in human leukemias, providing relevance of this model to human disease.18 This model recapitulates blast-crisis chronic myelogenous leukemia (bcCML)16 with very rapid disease progression as well as a lack of chronic disease. Therefore, it may closely represent the effects of AML on the normal marrow microenvironment. We used this in vivo model to define the progressive effects of AML on bone-forming and bone-resorbing cells.
Figure 1.
Murine model of bcCML. (A) Murine stem cell virus construct containing BCR/ABL and GFP. (B) MSCV construct containing Nup98/HoxA9 and YFP. (C) Schematic representation of the transplant strategy used to produce the leukemic mice used. (D) Flow cytometric gating strategy to identify leukemic cells as GFP+ and YFP+. (E) Representative anti-GFP immunohistochemistry of the femur's marrow space at the metaphysis. GFP is visualized by brown staining, with a hematoxylin counterstain. Top panel: 20× objective; bottom panel: 60× objective. (F) Flow cytometric data represent bcCML cells as a percentage of total marrow mononuclear cells. (G) Flow cytometric data representing bcCML cells as a percentage of total spleen. (H) Total peripheral blood mononuclear cells over the course of 10 days. *P ≤ .05. **P ≤ .01. ***P ≤ .001. n = 5 mice per time point. Bar represents SEM in this and subsequent experiments.
Methods
Mice
The Institutional Animal Care and Use Committee at the University of Rochester School of Medicine and Dentistry approved all animal studies.
Patient samples
Peripheral blood and marrow aspirates were collected from both patients with AML and healthy volunteer donors. Marrow aspirates were obtained from the posterior iliac crest. Blood and marrow plasma was isolated by centrifugation and analyzed for protein levels by ELISA. Bone marrow and peripheral blood mononuclear cells were isolated, and CD34+CD38− normal and leukemic cells were isolated as previously described.19 Relative gene expression was performed by real-time quantitative PCR. All patients and volunteers provided written, informed consent in accordance with the Declaration of Helsinki on protocols approved by the Research Subjects Review Board of the University of Rochester.
Model of AML
The MSCV-BCR/ABL-IRES-green fluorescent protein (GFP) and MSCV-Nup98/HoxA9-yellow fluorescent protein (YFP) vectors (Figure 1A-B) were previously described.16 Marrow cells from 6- to 8-week-old male CD45.1 mice were enriched for hematopoietic stem and progenitor cells by FACS sorting to purify the population of lineage-negative, sca-1+, c-kit+ cells. Lineage-negative, sca-1+, c-kit+ cells were infected with both viral vectors simultaneously as previously described.16 Six- to 8-week-old male C57bl/6 primary recipients were sublethally irradiated (6 Gy) using a 137Cs source of radiation (GAMMACELL-40). Immediately after irradiation, the mice were injected by tail vein with 2 × 104 leukemic cells in 0.1 mL of PBS containing 2% heat inactivated FBS (FACS buffer). After 15 days, the spleens of primary recipient mice were harvested and crushed using the plunger of a 3-mL syringe. The resulting cell suspension was strained using a 40-μm pore size cell strainer, resuspended in Cryostor CS10 (Biolife Solutions) at a concentration of 2 × 107 cells/mL, and cryogenically stored in liquid nitrogen. To induce leukemia in animals used for all described experiments, cells were thawed and 2 × 105 cells in 0.1 mL FACS buffer were injected by tail vein into nonirradiated 6- to 8-week-old male C57bl/6 mice. These are referred to as leukemic mice and were killed at the times after secondary transplantation as described for each experiment. Normal controls were always age- and sex-matched to the leukemic mice in the same experiment.
Murine marrow, spleen, and peripheral blood cell collection
For hematopoietic analysis, marrow cells were flushed from the long bones of the hind limbs of mice using a 25-gauge needle. Spleen cells were collected by crushing the spleen inside of a 40-μm pore size cell strainer using the plunger from a 3-mL syringe. Peripheral blood cells were collected by submandibular bleeds followed by incubation for 20 minutes at room temperature in 2% 500 000 molecular weight dextran to precipitate the RBCs.
Flow cytometric analysis and FACS
Using cells collected as previously described RBCs were lysed in 1 mL RBC lysis buffer (156mM NH4Cl, 127μM EDTA, and 12mM NaHCO3) for 5 minutes at room temperature, 1 × 107 cells were suspended in 100 μL FACS buffer and stained with appropriate antibodies. The cells were washed, and data were collected on a LSR-II (BD Biosciences). The data were analyzed using FlowJo Version 8.8.6 software (TreeStar). For sorting, cells were prepared as described for flow cytometric analysis, and GFP+/YFP+ cells were sorted using a FACSAria cell sorter (BD Biosciences) into FACS buffer.
Histology and immunohistochemistry
Hind limbs were collected, cleaned of soft tissue, fixed in 10% neutral buffered formalin for 48 hours, and decalcified in 14% EDTA for 10 days. Tissues were then processed and embedded in paraffin, 5-μm-thick sections were cut and used for H&E staining, immunohistochemistry, or tartrate-resistant acid phosphatase (TRAP) staining. Immunohistochemical staining for GFP and osteopontin used the monoclonal JL-8 antibody (Clontech; 632380) and the AKm2A1 antibody (Santa Cruz Biotechnology; sc-21742), respectively. Both immunohistochemical stains were performed using the MOM kit (Vector Laboratories; PK-2200) and were counterstained with hematoxylin. TRAP staining was performed as previously reported20 and counterstained with fast-green.
Osteoblastic cell collection from long bones
Osteoblastic cells were collected from the long bones of the hind limbs according to a previously described protocol.21 In brief, the long bones of the hindlimbs were cleaned of soft tissue, and the bone marrow was flushed with a 25-gauge needle and discarded. The resulting cleaned and flushed bones were cut into less than 1-mm fragments and digested twice in collagenase (StemCell Technologies; 7902) for 30 and 60 minutes sequentially. Remaining fragments were removed from the collected digest by passing through a 40-μm pore size cell strainer. The cells obtained were seeded in 6-well cell culture dishes at 1 × 106 cells per well for cell culture and bone nodule formation, or magnetically separated according to CD45 expression using the IMagnet system (BD Biosciences; 552311) and a biotinylated CD45 antibody (eBioscience, 13-0451-82).
Bone nodule assay
After 4 days in culture, media were changed to mineralizing media and 50 μg/mL L-ascorbic acid 2-phosphate (Sigma-Aldrich), and 10mM gycerol 2-phosphate disodium salt hydrate (Sigma-Aldrich) were added. At specified times after the addition of osteogenic media, cells were fixed with 10% neutral buffered formalin for 30 minutes followed by detection of alkaline phosphatase activity using a staining buffer containing 100mM Tris-HCl (Sigma-Aldrich), 0.005% weight/volume naphthol AS MX-PO4 (Sigma-Aldrich), and 0.03% weight/volume Red Violet LB salt (Sigma-Aldrich), with a final pH of 8.3 for 45 minutes. After alkaline phosphatase staining, cells were von Kossa stained (2.5% weight/volume AgNO3 [Sigma-Aldrich] for 30 minutes).
Osteoprogenitor cultures from marrow
Whole marrow flushed from the long bones of normal or leukemic mice was sorted as described for GFP−/YFP− cells and seeded in 6-well tissue culture dishes at 4 × 106 cells per well in minimum essential medium (α-MEM) containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) and then used for bone nodule assays.
MicroCT analysis
The right hindlimb of each mouse was fixed in 10% formalin for 48 hours and then stored in 70% ethanol at 4°C. MicroCT analysis was performed as previously described.22
ELISAs
All ELISAs were performed as indicated by the manufacturer. C-terminal telopeptide (CTX) measurements were performed using the RatLaps ELISA (AC-06F1; Immunodiagnostic Systems). TRACP 5b protein measurement was performed using Mouse TRAP ELISA (SB-TR103; Immunodiagnostic Systems). Osteocalcin measurement was performed using the Mouse Osteocalcin EIA kit (BT-470; Biomedical Technologies). Bovine CTX from bone wafer culture media was measured using the Crosslaps for Culture ELISA (AC-07F1; Immunodiagnostic Systems). Murine CCL3 protein measurement was performed using the Mouse CCL3/MIP-1α Quantikine ELISA Kit (MMA00; R&D Systems). Human CCL3 protein measurement was performed using the Human CCL3/MIP-1α Quantikine ELISA Kit (DMA00; R&D Systems).
ZA treatment
Mice were given a 0.25-mg/kg intraperitoneal injection of ZA biweekly for 2 weeks before induction of disease and throughout the course of the disease.
Osteoclastogenic cultures
Spleen cells were collected and RBCs were lysed as previously described. The remaining cells were cultured at a concentration of 8.75 × 105 cells/mL in α-MEM containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen; complete α-MEM), and 30 ng/mL M-CSF (R&D Systems) for 2 days; 10 ng/mL RANKL (R&D Systems) was added to the media, and the cultures were continued for an additional 4 days and then TRAP stained. For osteoblastic cocultures, osteoblastic cells were isolated as described. An osteoblast feeder layer was established by culturing 1.4 × 104 cells/well in 96-well plates in complete α-MEM supplemented with 1 × 10−9M 1,25 dihydroxyvitamin D3 (Sigma-Aldrich; D1530). After 2 days, 8.75 × 104 whole marrow cells from either normal or leukemic mice were added to each well. Half-media changes were performed every 2 days. Cultures were ended and TRAP stained after 7 days.
For cultures using bovine bone wafers, an IsoMet low-speed saw with a diamond blade was used to cut 300-μm-thick wafers of bone from a 4-mm2 pillar of devitalized bovine cortical bone to produce 300 μm × 4 mm × 4 mm bone wafers.
Analysis of relative gene expression by real-time RT-PCR
Total mRNA was extracted using the RNeasy kit (QIAGEN) according to the manufacturer's instructions. The Quantitect Reverse Transcription kit (QIAGEN) was used to transcribe cDNA, which was then diluted 1:50 in water, combined with SYBR Green PCR master mix (Bio-Rad) and amplified using MyiQ Single Color PCR detection systems and software under the following conditions: 95°C for 3 minutes followed by 40 cycles of 95°C for 15 seconds and 58°C (osteocalcin) or 60°C (CCL-3) for 30 seconds. Data were analyzed using the relative standard curve method, normalized to β-actin: murine β-actin 5′ primer: GCCACTGCCGCATCCTCTT; 3′ primer: GGAACCGCTCGTTGCCAATAG; murine osteocalcin 5′ primer: CCGCCTACAAACGCATCTACG; 3′ primer: GAGAGAGGACAGGGAGGATCAAG; and murine CCL3 5′ primer: AAGGATACAAGCAGCAGCGAGTA; 3′ primer: TGCAGAGTGTCATGGTACAGAGAA.
Human gene expression was determined using TaqMan technology (Applied Biosystems). Specific TaqMan probes were obtained from Applied Biosystems: CCL3 (Hs00234142_m1) and GAPDH (Hs02758991_g1) cDNA amplification was performed according to manufacturer's specifications using a LightCycler 480 II (Roche Diagnostics).
Cocultures and conditioned media
CFU-OB cultures were started as previously described. After 4 days in culture, osteogenic media were added, and 1 × 106 marrow cells from either normal or leukemic animals were added to the cultures. In addition, separate cultures containing normal CFU-OB cultures and 1 × 106 bone marrow cells from leukemic mice were used to produce conditioned media that was added to normal CFU-OB cultures starting on the same day as the bone marrow cells from leukemic mice were added to the cocultures.
Scanning electron microscopy
Bovine bone wafers were removed from culture media and fixed with 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer for 24 hours at 4°C. The next day, the wafers were postfixed in cacodylate buffered 1.0% osmium tetroxide, processed through a graded series of ethanol to 100% (3 times), and processed into a series of mixtures of 100% ethanol and hexamethyldisilazane before transitioning to 100% hexamethyldisilazane (3 times). The last exchange of 100% hexamethyldisilazane covering the wafers was allowed to evaporate at room temperature overnight in an uncovered 12-well plate in a fume hood. The dried wafers were mounted onto aluminum stubs, sputter coated for 90 seconds with gold, and imaged using a Zeiss Supra 40VP Field Emission scanning electron microscope.
Light microscopy
Histology slides were viewed at room temperature with a CKX41 upright microscope (Olympus). Objectives used were UPlan Fl 4×/0.13 UPlan Fl 20×/0.50, and UPlan FLN 60×/0.90 (Olympus). Cell culture dishes were viewed at room temperature with a BX41 inverted microscope (Olympus). Objective used was Plan N 2×/0.06 (Olympus). All images were obtained with a DP70 digital microscope camera and DPController Version 3.3.1.292 software (Olympus).
Statistical analysis
For quantitative assays, treatment groups were reported as mean ± SEM and compared using the Student t test in GraphPad Prism Version 5.0b (GraphPad Software). Statistical significance was established at P ≤ .05.
Results
An immunocompetent in vivo model where progression of leukemia is identified first in marrow, then spleen, and peripheral blood
We used a previously characterized model of acute leukemia (Figure 1A-C).16,17 Leukemic cells can be identified and quantified based on their expression of GFP and YFP (Figure 1D). In this model, secondary transplantation of the leukemia does not require irradiation of the recipient mice. Because our goal was to study the intact microenvironment, we chose to use as our experimental mice (heretofore designated as leukemic mice) the secondary transplantation recipients (Figure 1C) and as controls sex- and age-matched normal mice. Leukemic mice have rapid progression of disease that results in the accumulation of leukemic cells in the marrow identified by morphology, immunohistochemistry (Figure 1E), and flow cytometric analysis (Figure 1F), although total cellularity of the marrow is unchanged (29.94 ± 2.48 vs 27.18 ± 2.69, mononuclear marrow cells/hindlimb, normal vs leukemic, n = 13 from 3 separate experiments P = .4595). Leukemic cells also accumulate rapidly in the spleen (Figure 1G), followed by the blood (Figure 1H). This sequence suggests initial engraftment of leukemic cells in the marrow and spleen followed by migration into the bloodstream.
Leukemia decreases osteoblastic cells
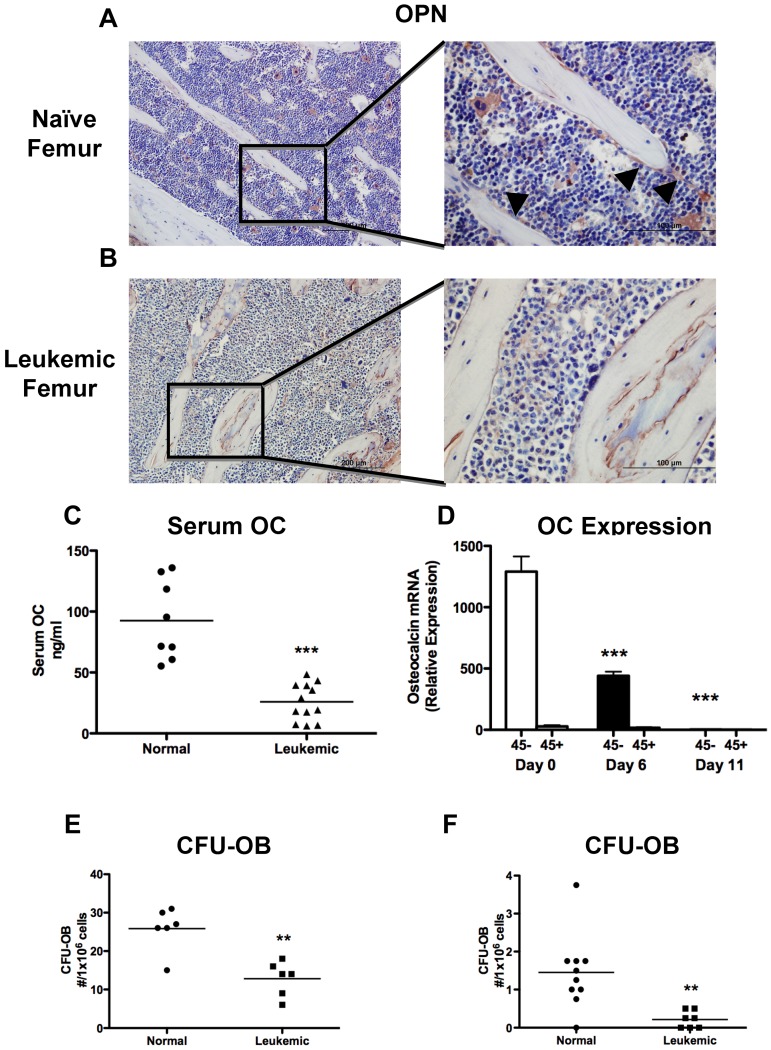
Examination by immunohistochemistry of sections from the long bones demonstrated a severe decrease in osteopontin+ endosteal osteoblastic cells in leukemic compared with normal mice (Figure 2A-B). Osteoblastic cells actively lay down bone; therefore, a decrease in bone formation would be expected with osteoblastic inhibition. Global bone formation correlates well with serum osteocalcin levels, which were strongly suppressed in leukemic mice compared with sex- and age-matched controls (Figure 2C). Because osteocalcin is specifically expressed in mature osteoblastic cells, we measured osteocalcin expression in cells obtained from the long bones (femur and tibia) by collagenase digestion after bulk hematopoietic cells are removed by flushing. As expected, osteocalcin expression was present only in nonhematopoietic, CD45− cells, and in leukemic mice there was a nearly 400-fold decrease in osteocalcin expression (Figure 2D), demonstrating loss of osteocalcin+ cells from the marrow microenvironment. Osteocalcin expression was already decreased at day 6, when only 10% to 15% of marrow mononuclear cells are leukemic (Figure 1F). To quantify the presence of osteoprogenitors in this cell pool, we performed bone nodule analysis and found that cells from the leukemic mice had reduced capacity to form alkaline phosphatase and von Kossa-positive colonies (Figure 2E). Notably, a small number of bone-attached CD45+ cells were still present, and in the cultures from leukemic mice, leukemic cells were detected by fluorescence, although they represented only a very small proportion of the total cells (data not shown). To identify more primitive osteoprogenitors and exclude leukemic cells from the bone nodule cultures from leukemic mice, marrow from leukemic mice was depleted of leukemic cells by FACS before plating. Marrow from leukemic mice had reduced osteoprogenitor numbers compared with normal sorted marrow (Figure 2F). These data suggest that, in addition to impaired function, osteoblastic and, to a lesser degree, osteoprogenitor numbers are decreased in the long bones of leukemic mice.
Figure 2.
Leukemia decreases osteoblastic number and function. (A-B) Osteopontin immunohistochemistry was performed on paraffin-embedded sections. Representative images are shown of (A) a naive femur and (B) a leukemic femur. Left panels 20×, and right panels 60× objectives. Osteopontin+ cells are stained brown, and sections were counterstained with hematoxylin (blue). Arrowheads indicate osteopontin+ cells. (C) Quantification of serum osteocalcin measured by ELISA. (D) Real-time RT-PCR quantifying osteocalcin RNA expression in osteoblast-like cells collected from the long bones of normal or leukemic mice at day 6 or 11 and magnetically separated based on CD45 expression. Statistical significance was determined compared with naive mice (day 0). n = 5 samples per experimental group. (E-F) CFU-OBs formed per well from (E) whole marrow after 28 days in culture and (F) cells collected by collagenase digestion of bone fragments after 15 days in culture. **P ≤ .01. ***P ≤ .001.
Leukemia induces bone loss
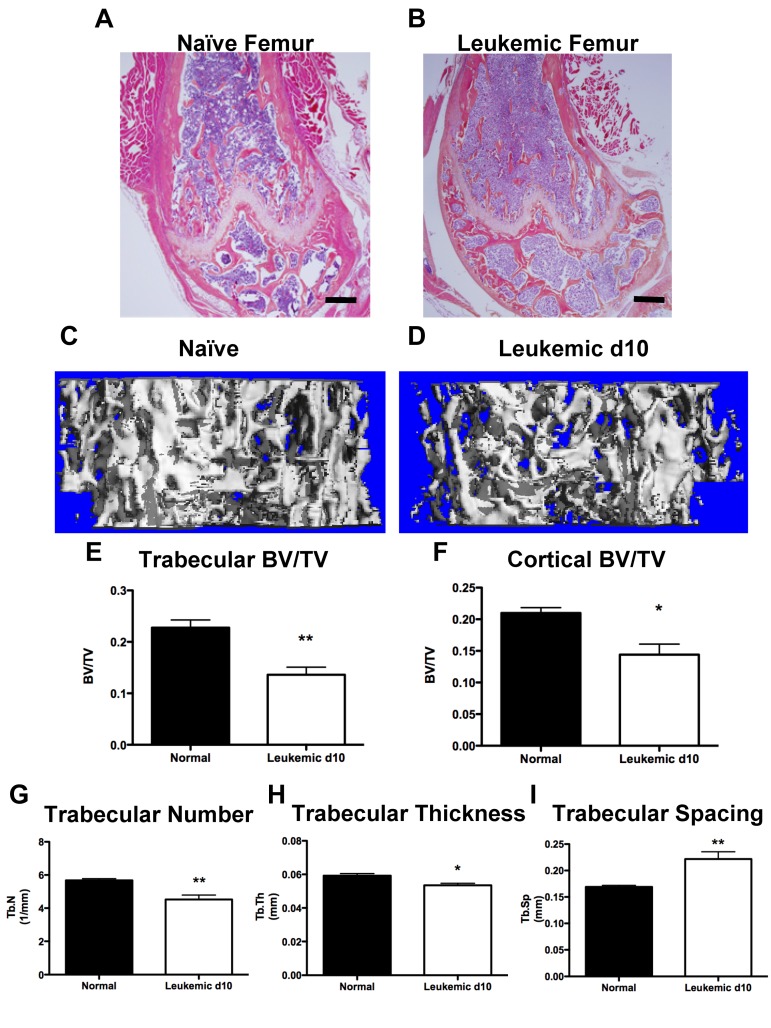
Functional inhibition of osteoblastic cells would be expected to result in a decrease in bone structures. Leukemic mice exhibited qualitative loss of trabecular structures, both in the metaphyseal region and the secondary ossification centers of the long bones, as well as cortical thinning (Figure 3A-B). At day 10, quantification of mineralized bone volume by micro-CT analysis demonstrated severe loss of trabecular (Figure 3C-E) and cortical (Figure 3F) bone in femora and tibiae of leukemic mice compared with age- and sex-matched controls. Trabecular number and thickness were decreased (Figure 3G-H) and trabecular spacing was increased (Figure 3I) in leukemic mice, consistent with alterations in the bone microenvironment and loss of trabecular bone volume. Therefore, in mice, development of AML results in net loss of mineralized bone.
Figure 3.
Leukemic environment induces bone loss. (A-B) Representative images of H&E-stained paraffin sections of the distal femur from a (A) naive and (B) leukemic mouse 10 days after transplantation, 4× objective. (C-D) Representative micro-CT images from the metaphysis of the femur from (C) naive and (D) leukemic mice. (E) Micro-CT analysis of femur trabecular bone volume/total volume. (F) Femur cortical bone volume/total volume. (G) Trabecular number. (H) Trabecular thickness. (I) Trabecular spacing. *P ≤ .05. **P ≤ .01. n = 4 mice per experiment.
Leukemic environment transiently increases osteoclast numbers in vivo
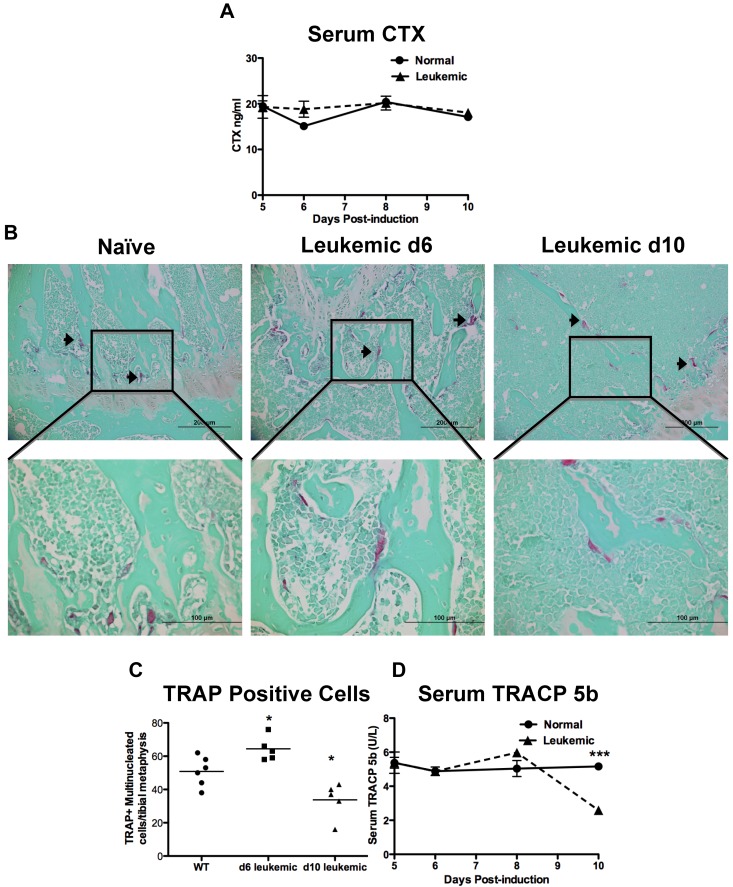
Bone formation and resorption are closely linked; therefore, bone loss could result from increased bone resorption, in addition to decreased bone formation. CTX are released from the bone matrix when it is resorbed and are an established measure of global bone resorption.23 When serum CTX levels were measured in the sera from leukemic and normal mice, there was no measured change in CTX levels from leukemic mice at any time during disease progression (Figure 4A).
Figure 4.
Leukemic environment mildly and transiently increases osteoclastic numbers in vivo. (A) Serum levels of carboxy-terminal collagen cross-link CTX, a marker of bone resorption as measured by ELISA. (B) Representative images of paraffin sections stained for the osteoclastic marker TRAP. TRAP+ cells are pink and highlighted by arrowheads. Top panels 20×, and bottom panels 60× objectives. (C) Quantification of multinucleated TRAP+ cells in a 1-mm2 area just proximal of the distal growth plate in the femur from sections represented by the panels in B. (D) Serum levels of TRACP 5b, the osteoclast specific TRAP enzyme, measured by ELISA. *P ≤ .05. ***P ≤ .001. Each point indicates an individual mouse in this and subsequent experiments; n = 5 mice per experiment.
To quantify osteoclasts, histologic sections from normal and leukemic mice were stained for TRAP activity. Multinucleated TRAP+ cells were scored as osteoclasts (Figure 4B). In leukemic mice, osteoclasts were mildly increased 6 days after induction of disease but were decreased by 10 days in leukemic mice compared with controls (Figures 4B-C). Measurement of serum TRACP 5b has been validated as a measure of global osteoclast numbers.24 In normal mice, the TRACP 5b serum levels were stable, contrasting with leukemic mice in which global TRACP 5b levels were initially mildly increased and fell below normal by day 10, mirroring the histologic findings (Figure 4D). These data suggest that, in our model, there is an initial and transient increase in osteoclastic cells, but as the disease progresses to overt leukemia, osteoclastic cell numbers decline.
Leukemic cells do not differentiate into osteoclasts and do not resorb bone matrix
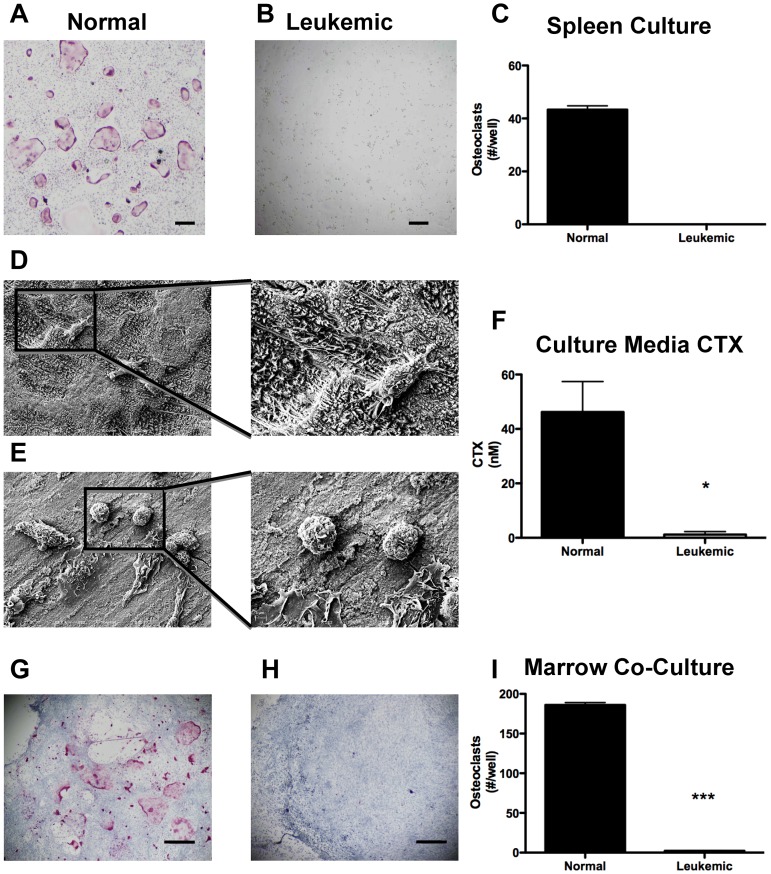
As osteoclasts are derived from the myeloid lineage, it is possible that leukemic cells may differentiate into osteoclasts. Therefore, we evaluated the ability of spleen-derived leukemic cells to differentiate into osteoclasts in vitro. Under osteoclastogenic conditions, cells from normal spleens produce abundant osteoclasts (Figure 5A,C). In striking contrast, GFP+/YFP+ leukemic cells isolated from spleens produce no TRAP+ osteoclasts (Figure 5B-C).
Figure 5.
Leukemia cells do not differentiate into osteoclasts and do not resorb bone. Representative light micrographs of (A) normal and (B) leukemic spleen cells under osteoclastogenic conditions in vitro. Pink cells are positive for TRAP activity. (C) Quantification of TRAP+ cells in panels A and B. (D) Low and high power scanning electron micrographs of osteoclasts on bovine bone wafers. (E) Low and high power scanning electron micrographs of leukemia cells on bovine bone wafers. (F) ELISA quantification of CTX released into culture media during culture of cells with bovine bone wafers. (G-H) Representative light micrographs of cocultures containing osteoblasts and (G) normal marrow cells and (H) leukemic marrow cells. (I) Quantification of TRAP+ cells formed per well from osteoblastic cocultures with normal and leukemic marrows. *P ≤ .05. ***P ≤ .001. n = 3 or 4 mice per treatment group.
Although leukemic cells do not differentiate into TRAP+ osteoclasts in vitro, they may be able to resorb bone matrix. To determine whether leukemic cells resorb bone directly, we sorted GFP+/YFP+ cells from the spleens of leukemic mice and cultured them on bovine bone wafers. Scanning electron microscopic images were obtained to visualize the surface of the bone wafers, where osteoclastic activity is identified by the presence of resorption pits with a rough appearance that were abundant in normal controls (Figure 5D). Whereas viable leukemic cells were observed to adhere to the surface of the bone wafers, the bone matrix in wells containing leukemic cells remained smooth and lacked resorption pits (Figure 5E). The amount of bone resorption was determined by the quantification of bovine CTX in the culture media. Normal osteoclast precursors under osteoclastogenic conditions readily resorb bone, as was demonstrated by the increase in bovine CTX. In contrast, leukemic cells under the same conditions did not increase bovine CTX (Figure 5F). These data suggest that GFP+/YFP+ leukemic cells do not directly contribute to bone resorption.
To determine whether the initial increase in osteoclasts in vivo was the result of pro-osteoclastogenic signals from the leukemic clone, we cocultured marrow cells from either leukemic or normal mice with a normal osteoblastic feeder layer in the presence of 1,25 dihydroxyvitamin D3. Under these culturing conditions, osteoclast formation was much more abundant in normal marrow cells compared with leukemic (Figure 5G-I). These data confirm the inability of leukemia cells to form mature osteoclasts in vitro and suggest that leukemic cells require a full leukemic microenvironment to stimulate the in vivo transient increase in osteoclasts.
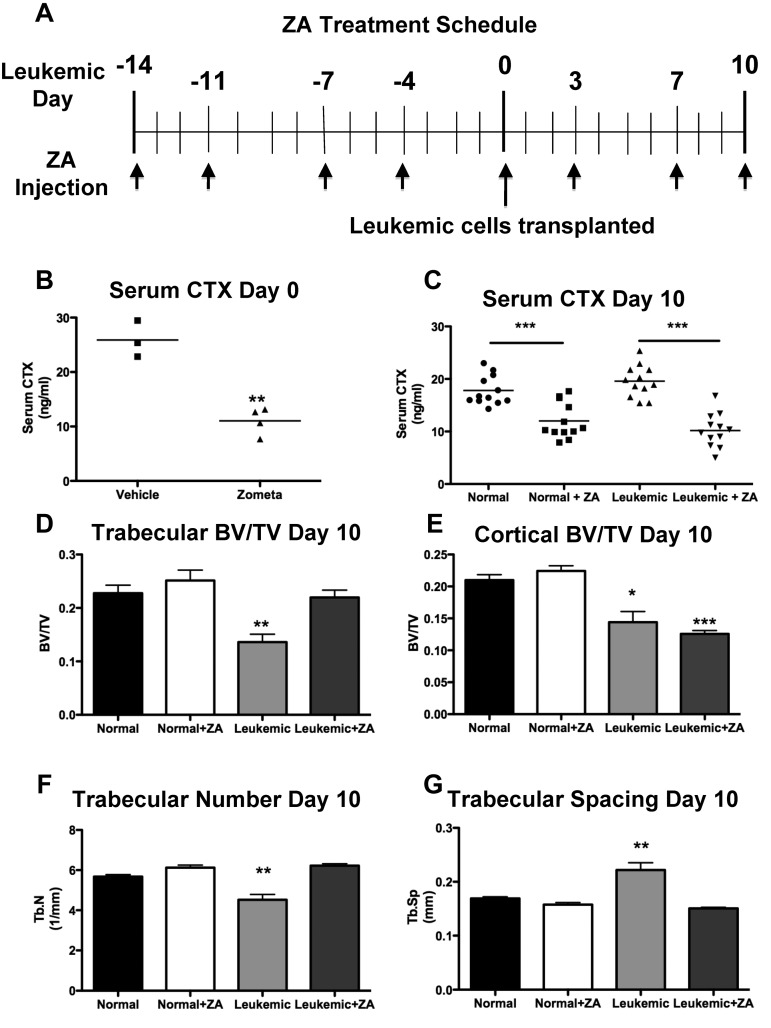
Treatment with ZA blocks bone resorption but only partially rescues bone loss in leukemic mice
To determine whether loss of bone formation or increased numbers of osteoclasts was responsible for the loss of bone in leukemic animals, we treated mice with zoledronic acid (ZA), a highly potent osteoclastic inhibitor,25 before transplantation with leukemic cells (Figure 6A). This treatment was sufficient to inhibit the activity of osteoclasts (Figure 6B). The serum levels of osteocalcin, a well-established bone formation marker, were significantly decreased in normal ZA-treated mice (142 ± 28 vs 49 ± 1 ng/mL, vehicle vs ZA-treated mice, P = .0296). This homeostatic compensatory decrease in bone formation in response to effective osteoclastic inhibition explains the lack of significant ZA-dependent increases in bone volumes in wild-type mice (Figure 6D-E). In leukemic mice treated with ZA simultaneous to control mice treated with PBS (Figure 4A), serum CTX dropped to subnormal levels (Figure 6C), confirming the inhibitory effect of ZA on osteoclasts in the setting of leukemia. Loss of trabecular bone volume was ameliorated by treatment with ZA (Figure 6D). Trabecular number (Figure 6F) and spacing (Figure 6G) were also rescued, consistent with ZA-dependent protection of the trabecular bone volume loss induced by leukemia. In contrast, cortical bone volume loss remained unchanged by inhibition of resorption (Figure 6E). When disease burden was quantified in leukemic mice treated with ZA, there was a mild decrease in disease burden in the marrow (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) but not in spleen (supplemental Figure 1B) or peripheral blood (supplemental Figure 1C). In this model, the phenotype of the leukemia stem cell has been previously defined16 (supplemental Figure 1D); however, treatment with ZA did not result in measurable changes in the frequency of leukemia stem cell in marrow (supplemental Figure 1E) or spleen (supplemental Figure 1F). Moreover, there were no changes in progression of disease or mortality in ZA-treated leukemic mice (data not shown). These data suggest that inhibition of bone resorption only partially reverses leukemic-induced bone loss without significantly changing disease progression.
Figure 6.
ZA rescues trabecular, but not cortical, bone loss. (A) Schematic for the treatment schedule of leukemic and normal mice with ZA. Leukemia was initiated on day 0 after 2 weeks of ZA treatment. Arrows indicate injection of ZA. (B) Serum CTX levels in normal mice after 2 weeks of treatment with ZA. (C) Serum CTX levels in normal and leukemic mice after the ZA treatment schedule. (D) Trabecular bone volume/total volume. (E) Cortical bone volume/total volume. *P < .05 (F) Trabecular number. (G) Trabecular spacing in normal or leukemic mice as quantified by micro-CT analysis after treatment with ZA. **P ≤ .01. ***P ≤ .001. (D-G) n = 4 mice per treatment group.
Expression of the chemokine CCL3 is increased in malignant marrow cells in leukemic mice
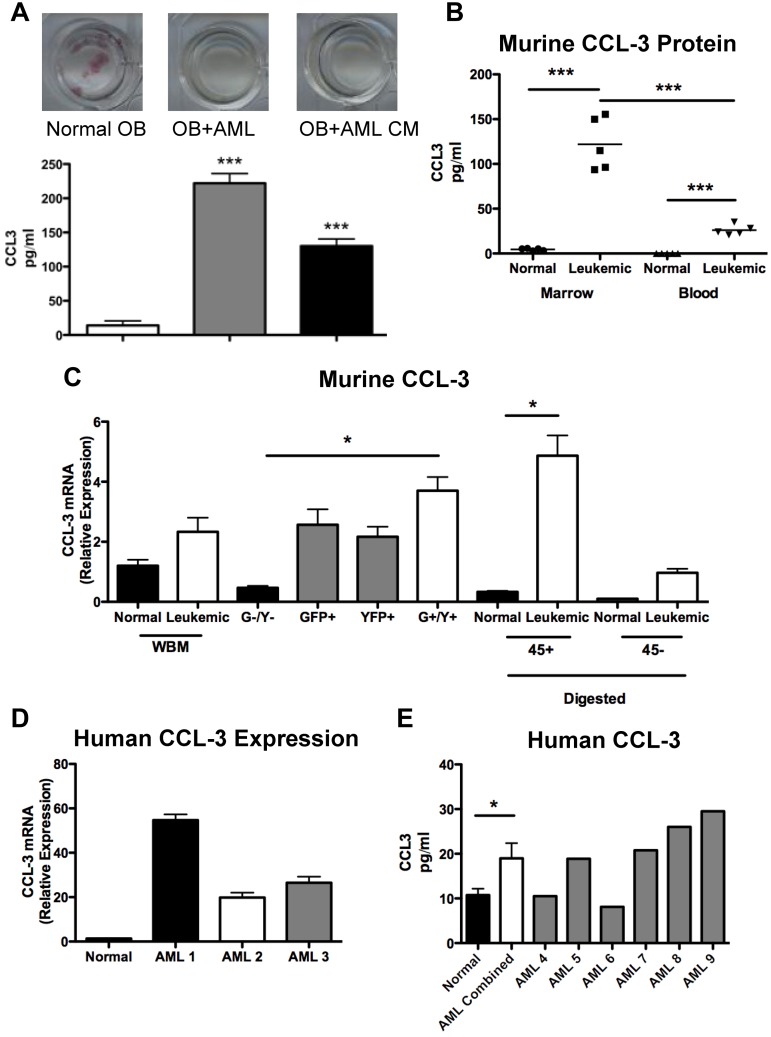
Osteoblastic inhibition by leukemia may be mediated by cell contact and/or by secreted molecules. CFU-OB cocultures of wild-type pre-osteoblasts with murine leukemic cells or with conditioned media generated from murine leukemic cells both showed a similar lack of alkaline phosphatase+ colonies (Figure 7A), suggesting that a secreted factor(s) is (are) sufficient to suppress colony formation. CCL3 is a proinflammatory cytokine in the CC chemokine family that has been implicated in the pro-osteoclastic actions observed in multiple myeloma.26,27 In addition, data have recently suggested that CCL3 inhibits osteoblastic cells in mice and in human in vitro studies.28 The level of CCL3 in the media was elevated in both cocultures and conditioned media (Figure 7A). CCL3 protein levels were also increased in the marrow plasma and blood serum from leukemic animals compared with normal controls (Figure 7B). To determine the cellular source of CCL3, mRNA was measured by quantitative real-time RT-PCR in total mRNA of marrow mononuclear cells, cells digested from bone fragments of the hind limbs that were separated based on surface CD45 positivity, and sorted GFP+/YFP+ leukemic cells. Bone-associated CD45+ cells from leukemic mice expressed higher levels of CCL3 mRNA compared with normal controls (Figure 7C). In sorted cells, GFP+/YFP+ leukemic cells demonstrated the highest level of CCL3 expression (Figure 7C). To assess whether CCL3 is also up-regulated in patients with AML, CCL3 expression was quantified in sorted CD34+CD38−CD123+ human marrow cells and found to be increased in marrow samples from AML patients compared with normal controls (Figure 7D). In addition, CCL3 protein levels were elevated in the marrow plasma from AML patients compared with normal controls (Figure 7F). These data are in agreement with the analysis of microarray data previously published by Bullinger et al in which 58 of 75 AML samples had elevated levels of CCL3 (supplemental Figure 2).29
Figure 7.
CCL-3 expression is increased in malignant cells from leukemic mice. (A) Top panel: Representative wells from CFU-OB cultures stained for alkaline phosphatase activity (pink). Bottom panel: CCL3 levels in culture media from CFU-OB cultures. (B) CCL3 protein levels in murine model of AML compared with normal controls. (C) Relative expression of CCL3 in bone marrow mononuclear cells isolated from whole bone marrow, cells sorted for GFP and YFP expression according to Figure 1D, and cells liberated from bone fragments by collagenase digestion and magnetically separated based on CD45 cell surface expression. (D) Relative expression of human CCL3 in primitive CD34+CD38−CD123+ AML cells compared with normal controls. Each bar represents a single AML sample normalized to 3 normal controls. (E) CCL3 protein levels in human AML patient marrow plasma. Each bar represents a single AML marrow sample compared with 7 normal controls. *P ≤ .05. ***P ≤ .001. (A-B,D) Each data point represents an individual mouse. (C) n = 3 mice per treatment group.
Discussion
It has long been known that osteoblastic cells support and expand HSCs in vitro30 and cotransplantation of osteoblastic cells with HSCs can increase engraftment rate.31 Work in our laboratory and others first identified osteoblastic cells as a regulatory component in the HSC niche through genetic means.2,3 It has become evident that osteoblastic cells can both stimulate2,3 and limit HSC expansion,32,33 promote quiescence,34–36 initiate HSC mobilization,37 and integrate sympathetic nervous system and HSC regulation.38 In addition, bone progenitor dysfunction is sufficient to induce myelodysplasia and secondary leukemia.39 Finally, recent data suggest that mesenchymal stem cells, which give rise to cells of the osteogenic lineage, regulate HSCs.40 In addition to increasing evidence that osteoblastic lineage cells act as orchestrators of HSC behavior, data strongly suggest that osteoblastic dysfunction results in pancytopenia.4,41
Pancytopenia is the cause of significant morbidity in leukemia, and yet the mechanisms by which leukemia causes anemias remain poorly understood. We have shown that induction of myeloid leukemia in an in vivo immunocompetent nonirradiated murine microenvironment induces severe functional inhibition of osteoblastic cells, even when the burden of disease is relatively low and when leukemic cells are undetectable in blood. Therefore, osteoblastic damage by leukemia may be the result of specific leukemic-initiated interactions rather than as a consequence of systemic disease.
Osteoblastic inhibition in leukemia resulted in decreased bone formation and net bone loss, particularly in cortical bone, where inhibition of osteoclast activity could not compensate for the leukemic-dependent bone loss. Although there are little data reporting the effects on bone in adult AML, these data are consistent with findings at diagnosis in pediatric acute leukemia, where decreased markers of bone formation before corticosteroid treatment have been documented in numerous studies.42,43 This disruption in bone formation may be reversible as, despite corticosteroid treatment in this disease, bone formation markers improve with reduction of disease burden after chemotherapy.44
Bone turnover is a closely regulated event in which bone formation and bone resorption are tightly coupled. In the setting of dramatic loss of bone formation, a compensatory loss of osteoclastic bone resorption would be expected. In contrast, there was a mild and transient increase in osteoclastic cells in mice with leukemia. Moreover, leukemic-induced trabecular bone loss was completely blocked by osteoclast inhibition. Therefore, leukemia initiation results in uncoupling of bone formation and bone resorption. This effect is probably responsible for the trabecular loss and may be important in the early stages of disease. We speculate that the decrease in osteoclastic number seen at later time points may be the result, in part, of the leukemia-induced block in hematopoietic differentiation that could limit the pool of hematopoietic osteoclast precursors. This result also indicates a potential role for osteoclastic inhibition, particularly at the time of recurrence when the percentage of blasts present in the marrow is relatively low and hematopoietic differentiation is preserved.
The mechanisms by which AML inhibits osteoblastic cells may include both direct and indirect actions of leukemia on their microenvironment. However, review of microarray data comparing gene expression in total RNA obtained from sorted lin−GFP+YFP+ cells with lin−GFP−YFP− cells from this mouse model indicated no significant changes in the expression of genes encoding cytokines with potent inhibitory activity against osteoblastic cells, such as dkk1, CCL2, SOST, and noggin (supplemental Table 1), suggesting they are not likely direct mediators of the leukemia-induced osteoblastic defect. In contrast, we have shown significantly increased expression of the gene encoding the chemokine CCL3. Receptors for CCL3 (CCR1 and CCR5) are present on osteoblastic cells,45,46 and recent data have demonstrated the inhibitory effects of CCL3 on both murine and human osteoblastic cells.28 Our findings reveal that CCL3 is highly expressed both by our leukemic model as well as in human AML. Further experiments are needed to determine whether CCL3 is indeed the key mediator of AML-dependent effects on the marrow microenvironment. Although initial studies identified CCL3 as an inhibitor of proliferation that may have differential effects on benign compared with malignant primitive hematopoietic cells,47 its role in AML remains unclear. This chemokine also has strong pro-osteoclastic effects26,27 that may at least in part explain the transient increase in TRAP+ cells found in our model, consistent with the uncoupling of bone formation and bone resorption found in our model. Of note, strategies for CCL3 inhibition have already been developed for the treatment of myeloma,48 where CCL3 is an important mediator of osteolytic disease. Therefore, our findings, if verified in human AML, may suggest a therapeutic approach in which CCL3 inhibition may be used to ameliorate osteoblastic dysfunction and accelerate recovery of normal hematopoiesis in the setting of leukemia treatment.
In conclusion, findings in our model support the concept that leukemia disrupts the normal marrow microenvironment and particularly targets cells that have been demonstrated to support and regulate HSCs. Because the loss of normal hematopoietic function during human leukemia is a major determinant of morbidity and mortality,1,49 our data support investigation of a human osteoblastic defect as a result of AML. If osteoblastic dysfunction is confirmed in human AML, mitigation of osteoblastic defects in the setting of chemotherapy may provide a novel therapeutic strategy to accelerate hematopoietic recovery.
Acknowledgments
The authors thank Drs Marshall Lichtman and James Palis for helpful discussion, Karen Bentley of the University of Rochester School of Medicine and Dentistry Electron Microscope Research Core facility for the SEM analysis, Michael Thullen for micro-CT analysis, and Mark LaMere for assistance in all studies involving human samples.
This work was supported by the Wilmot Scholar Cancer Research Award and the Pew Scholar in Biomedical Sciences Award (L.M.C.).
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.J.F. and J.M.A. performed experiments; L.P.X., M.W.B., and C.T.J. provided advice; and B.J.F. and L.M.C. designed the research, analyzed experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laura M. Calvi, Endocrine Division, Department of Medicine, University of Rochester School of Medicine, 601 Elmwood Ave, Box 693 Rochester, NY 14642; e-mail: laura_calvi@urmc.rochester.edu.
References
- 1.Lichtman MA. Interrupting the inhibition of normal hematopoiesis in myelogenous leukemia: a hypothetical approach to therapy. Stem Cells. 2000;18(5):304–306. doi: 10.1634/stemcells.18-5-304. [DOI] [PubMed] [Google Scholar]
- 2.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425(6960):836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 4.Visnjic D, Kalajzic Z, Rowe DW, Katavic V, Lorenzo J, Aguila HL. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103(9):3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- 5.Walkley CR, Olsen GH, Dworkin S, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129(6):1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollet O, Dar A, Shivtiel S, et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12(6):657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 7.Lymperi S, Horwood N, Marley S, Gordon MY, Cope AP, Dazzi F. Strontium can increase some osteoblasts without increasing hematopoietic stem cells. Blood. 2008;111(3):1173–1181. doi: 10.1182/blood-2007-03-082800. [DOI] [PubMed] [Google Scholar]
- 8.Porter RL, Calvi LM. Key endothelial signals required for hematopoietic recovery. Cell Stem Cell. 2009;4(3):187–188. doi: 10.1016/j.stem.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 10.Ninomiya M, Abe A, Katsumi A, et al. Homing, proliferation and survival sites of human leukemia cells in vivo in immunodeficient mice. Leukemia. 2007;21(1):136–142. doi: 10.1038/sj.leu.2404432. [DOI] [PubMed] [Google Scholar]
- 11.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 12.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12(10):1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 13.Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322(5909):1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- 14.Rombouts EJ, Pavic B, Lowenberg B, Ploemacher RE. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood. 2004;104(2):550–557. doi: 10.1182/blood-2004-02-0566. [DOI] [PubMed] [Google Scholar]
- 15.Sipkins DA, Wei X, Wu JW, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435(7044):969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neering SJ, Bushnell T, Sozer S, et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110(7):2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dash AB, Williams IR, Kutok JL, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99(11):7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto K, Nakamura Y, Saito K, Furusawa S. Expression of the NUP98/HOXA9 fusion transcript in the blast crisis of Philadelphia chromosome-positive chronic myelogenous leukaemia with t(7;11)(p15;p15). Br J Haematol. 2000;109(2):423–426. doi: 10.1046/j.1365-2141.2000.02003.x. [DOI] [PubMed] [Google Scholar]
- 19.Majeti R, Becker MW, Tian Q, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009;106(9):3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvi LM, Sims NA, Hunzelman JL, et al. Activated parathyroid hormone/parathyroid hormone-related protein receptor in osteoblastic cells differentially affects cortical and trabecular bone. J Clin Invest. 2001;107(3):277–286. doi: 10.1172/JCI11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chitteti BR, Cheng YH, Poteat B, et al. Impact of interactions of cellular components of the bone marrow microenvironment on hematopoietic stem and progenitor cell function. Blood. 2010;115(16):3239–3248. doi: 10.1182/blood-2009-09-246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frisch BJ, Porter RL, Gigliotti BJ, et al. In vivo prostaglandin E2 treatment alters the bone marrow microenvironment and preferentially expands short-term hematopoietic stem cells. Blood. 2009;114(19):4054–4063. doi: 10.1182/blood-2009-03-205823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risteli J, Elomaa I, Niemi S, Novamo A, Risteli L. Radioimmunoassay for the pyridinoline cross-linked carboxy-terminal telopeptide of type I collagen: a new serum marker of bone collagen degradation. Clin Chem. 1993;39(4):635–640. [PubMed] [Google Scholar]
- 24.Aya K, Alhawagri M, Hagen-Stapleton A, Kitaura H, Kanagawa O, Novack DV. NF-kappaB-inducing kinase controls lymphocyte and osteoclast activities in inflammatory arthritis. J Clin Invest. 2005;115(7):1848–1854. doi: 10.1172/JCI23763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green JR, Muller K, Jaeggi KA. Preclinical pharmacology of CGP 42′446, a new, potent, heterocyclic bisphosphonate compound. J Bone Miner Res. 1994;9(5):745–751. doi: 10.1002/jbmr.5650090521. [DOI] [PubMed] [Google Scholar]
- 26.Han JH, Choi SJ, Kurihara N, Koide M, Oba Y, Roodman GD. Macrophage inflammatory protein-1alpha is an osteoclastogenic factor in myeloma that is independent of receptor activator of nuclear factor kappaB ligand. Blood. 2001;97(11):3349–3353. doi: 10.1182/blood.v97.11.3349. [DOI] [PubMed] [Google Scholar]
- 27.Lentzsch S, Gries M, Janz M, Bargou R, Dorken B, Mapara MY. Macrophage inflammatory protein 1-alpha (MIP-1 alpha) triggers migration and signaling cascades mediating survival and proliferation in multiple myeloma (MM) cells. Blood. 2003;101(9):3568–3573. doi: 10.1182/blood-2002-08-2383. [DOI] [PubMed] [Google Scholar]
- 28.Vallet S, Pozzi S, Patel K, et al. A novel role for CCL3 (MIP-1alpha) in myeloma-induced bone disease via osteocalcin downregulation and inhibition of osteoblast function. Leukemia. 2011;25(7):1174–1181. doi: 10.1038/leu.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 30.Taichman RS, Emerson SG. Human osteoblasts support hematopoiesis through the production of granulocyte colony-stimulating factor. J Exp Med. 1994;179(5):1677–1682. doi: 10.1084/jem.179.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Badri NS, Wang BY, Cherry Good RA. Osteoblasts promote engraftment of allogeneic hematopoietic stem cells. Exp Hematol. 1998;26(2):110–116. [PubMed] [Google Scholar]
- 32.Nilsson SK, Johnston HM, Whitty GA, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106(4):1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 33.Stier S, Ko Y, Forkert R, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201(11):1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Yoshihara H, Arai F, Hosokawa K, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1(6):685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 36.Fleming HE, Janzen V, Lo Celso C, et al. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell. 2008;2(3):274–283. doi: 10.1016/j.stem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayack SR, Wagers AJ. Osteolineage niche cells initiate hematopoietic stem cell mobilization. Blood. 2008;112(3):519–531. doi: 10.1182/blood-2008-01-133710. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Katayama Y, Battista M, Kao WM, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124(2):407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 39.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464(7290):852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shono Y, Ueha S, Wang Y, et al. Bone marrow graft-versus-host disease: early destruction of hematopoietic niche after MHC-mismatched hematopoietic stem cell transplantation. Blood. 2010;115(26):5401–5411. doi: 10.1182/blood-2009-11-253559. [DOI] [PubMed] [Google Scholar]
- 42.Sala A, Barr RD. Osteopenia and cancer in children and adolescents: the fragility of success. Cancer. 2007;109(7):1420–1431. doi: 10.1002/cncr.22546. [DOI] [PubMed] [Google Scholar]
- 43.Sinigaglia R, Gigante C, Bisinella G, Varotto S, Zanesco L, Turra S. Musculoskeletal manifestations in pediatric acute leukemia. J Pediatr Orthop. 2008;28(1):20–28. doi: 10.1097/BPO.0b13e31815ff350. [DOI] [PubMed] [Google Scholar]
- 44.Crofton PM, Ahmed SF, Wade JC, et al. Effects of intensive chemotherapy on bone and collagen turnover and the growth hormone axis in children with acute lymphoblastic leukemia. J Clin Endocrinol Metab. 1998;83(9):3121–3129. doi: 10.1210/jcem.83.9.5133. [DOI] [PubMed] [Google Scholar]
- 45.Abbas S, Zhang YH, Clohisy JC, Abu-Amer Y. Tumor necrosis factor-alpha inhibits pre-osteoblast differentiation through its type-1 receptor. Cytokine. 2003;22(1):33–41. doi: 10.1016/s1043-4666(03)00106-6. [DOI] [PubMed] [Google Scholar]
- 46.Yano S, Mentaverri R, Kanuparthi D, et al. Functional expression of beta-chemokine receptors in osteoblasts: role of regulated upon activation, normal T cell expressed and secreted (RANTES) in osteoblasts and regulation of its secretion by osteoblasts and osteoclasts. Endocrinology. 2005;146(5):2324–2335. doi: 10.1210/en.2005-0065. [DOI] [PubMed] [Google Scholar]
- 47.Eaves CJ, Cashman JD, Wolpe SD, Eaves AC. Unresponsiveness of primitive chronic myeloid leukemia cells to macrophage inflammatory protein 1 alpha, an inhibitor of primitive normal hematopoietic cells. Proc Natl Acad Sci U S A. 1993;90(24):12015–12019. doi: 10.1073/pnas.90.24.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallet S, Raje N, Ishitsuka K, et al. MLN3897, a novel CCR1 inhibitor, impairs osteoclastogenesis and inhibits the interaction of multiple myeloma cells and osteoclasts. Blood. 2007;110(10):3744–3752. doi: 10.1182/blood-2007-05-093294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114(6):1150–1157. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]