Abstract
With the recent explosion of scientific data of unprecedented size and complexity, feature ranking and screening are playing an increasingly important role in many scientific studies. In this article, we propose a novel feature screening procedure under a unified model framework, which covers a wide variety of commonly used parametric and semiparametric models. The new method does not require imposing a specific model structure on regression functions, and thus is particularly appealing to ultrahigh-dimensional regressions, where there are a huge number of candidate predictors but little information about the actual model forms. We demonstrate that, with the number of predictors growing at an exponential rate of the sample size, the proposed procedure possesses consistency in ranking, which is both useful in its own right and can lead to consistency in selection. The new procedure is computationally efficient and simple, and exhibits a competent empirical performance in our intensive simulations and real data analysis.
Keywords: Feature ranking, feature screening, ultrahigh-dimensional regression, variable selection
1 Introduction
High-dimensional data have frequently been collected in a large variety of areas such as biomedical imaging, functional magnetic resonance imaging, tomography, tumor classifications, and finance. In high-dimensional data, the number of variables or parameters p can be much larger than the sample size n. Such a “large p, small n” problem has imposed many challenges for statistical analysis, and calls for new statistical methodologies and theories (Donoho, 2000; Fan and Li, 2006). The sparsity principle, which assumes that only a small number of predictors contribute to the response, is frequently adopted and deemed useful in the analysis of high-dimensional data. Following this general principle, a large number of variable selection approaches have been developed in the recent literature to estimate a sparse model and select significant variables simultaneously. Examples include Lasso (Tibshirani, 1996), SCAD (Fan and Li, 2001), nonnegative garrote (Breiman, 1995), group Lasso (Yuan and Lin, 2006), adaptive Lasso (Zou, 2006), and Dantzig selector (Candes and Tao, 2007). See Fan and Lv (2010) for an overview.
While those variable selection methods have been successfully applied in many high-dimensional analysis, modern applications in areas such as genomics, proteomics, and high-frequency finance further push the dimensionality of data to an even larger scale, where p may grow exponentially with n. Such ultrahigh-dimensional data present simultaneous challenges of computational expediency, statistical accuracy and algorithm stability (Fan, Samworth and Wu, 2009). It is difficult to directly apply the aforementioned variable selection methods to those ultrahigh-dimensional statistical learning problems due to the computational complexity inherent in those methods. To address those challenges, Fan and Lv (2008) emphasized the importance of feature screening in ultrahigh-dimensional data analysis, and proposed sure independence screening (SIS) and iterated sure independence screening (ISIS) in the context of linear regression models. Furthermore, Fan, Samworth and Wu (2009) and Fan and Song (2010) extended SIS and ISIS from a linear model to a generalized linear model. Each of those proposals focuses on a specific model, and its performance is based upon the belief that the imposed working model is close to the true model.
In this article, we propose a model-free feature screening approach for ultrahigh-dimensional data. Compared with the SIS, the most distinguishable feature of our proposal is that we only impose a very general model framework instead of a specific model. It is so general that the newly proposed procedure can be viewed as a model-free screening method, and it covers a wide range of commonly used parametric and semiparametric models. This feature makes our proposed procedure particularly appealing for feature screening when there are a huge number of candidate variables, but little information suggesting that the actual model is linear or follows any other specific parametric form. This flexibility is achieved by using the newly proposed marginal utility measure that is concerned with the entire conditional distribution of the response given the predictors. In addition, our method is robust to outliers and heavy-tailed responses in that it only uses the ranks of the observed response values. Theoretically, we establish that the proposed method possesses a consistency in ranking (CIR) property. That is, in probability, our marginal utility measure always ranks an active predictor above an inactive one, and thus guarantees a clear separation between the active and inactive predictors. The CIR property can be particularly useful in some genomic studies (Choi, Shedden, Sun and Zhu, 2009) where ranking is more of a concern than selection. Moreover, it leads to consistency in selection; that is, it simultaneously selects all active predictors and excludes all inactive predictors in probability, provided an ideal cutoff of the utility measure is available. The proposed procedure is valid provided that the total number of predictors p grows slower than exp(an) for any fixed a > 0. This rate is similar to the exponential rate achieved by the SIS procedures. Given a rank of all candidate features, we further propose a combination of hard and soft thresholding strategies to obtain the cutoff point that separates the active and inactive predictors. The soft threshold is constructed by adding a series of auxiliary variables, motivated by the idea of adding pseudo variables in model selection proposed by Luo, Stefanski and Boos (2006) and Wu, Boos and Stefanski (2007). Similar to the iterative SIS procedures, we also propose an iterative version of our new screening method. This is due to the fact that the marginal utility measure may miss an active predictor that is marginally independent of the response, a phenomenon also observed in the SIS procedures. The iterative procedure is shown to resolve this issue effectively. Computationally, the proposed screening procedure does not require any complicated numerical optimization and is very simple and fast to implement.
The rest of the article is organized as follows. In Section 2, we first present our general model framework, then develop the new feature ranking and screening approach. Section 3 illustrates the finite sample performance by both Monte Carlo simulations and a real data analysis. All technical proofs are given in the Appendix.
2 A Unified Feature Screening Approach
2.1 A General Model Framework
Let Y be the response variable with support Ψy, and Y can be both univariate and multivariate. Let x = (X1, · · ·, Xp)T be a covariate vector. Here we adopt the same notation system as Fan and Lv (2008) where a boldface lower case letter denotes a vector and a boldface capital letter denotes a matrix. We first develop the notion of active predictors and inactive predictors without specifying a regression model. We consider the conditional distribution function of Y given x, denoted by F(y | x) = P(Y < y | x). Define two index sets:
 = {k : F(y | x) functionally depends on Xk for some y ∈ Ψy},
= {k : F(y | x) functionally depends on Xk for some y ∈ Ψy}, = {k : F(y | x) does not functionally depend on Xk for any y ∈ Ψy}.
= {k : F(y | x) does not functionally depend on Xk for any y ∈ Ψy}.
If k ∈
 , Xk is referred to as an active predictor, whereas if k ∈
, Xk is referred to as an active predictor, whereas if k ∈
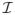 , Xk is referred to as an inactive predictor. Let x
, Xk is referred to as an inactive predictor. Let x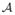 , a p1 × 1 vector, consist of all Xk with k ∈
, a p1 × 1 vector, consist of all Xk with k ∈
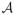 . Similarly, let x
. Similarly, let x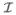 , a (p − p1) ×1 vector, consist of all inactive predictors Xk with k ∈
, a (p − p1) ×1 vector, consist of all inactive predictors Xk with k ∈
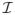 .
.
Next we consider a general model framework under which we are to develop our unified screening approach. Specifically, we consider that F(y | x) depends on x only through βTx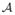 for some p1 × K constant matrix β. In other words, we assume that
for some p1 × K constant matrix β. In other words, we assume that
| (2.1) |
where F0(· | βTx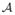 ) is an unknown distribution function for a given βTx
) is an unknown distribution function for a given βTx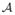 . We make the following remarks. First, β may not be identifiable; what is identified is the space spanned by the columns of β. However, the identifiability of β is of no concern here because our primary goal is to identify active variables rather than to estimate β itself. Actually, our screening procedure does not require an explicit estimation of β. Second, the form of (2.1) is fairly common in a large variety of parametric and semiparametric models where the response Y depends on the predictors x through a number of linear combinations βTx
. We make the following remarks. First, β may not be identifiable; what is identified is the space spanned by the columns of β. However, the identifiability of β is of no concern here because our primary goal is to identify active variables rather than to estimate β itself. Actually, our screening procedure does not require an explicit estimation of β. Second, the form of (2.1) is fairly common in a large variety of parametric and semiparametric models where the response Y depends on the predictors x through a number of linear combinations βTx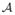 . As we will show next, (2.1) covers a wide range of existing models and, in many cases, K is as small as just one, two, or three.
. As we will show next, (2.1) covers a wide range of existing models and, in many cases, K is as small as just one, two, or three.
Before we continue the pursuit of feature screening, we examine some special cases of model (2.1) to show its generality. Note that many existing regression models for a continuous response can be written in the following form:
| (2.2) |
where h(·) is a monotone function, f2(·) is a nonnegative function, α1, α2, and α3 are unknown coefficients, and it is assumed that ε is independent of x. Here h(·), f1(·)and f2(·) may be either known or unknown. Clearly model (2.2) is a special case of (2.1) if we choose β to be a basis of the column space spanned by α1, α2 and α3. Meanwhile, it is seen that model (2.2) with h(Y) = Y includes the following special cases: the linear regression model, the partially linear model (Härdle, Liang and Gao, 2000), the single-index model (Härdle, Hall and Ichimura, 1993), and the partially linear single-index model (Carroll, Fan, Gijbels and Wand, 1997). Model (2.2) also includes the transformation regression model for a general transformation h(Y).
In survival data analysis, the response Y is the time to event of interest, and a commonly used model for Y is the accelerated failure time model:
where ε is independent of x. Different choices for the error distribution of ε lead to models that are frequently seen in survival analysis; that is, the extreme value distribution for ε yields the proportional hazards model (Cox, 1972), and the logistic distribution for ε yields the proportional odds model (Pettitt, 1982). It can again be easily verified that all those survival models are special cases of model (2.1).
Various existing models for discrete responses such as binary outcomes and count responses can be treated as a generalized partially linear single-index model (Carroll, Fan, Gijbels and Wand, 1997)
| (2.3) |
where the conditional distribution of Y given x belongs to the exponential family, g1(·) is a link function, g2(·) is an unknown function, and α1 and α2 are unknown coefficients. While model (2.3) includes the generalized linear model and the generalized single-index model as special cases, (2.3) itself is a special case of (2.1), which allows an unknown link function g1(·) as well.
In summary, a large variety of existing models with various types of response variables can be cast into the common model framework of (2.1). As a consequence, our feature screening approach developed under (2.1) offers a unified approach that works for a wide range of existing models.
2.2 A New Screening Procedure
To facilitate presentation, we assume throughout this article that E(Xk) = 0 and var(Xk) = 1 for k = 1, …, p. Define Ω(y) = E{xF (y | x)}. It then follows by the law of iterated expectations that Ω(y) = E[xE{1(Y < y) | x}] = cov{x, 1(Y < y)}. Let Ωk(y) be the k-th element of Ω(y), and define
| (2.4) |
Then ωk is to serve as the population quantity of our proposed marginal utility measure for predictor ranking. Intuitively, one can see that, if Xk and Y are independent, then Xk and the indicator function 1(Y < y) change independently. Consequently Ωk(y) = 0 for any y ∈ Ψy and ωk = 0. On the other hand, if Xk and Y are related, then there exists some y ∈ Ψy such that Ωk(y) ≠ 0, and hence ωk must be positive. This observation motivates us to employ the sample estimate of ωk to rank all the predictors. We will summarize this intuitive observation more rigorously in Corollary 1 in the next section.
Given a random sample {(xi, Yi), i = 1, · · ·, n} from {x, Y}, we next derive a sample estimator of ωk. For ease of presentation, we assume that the sample predictors are all standardized; that is, and for k = 1, · · ·, p. A natural estimator for ωk is
where Xik denotes the k-th element of xi. As shown in the proof of Theorem 2,
is a U-statistics. This enables us to directly use the theory of U-statistics to establish asymptotic property of ω̂k. Note that ω̂k is a scaled version of ω̃k. They lead to the same result of feature ranking and screening.
In sum, we propose to rank all the candidate predictors Xk, k = 1, …, , p, according to ω̂k from the largest to smallest. We then select the top ones as the active predictors. Later we will propose a thresholding rule for obtaining the cutoff value that separates the active and inactive predictors.
Before we turn to the theoretical properties of the proposed procedure, we will examine some simple settings to get more insight into our proposal. First, we consider a case where K = 1 and x ~ Np(0, σ2Ip) with unknown σ2. Note that the normality assumption on x is not necessary and will be relaxed later, to derive the measure’s properties. For ease of presentation, we write , and define b = (b1, …, bp)T = (βT, 0T)T. It follows by a direct calculation that
where with φ(v; 0, σ2) being the density function of N(0, σ2) at v. Then . If E{c2(Y)} > 0, then
| (2.5) |
and ωk = 0 if and only if k ∈
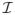 . This implies that the quantity ωk may be used for feature screening in this setting.
. This implies that the quantity ωk may be used for feature screening in this setting.
2.3 Theoretical Properties
The property (2.5) allows us to perform feature ranking and feature screening. To ensure this property in general, we impose the following conditions. It is interesting to note that all the conditions are placed on the distribution of x only.
-
(C1)The following inequality condition holds uniformly for p:
(2.6) where , Ω
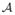 (y) = {Ω1(y), · · ·, Ωp1(y)}T, and λmax{B} and λmin{B} denote the largest and smallest eigenvalues of a matrix B, respectively. Note that λmin(B) and λmax(B) may depend on the dimension of B. Throughout this article, when we say that “a < b holds uniformly for p”, it means that
.
(y) = {Ω1(y), · · ·, Ωp1(y)}T, and λmax{B} and λmin{B} denote the largest and smallest eigenvalues of a matrix B, respectively. Note that λmin(B) and λmax(B) may depend on the dimension of B. Throughout this article, when we say that “a < b holds uniformly for p”, it means that
. -
(C2)The linearity condition:
(2.7) -
(C3)The moment condition: there exists a positive constant t0 such that
Condition (C1) dictates the correlations among the predictors, and is the key assumption to ensure that the proposed screening procedure works properly. We make the following remarks about this condition. First, as the dimension K of β in (2.1) increases, the condition becomes more stringent. Therefore, a model with a small K is favored by our procedure. In many commonly used models, however, K is indeed small, as partially shown in Section 2.1. Second, for the left hand side of (2.6), the numerator measures the correlation between the active predictors x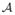 and the inactive ones x
and the inactive ones x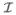 , while the denominator measures the correlation among the active predictors themselves. When x
, while the denominator measures the correlation among the active predictors themselves. When x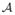 and x
and x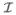 are uncorrelated, (C1) holds automatically. For the proposed screening method to work well, this condition rules out the case in which there is strong collinearity between the active and inactive predictors, or among the active predictors themselves. This is very similar to Condition 4 of Fan and Lv (2008, page 870). Third, the quantity
on the right hand side of (2.6) reflects the signal strength of individual active predictors, which in turn controls the rate of probability error in selecting the active predictors. This aspect is similar to Condition 3 of Fan and Lv (2008, page 870), which requires the contribution of an active predictor to be sufficiently large. Finally, we note that (2.6) is not scale invariant, since Σ = cov(x, xT) is not taken into account. This is similar to the linear SIS procedure of Fan and Lv (2008), which is based upon the covariance vector cov(x, Y) alone without the term Σ. Fan and Lv (2008) imposed the concentration property (Fan and Lv, 2008, Equation (16) on page 870) that implicitly requires the marginal variances of all predictors be of the same order. In our setup, we always marginally standardize all the predictors to have sample variance equal to one.
are uncorrelated, (C1) holds automatically. For the proposed screening method to work well, this condition rules out the case in which there is strong collinearity between the active and inactive predictors, or among the active predictors themselves. This is very similar to Condition 4 of Fan and Lv (2008, page 870). Third, the quantity
on the right hand side of (2.6) reflects the signal strength of individual active predictors, which in turn controls the rate of probability error in selecting the active predictors. This aspect is similar to Condition 3 of Fan and Lv (2008, page 870), which requires the contribution of an active predictor to be sufficiently large. Finally, we note that (2.6) is not scale invariant, since Σ = cov(x, xT) is not taken into account. This is similar to the linear SIS procedure of Fan and Lv (2008), which is based upon the covariance vector cov(x, Y) alone without the term Σ. Fan and Lv (2008) imposed the concentration property (Fan and Lv, 2008, Equation (16) on page 870) that implicitly requires the marginal variances of all predictors be of the same order. In our setup, we always marginally standardize all the predictors to have sample variance equal to one.
Condition (C2) holds if x follows a normal or an elliptical distribution (Fang, Kotz and Ng, 1989). This condition was first proposed by Li (1991) and has been widely used in the dimension-reduction literature. It is remarkable though that Condition (C2) is itself weaker than both the normality and the elliptical symmetry conditions because we only require it to hold for the true value of β. Furthermore, Hall and Li (1993) showed that the linearity condition holds asymptotically if the number of predictors p diverges while the dimension K remains fixed. For this reason, we view the linearity condition as a mild assumption in ultrahigh-dimensional regressions, where p is essentially very large and grows at a fast rate towards infinity.
Condition (C3) is concerned with the moments of the predictors, which assumes that all moments of the predictors are uniformly bounded. This condition holds for a variety of distributions, including the normal distribution and the distributions with bounded support. Compared with the usual conditions imposed in the feature screening literature, (C3) relaxed the normality assumption assumed by Fan and Lv (2008), in which both x and Y | x are assumed to be normally distributed.
Next we present the theoretical properties of the proposed screening measure. The proof is given in the Appendix. It is the main theoretical foundation for our feature screening procedure.
Theorem 1
Under Conditions (C1)–(C3), the following inequality holds uniformly for p:
| (2.8) |
The following corollary reveals that the quantity ωk is in fact a measure of the correlation between the marginal covariate Xk and the linear combinations βTx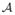 .
.
Corollary 1
Under the linearity condition (C2) and for k = 1, · · ·, p, ωk = 0 if and only if cov(βTx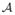 , Xk) = 0.
, Xk) = 0.
Theorem 1 and Corollary 1 together offer more insights into the newly proposed utility measure ωk. First, it is easy to see that, when Xk is independent of Y, ωk = 0. On the other hand, k ∈
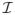 alone does not necessarily imply that ωk = 0. The quantity is zero only if Xk is uncorrelated with βTx
alone does not necessarily imply that ωk = 0. The quantity is zero only if Xk is uncorrelated with βTx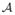 . Theorem 1, however, ensures that ωk of an inactive predictor is always smaller than ωk of an active predictor, which is sufficient for the purpose of predictor ranking.
. Theorem 1, however, ensures that ωk of an inactive predictor is always smaller than ωk of an active predictor, which is sufficient for the purpose of predictor ranking.
We next present the main theoretical result on feature ranking in terms of the utility measure ω̂k.
Theorem 2. (Consistency in Ranking)
In addition to the conditions in Theorem 1, we further assume that p = o {exp(an)} for any fixed a > 0. Then, for any ε > 0, there exists a sufficiently small constant sε ∈ (0, 2/ε) such that
In addition, if we write , then there exists a sufficiently small constant sδ/2 ∈ (0, 4/δ;) such that
Note that p = o {exp(an)}. Thus, the right-hand side of the above equation approaches 1 with an exponential rate as n → ∞. Theorem 2 justifies using ω̂k to rank the predictors, and it establishes the consistency in ranking. That is, ω̂k always ranks an active predictor above an inactive one in probability, and so guarantees a clear separation between the active and inactive predictors. Provided an ideal cutoff is available, this property would lead to consistency in selection in the ultrahigh-dimensional setup. Next we propose a thresholding rule to obtain a cutoff value to separate the active and inactive predictors.
2.4 Thresholding Rule
The thresholding rule is based upon a combination of a soft cutoff value obtained by adding artificial auxiliary variables to the data, and a hard cutoff that retains a fixed number of predictors after ranking.
The idea of introducing auxiliary variables for thresholding was first proposed by Luo, Stefanski and Boos (2006) to tune the entry significance level in forward selection, and then extended by Wu, Boos and Stefanski (2007) to control the false selection rate of forward regression in the linear model. We adopt this idea in our setup as follows. We independently and randomly generate d auxiliary variables z ~ Nd(0, Id) such that z is independent of both x and Y. The normality is not critical here, as we shall see later. Regard the (p + d) dimensional vector (xT, zT)T as the predictors and Y as the response. We calculate ωk for k = 1, · · ·, p + d. Since z is truly inactive by construction, we have by Theorem 1, and given a random sample {(xi, zi, Yi), i = 1, …, n}, it holds in probability that by Theorem 2. Define , which can be viewed as a benchmark that separates the active predictors from the inactive ones. This leads to the selection,
| (2.9) |
We call (2.9) the soft thresholding selection.
The next theorem gives an upper bound on the probability of recruiting any inactive variables by the above soft thresholding selection. It can be viewed as an analogue of Theorem 1 of Fan, Samworth and Wu (2009), while the exchangeability condition imposed in this theorem is similar in spirit to their condition (A1). This result shows how the soft thresholding rule performs.
Theorem 3
Let r ∈ ℕ, the set of natural numbers. We assume the exchangeability condition, that is, the inactive predictors {Xj, j ∈
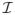 } and the auxiliary variables {Zj, j = 1, …, d} are exchangeable in the sense that both the inactive and auxiliary variables are equally likely to be recruited by the soft thresholding procedure. Then
} and the auxiliary variables {Zj, j = 1, …, d} are exchangeable in the sense that both the inactive and auxiliary variables are equally likely to be recruited by the soft thresholding procedure. Then
where |·| denotes the cardinality of a set.
An issue of practical interest in soft thresholding is the choice of number of auxiliary variables d. Intuitively, a small d value may introduce much variability, whereas a large d value requires heavier computation. Empirically, we choose d = p, and our numerical experience has suggested that this choice works quite well. Choosing an optimal d, however, is out of the scope of this paper and is a potential direction for future research.
In addition to soft thresholding, we also consider a hard thresholding rule proposed by Fan and Lv (2008), which retains a fixed number of predictors with the largest N values of ωk’s; that is,
| (2.10) |
where N is usually chosen to be [n/log n] and ω̂(N) denotes the N-th largest value among all ω̂k’s.
In practice, the data determine whether the soft or hard thresholding comes into play. To better understand the two thresholding rules, we conducted a simulation study. The results are not reported here but in an earlier version of this paper available at the authors’ websites. We make the following observations from our simulation study. When the signal in the data is sparse (a small p1), the hard thresholding rule often dominates the soft selection rule. On the other hand, when there are many active predictors (a large p1), the soft thresholding becomes more dominant. While the hard thresholding is fully determined by the sample size, soft thresholding takes into account the effect of signals in the data, which is helpful when p1 is relatively large. Consequently, we propose to combine the soft and hard thresholding, and construct the final active predictor index set as
| (2.11) |
where the union of the two sets is taken.
2.5 Iterative Feature Screening
An inherent issue with any feature screening procedure based on a marginal utility measure is that the method may miss those predictors which are marginally unrelated but jointly related to the response. To overcome this problem, we develop an iterative version of our proposed screening method. It is similar in spirit to the family of iterative SIS methods. However, unlike iterative SIS which breaks the correlation structure among predictors through the correlation between the residuals of the response and the remaining predictors, our method computes the correlation between the original response Y and the residual of the remaining x. This is because, the residual of Y is not available in a model-free context. However, we can compute the residual of x, where the residual is defined as the projection of the remaining of x onto the orthogonal complement space of the predictors selected in the previous steps. More specifically, our iterative procedure is given as follows.
-
Step 1
We first apply our proposed screening procedure for y and X, where X denotes the n × p data matrix that stacks n sample observations x1, …, xn and y = (Y1, …, Yn)T. Suppose p(1) predictors are selected, where p(1) < N = [n/log n]. We denote the set of indices of the selected predictors by
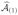 , and the associated n × p(1) data matrix by X
, and the associated n × p(1) data matrix by X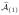 .
. -
Step 2Let
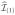 denote the complement of
denote the complement of
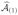 , and X
, and X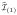 denote the remaining n × (p − p(1)) data matrix. Next, we define the predictor residual matrix
denote the remaining n × (p − p(1)) data matrix. Next, we define the predictor residual matrix
Apply again our proposed screening procedure for y and Xr. Suppose p(2) predictors are selected, and the resulting index set is denoted by
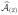 . Update the total selected predictor set by
. Update the total selected predictor set by
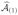 ∪
∪
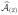
-
Step 3
Repeat Step 2 M − 1 times until the total selected number of predictors p(1) + … + p(M) exceeds the pre-specified number N = [n/log n]. The final selected predictor set is
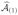 ∪ … ∪
∪ … ∪
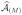 .
.
For the iterative procedure, we fix the number of total selected predictors N =[n/log n]. In our simulations, we consider an M = 2 iterative procedure and choose p(1) = [N/2], which works well for our example. Some guidelines on selecting these parameters in an iterative feature screening procedure can be found in Fan, Samworth and Wu (2009).
3 Numerical Studies
3.1 General Setup
In this section we assess the finite sample performance of the proposed method and compare it with existing competitors via Monte Carlo simulations. For brevity, we refer our approach as sure independent ranking and screening (SIRS). Throughout, we set the sample size n = 200 and the total number of predictors p = 2000. We repeat each scenario 1000 times. For the soft thresholding, we set the number of auxiliary variables d = p. We generate the predictors x from a normal distribution with mean zero. Unless otherwise specified, we consider two covariance structures of x:Σ1 = (σij)p×p with σij = 0.8|i−j|; and Σ2 = (σij)p×p with σii = 1, σij = 0.4 if both i, j ∈
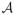 or i, j ∈
or i, j ∈
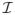 , and σij = 0.1 otherwise.
, and σij = 0.1 otherwise.
To evaluate the performance of the proposed method, we employ mainly two criteria. The first criterion measures accuracy of ranking the predictors (with no thresholding). For that purpose, we record the minimum number of predictors in a ranking that is required to ensure the inclusion of all the truly active predictors. We denote this number by
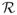 . The second criterion focuses on accuracy of feature screening when applying the proposed thresholding rule to the ranked predictors. Unlike feature selection, where it is important to simultaneously achieve both a high true positive and a low false positive, feature screening is more concerned with retaining all the truly active predictors. This is because screening usually serves as a preliminary massive reduction step, and is often followed by a conventional feature selection for further refinement. For that reason, we record the proportion that all the truly active predictors are correctly identified after thresholding in 1000 repetitions, and denote this proportion by
. The second criterion focuses on accuracy of feature screening when applying the proposed thresholding rule to the ranked predictors. Unlike feature selection, where it is important to simultaneously achieve both a high true positive and a low false positive, feature screening is more concerned with retaining all the truly active predictors. This is because screening usually serves as a preliminary massive reduction step, and is often followed by a conventional feature selection for further refinement. For that reason, we record the proportion that all the truly active predictors are correctly identified after thresholding in 1000 repetitions, and denote this proportion by
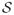 . A ranking and screening procedure is deemed competent if it yields an
. A ranking and screening procedure is deemed competent if it yields an
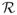 value that is close to the true number of active predictors p1, and an
value that is close to the true number of active predictors p1, and an
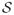 value that is close to one.
value that is close to one.
3.2 Linear Models
A large number of well known variable screening and selection approaches, such as linear SIS (Fan and Lv, 2008), Lasso (Tibshirani, 1996), stepwise regression, and forward regression (Wang, 2009). We thus begin with a class of linear models. Our simulations reveal the following two key observations. First, when the model is indeed linear homoscedastic with a normal error, SIRS has a comparable performance to the model-based methods which correctly specify the model. Second, when the true model deviates from the imposed model assumptions (e.g., the variance is heteroscedastic or the error distribution is heavily tailed), our method clearly outperforms the model-based methods.
Example 1
In the first example, we consider a classical linear model with varying squared multiple correlation coefficient R2, variance structure and error distribution:
| (3.1) |
where β = (1, 0.8, 0.6, 0.4, 0.2, 0, · · ·, 0)T takes grid values. We consider two predictor covariances Σ1 and Σ2 as specified in Section 3.1. We also examine two variance structures: σ = σ1, a constant, and σ = σ2 = exp(γTx), with γ = (0, · · ·, 0, 1, 1, 1, 0, · · ·, 0)T and ones appear in the 20th, 21st and 22nd positions. Thus, σ1 leads to a constant variance model, and we choose σ1 = 6.83 for Σ1, and σ1 = 4.92 for Σ2, which equals var(βTx) at the population level for the corresponding x. σ2 leads to a non-constant variance model. We consider two error ε distributions, a standard normal N(0, 1), and a t-distribution with one degree of freedom that has a heavy tail. We vary the constant c in front of βTx to control the signal-to-noise ratio. For the constant variance model σ1, we choose c = 0.5, 1 and 2, with the corresponding R2 = 20%, 50% and 80% respectively. For the non-constant variance model σ2, R2 are all very small (< 0.01%).
We first evaluate our proposed utility measure in terms of accuracy in ranking the predictors. We also compare our method (SIRS) with another ranking procedure, linear SIS of Fan and Lv (2008). Table 1 reports the median of the
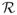 values. For σ= σ1, the number of truly actives p1 = 5 and for σ = σ2, p1 = 8. It is seen that, when the model is linear, homoscedastic (σ1), and the error follows a standard normal distribution N(0, 1), linear SIS performs the best, with the
values. For σ= σ1, the number of truly actives p1 = 5 and for σ = σ2, p1 = 8. It is seen that, when the model is linear, homoscedastic (σ1), and the error follows a standard normal distribution N(0, 1), linear SIS performs the best, with the
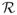 measure being very close to p1. However, the method breaks down for the heteroscedastic variance (σ2) or the heavy-tailed error distribution (t1). By contrast, our proposed procedure is comparable to linear SIS for the homoscedastic normal error, but is consistently superior with either the heteroscedastic variance or the heavy-tailed error distribution. Notably, our screening measure uses only the ranks of the observed response values, which partly explains why our method performs well for a heavy-tailed error (t1). In addition, we observe that our method performs well across a wide range of signal-to-noise ratios (σ1 with varying c), and the results for Σ1 and Σ2 are similar.
measure being very close to p1. However, the method breaks down for the heteroscedastic variance (σ2) or the heavy-tailed error distribution (t1). By contrast, our proposed procedure is comparable to linear SIS for the homoscedastic normal error, but is consistently superior with either the heteroscedastic variance or the heavy-tailed error distribution. Notably, our screening measure uses only the ranks of the observed response values, which partly explains why our method performs well for a heavy-tailed error (t1). In addition, we observe that our method performs well across a wide range of signal-to-noise ratios (σ1 with varying c), and the results for Σ1 and Σ2 are similar.
Table 1.
The ranking criterion
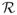 for Example 1 – minimum number of predictors required to ensure the inclusion of all the truly active predictors. The numbers reported are the median of
for Example 1 – minimum number of predictors required to ensure the inclusion of all the truly active predictors. The numbers reported are the median of
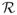 out of 1000 replications.
out of 1000 replications.
| ε | σ | Method | c = .5 | c = 1 | c = 2 | c = .5 | c = 1 | c = 2 |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Σ1 | Σ2 | |||||||
| N(0, 1) | σ1 | SIRS | 5 | 5 | 5 | 7 | 5 | 5 |
| SIS | 5 | 5 | 5 | 6 | 5 | 5 | ||
|
| ||||||||
| σ2 | SIRS | 9 | 11 | 18 | 8 | 9 | 8 | |
| SIS | 1739 | 1735 | 1646 | 1571 | 1447 | 1210 | ||
|
| ||||||||
| t1-dist | σ1 | SIRS | 5 | 5 | 5 | 5 | 5 | 5 |
| SIS | 1358 | 566 | 31 | 1608 | 1257 | 337 | ||
|
| ||||||||
| σ2 | SIRS | 10 | 9 | 12 | 10 | 9 | 9 | |
| SIS | 1735 | 1732 | 1757 | 1687 | 1678 | 1666 | ||
Next we evaluate our feature screening method with the proposed thresholding rule (2.11). We also compare with some commonly used and linear-model-based feature selection approaches, including linear SIS, Lasso, stepwise regression and forward regression. For stepwise regression, we use 0.05 as the inclusion probability and 0.10 as the exclusion probability. For Lasso and forward regression, we find that the BIC criterion proposed in the literature does not yield a satisfactory performance in our setup. Therefore, for those two methods, as well as linear SIS, we choose the same number of predictors as our proposed screening using the thresholding rule (2.11). The proportion
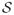 is reported in Table 2, which indicates that the SIRS performs competently across different scenarios, with the proportion
is reported in Table 2, which indicates that the SIRS performs competently across different scenarios, with the proportion
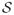 close to one. As expected, SIRS outperforms other methods for error being t-distribution with one degree of freedom (i.e., the Cauchy distribution) since other methods require finite error variance. It is also expected that all the selection methods except for SIRS cannot identify the active predictors in the variance of random error. Thus, when the error is heteroscedastic, the proportions shown in Table 2 for all methods except SIRS are almost zero. To make favorable comparison toward the model-based methods when the error is heteroscedastic, we further summarize the proportion that all active predictors (X1–X5) contained in the regression function are correctly identified out of 1000 replications in Table 3, from which it can be seen that SIRS performs very well, while all other methods perform unsatisfactorily. This is because the random error in this case contains some very extreme values (outliers), and the SIRS is robust to the outliers because it only uses the ranks of the observed response values.
close to one. As expected, SIRS outperforms other methods for error being t-distribution with one degree of freedom (i.e., the Cauchy distribution) since other methods require finite error variance. It is also expected that all the selection methods except for SIRS cannot identify the active predictors in the variance of random error. Thus, when the error is heteroscedastic, the proportions shown in Table 2 for all methods except SIRS are almost zero. To make favorable comparison toward the model-based methods when the error is heteroscedastic, we further summarize the proportion that all active predictors (X1–X5) contained in the regression function are correctly identified out of 1000 replications in Table 3, from which it can be seen that SIRS performs very well, while all other methods perform unsatisfactorily. This is because the random error in this case contains some very extreme values (outliers), and the SIRS is robust to the outliers because it only uses the ranks of the observed response values.
Table 2.
The selection criterion
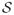 for Example 1 – proportion that all the truly active predictors (X1–X5 for σ = σ1 and X1–X5, X20–X22 for σ = σ2) are correctly identified out of 1000 replications. Reported are our proposal (SIRS), linear SIS, Lasso, stepwise regression (Step) and forward regression (FR).
for Example 1 – proportion that all the truly active predictors (X1–X5 for σ = σ1 and X1–X5, X20–X22 for σ = σ2) are correctly identified out of 1000 replications. Reported are our proposal (SIRS), linear SIS, Lasso, stepwise regression (Step) and forward regression (FR).
| ε | σ | Method | c = .5 | c = 1 | c = 2 | c = .5 | c = 1 | c = 2 |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Σ1 | Σ2 | |||||||
| N(0, 1) | σ1 | SIRS | 0.953 | 1.000 | 1.000 | 0.778 | 0.998 | 1.000 |
| SIS | 0.965 | 1.000 | 1.000 | 0.832 | 0.999 | 1.000 | ||
| Lasso | 0.032 | 0.230 | 0.618 | 0.197 | 0.576 | 0.926 | ||
| Step | 0.001 | 0.007 | 0.066 | 0.002 | 0.034 | 0.306 | ||
| FR | 0.015 | 0.111 | 0.382 | 0.000 | 0.015 | 0.307 | ||
|
| ||||||||
| σ2 | SIRS | 0.993 | 0.989 | 0.814 | 0.918 | 0.900 | 0.891 | |
| SIS | 0.000 | 0.000 | 0.001 | 0.010 | 0.033 | 0.058 | ||
| Lasso | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | ||
| Step | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| FR | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
|
| ||||||||
| t1-dist | σ1 | SIRS | 0.996 | 1.000 | 1.000 | 0.883 | 0.992 | 1.000 |
| SIS | 0.052 | 0.231 | 0.513 | 0.014 | 0.118 | 0.357 | ||
| Lasso | 0.002 | 0.004 | 0.036 | 0.002 | 0.025 | 0.080 | ||
| Step | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.001 | ||
| FR | 0.000 | 0.007 | 0.016 | 0.000 | 0.000 | 0.003 | ||
|
| ||||||||
| σ2 | SIRS | 0.932 | 0.990 | 0.974 | 0.844 | 0.895 | 0.887 | |
| SIS | 0.000 | 0.000 | 0.000 | 0.004 | 0.005 | 0.006 | ||
| Lasso | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| Step | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| FR | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
Table 3.
The selection criterion
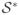 for Example 1 with heteroscedastic error – proportion that all active predictors (X1–X5) contained in the regression function are correctly identified out of 1000 replications. Reported are linear SIS, Lasso, stepwise regression (Step) and forward regression (FR).
for Example 1 with heteroscedastic error – proportion that all active predictors (X1–X5) contained in the regression function are correctly identified out of 1000 replications. Reported are linear SIS, Lasso, stepwise regression (Step) and forward regression (FR).
| ε | Method | c = .5 | c = 1 | c = 2 | c = .5 | c = 1 | c = 2 |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Σ1 | Σ2 | ||||||
| N(0, 1) | SIRS | 0.993 | 0.999 | 1.000 | 0.931 | 0.970 | 0.994 |
| SIS | 0.000 | 0.004 | 0.012 | 0.013 | 0.042 | 0.091 | |
| Lasso | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | |
| Step | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| FR | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
|
| |||||||
| t1-dist | SIRS | 0.932 | 0.990 | 1.000 | 0.848 | 0.944 | 0.980 |
| SIS | 0.000 | 0.000 | 0.000 | 0.004 | 0.005 | 0.007 | |
| Lasso | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Step | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| FR | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
Example 2
In this example, we continue to employ the linear model (3.1). In addition, we set σ = 1, c = 1 and β = (1, 1, 1, 0, · · ·, 0)T, so that there are p1 = 3 truly active predictors. What differs in this example is that we consider a more challenging covariance structure for the normally distributed x where cov(x) = Σ3 = (σij)p×p with entries σii = 1, i = 1, · · ·, p, and σij = 0.4, i ≠ j. We note that condition (C1) is not satisfied in this setup. In addition, we generate the error ε from a t distribution with 1, 2, 3 and 30 degrees of freedom. We remark that t1 is the Cauchy distribution, t1 and t2 have infinite variance, t3 has finite variance and t30 is almost indistinguishable from a standard normal distribution. As such we have a model that gradually approaches a normal distribution when the degrees of freedom increase.
Table 4 reports the ranking criterion
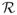 and Table 5 reports the selection criterion
and Table 5 reports the selection criterion
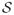 . Again we observe a qualitative pattern similar to Example 1. That is, when the error is close to normal (t30), the model-based SIS, Lasso, stepwise regression and forward regression perform very well, and our model-free procedure yields a comparable outcome. When the error deviates from a normal distribution (t with decreasing degrees of freedom), however, the performance of all the model-based alternatives quickly deteriorates, while our method continues to perform well.
. Again we observe a qualitative pattern similar to Example 1. That is, when the error is close to normal (t30), the model-based SIS, Lasso, stepwise regression and forward regression perform very well, and our model-free procedure yields a comparable outcome. When the error deviates from a normal distribution (t with decreasing degrees of freedom), however, the performance of all the model-based alternatives quickly deteriorates, while our method continues to perform well.
Table 4.
The ranking criterion
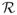 for Example 2. The quintuplet in each parenthesis consists of the minimum, the first quartile, median, third quartile and maximum value of
for Example 2. The quintuplet in each parenthesis consists of the minimum, the first quartile, median, third quartile and maximum value of
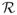 out of 1000 data replications.
out of 1000 data replications.
| ε | SIRS | SIS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| t1-dist | (3 | 4 | 9 | 28 | 1368) | (4 | 623 | 1126 | 1593 | 1999) |
| t2-dist | (3 | 3 | 3 | 5 | 680) | (3 | 3 | 7 | 36 | 1935) |
| t3-dist | (3 | 3 | 3 | 3 | 210) | (3 | 3 | 3 | 4 | 650) |
| t30-dist | (3 | 3 | 3 | 3 | 30) | (3 | 3 | 3 | 3 | 7) |
Table 5.
The selection criterion
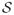 for Example 2. The caption is the same as Table 2.
for Example 2. The caption is the same as Table 2.
| ε | SIRS | SIS | Lasso | Step | FR |
|---|---|---|---|---|---|
| t1-dist | 0.961 | 0.076 | 0.027 | 0.002 | 0.004 |
| t2-dist | 0.997 | 0.913 | 0.849 | 0.640 | 0.647 |
| t3-dist | 0.998 | 0.995 | 0.995 | 0.982 | 0.987 |
| t30-dist | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
As shown above, when the model is correctly specified (e.g., Example 1 with c = 0.5 and a normal error), or sufficiently close to the true model (as seen in the trend of Example 2 as the error degree of freedom increases), the model-based solution is more competent than our model-free solution. This is not surprising because the former is equipped with additional model information. In practice, which solution to resort to depends on the amount of knowledge and confidence of an analyst has about the model. Our approach can be used in conjunction with, rather than as an alterative to, many model-based feature screening and selection solutions.
3.3 Nonlinear Models and Discrete Response
Our next goal is to demonstrate that the proposed model-free approach offers a useful and robust procedure in the sense that it works for a large variety of different models when there is little knowledge about the underlying true model. Toward that end, we consider two sets of examples that cover a wide range of commonly used parametric and semiparametric models. The first set involves a continuous response, including the transformation model, the multiple-index model and the heteroscedastic model.
Example 3
The response is continuous. The error ε follows a standard normal distribution. β = (2 − U1, …, 2 − Up1, 0, …, 0)T, β1 = (2 − U1, …, 2 − Up1/2, 0, …, 0)T, β 2 = (0, …, 0, 2 + Up1/2+1, …, 2 + Up1, 0, …, 0)T, and Uk’s follow a uniform distribution on [0, 1]. We vary the number of active predictors p1 to reflect different sparsity levels. The predictor x follows a mean zero normal distribution with two covariances Σ1 and Σ2 as given in Section 3.1.
-
3.a.
A transformation model: Y = exp {βTx/2 + ε}.
-
3.b.
A multiple-index model: .
-
3.c.
A heteroscedastic model: .
Table 6 reports the ranking criterion
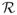 and Table 7 reports the selection proportion criterion
and Table 7 reports the selection proportion criterion
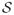 after applying the thresholding rule (2.11) to the ranked predictors. For a wide range of models under investigation,
after applying the thresholding rule (2.11) to the ranked predictors. For a wide range of models under investigation,
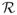 is often equal or close to the actual number of truly active predictors p1, whereas
is often equal or close to the actual number of truly active predictors p1, whereas
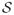 is equal or close to one, indicating a very high accuracy in both ranking and selection. In addition, our method clearly outperforms the alternative approaches which assume the linear homoscedastic model while the true models are not linear homoscedastic in this example.
is equal or close to one, indicating a very high accuracy in both ranking and selection. In addition, our method clearly outperforms the alternative approaches which assume the linear homoscedastic model while the true models are not linear homoscedastic in this example.
Table 6.
The ranking criterion
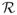 for Example 3. The caption is the same as Table 4.
for Example 3. The caption is the same as Table 4.
| p1 | Model | Method | Σ1 | Σ2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 3.a. | SIRS | (4 | 4 | 4 | 4 | 5) | (4 | 4 | 4 | 4 | 4) |
| SIS | (4 | 4 | 4 | 6 | 690) | (4 | 4 | 4 | 12 | 1808) | ||
| 3.b. | SIRS | (4 | 4 | 4 | 4 | 5) | (4 | 4 | 4 | 4 | 4) | |
| SIS | (4 | 4 | 6 | 12 | 1962) | (4 | 4 | 6 | 60 | 1996) | ||
| 3.c. | SIRS | (4 | 4 | 4 | 4 | 5) | (4 | 4 | 4 | 4 | 4) | |
| SIS | (4 | 5 | 7 | 23 | 1739) | (4 | 4 | 25 | 207 | 1998) | ||
|
| ||||||||||||
| 8 | 3.a. | SIRS | (8 | 8 | 8 | 8 | 10) | (8 | 8 | 8 | 8 | 8) |
| SIS | (8 | 25 | 78 | 214 | 1784) | (8 | 48 | 177 | 518 | 2000) | ||
| 3.b. | SIRS | (8 | 8 | 8 | 8 | 11) | (8 | 8 | 8 | 8 | 8) | |
| SIS | (8 | 147 | 458 | 1061 | 1997) | (8 | 99 | 349 | 825 | 1981) | ||
| 3.c. | SIRS | (8 | 8 | 8 | 8 | 10) | (8 | 8 | 8 | 8 | 8) | |
| SIS | (9 | 171 | 496 | 1097 | 1999) | (8 | 113 | 398 | 896 | 1988) | ||
|
| ||||||||||||
| 16 | 3.a. | SIRS | (16 | 16 | 16 | 16 | 22) | (16 | 16 | 16 | 16 | 16) |
| SIS | (29 | 463 | 845 | 1358 | 2000) | (18 | 456 | 881 | 1310 | 2000) | ||
| 3.b. | SIRS | (16 | 16 | 17 | 18 | 34) | (16 | 16 | 16 | 16 | 16) | |
| SIS | (35 | 1207 | 1676 | 1881 | 2000) | (25 | 559 | 1019 | 1517 | 1999) | ||
| 3.c. | SIRS | (16 | 16 | 17 | 18 | 34) | (16 | 16 | 16 | 16 | 16) | |
| SIS | (70 | 1286 | 1705 | 1890 | 2000) | (20 | 560 | 1047 | 1500 | 2000) | ||
Table 7.
The selection criterion
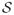 for Example 3. The caption is the same as Table 2.
for Example 3. The caption is the same as Table 2.
| Model | Method | Σ1 | Σ2 | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| p1 = 4 | p1 = 8 | p1 = 16 | p1 = 4 | p1 = 8 | p1 = 16 | ||
| 3.a. | SIRS | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| SIS | 0.963 | 0.330 | 0.002 | 0.878 | 0.310 | 0.034 | |
| Lasso | 0.118 | 0.000 | 0.000 | 0.475 | 0.003 | 0.000 | |
| Step | 0.008 | 0.000 | 0.000 | 0.014 | 0.000 | 0.000 | |
| FR | 0.035 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | |
|
| |||||||
| 3.b. | SIRS | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| SIS | 0.868 | 0.084 | 0.001 | 0.741 | 0.191 | 0.025 | |
| Lasso | 0.082 | 0.000 | 0.000 | 0.247 | 0.002 | 0.000 | |
| Step | 0.004 | 0.000 | 0.000 | 0.043 | 0.000 | 0.000 | |
| FR | 0.058 | 0.000 | 0.000 | 0.031 | 0.000 | 0.000 | |
|
| |||||||
| 3.c. | SIRS | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| SIS | 0.810 | 0.065 | 0.000 | 0.603 | 0.169 | 0.024 | |
| Lasso | 0.041 | 0.000 | 0.000 | 0.151 | 0.000 | 0.000 | |
| Step | 0.003 | 0.000 | 0.000 | 0.011 | 0.000 | 0.000 | |
| FR | 0.028 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | |
We have also examined a set of models with a discrete response, including the logistic model, the probit model, the Poisson log-linear model and the proportional hazards model (with a binary censoring indicator). Due to the space limitation, we only reported those results in an earlier version of this paper. Again, our extensive simulations show that the SIRS performs very well for the variety of discrete response models we have examined.
3.4 Iterative Screening
We next briefly examine the proposed iterative version of our marginal screening approach. The example is based upon a configuration in Fan and Lv (2008).
Example 4
We employ the linear model (3.1), with β = (5, 5, 5, −15ρ1/2, 0, · · ·, 0)T, c = 1, σ = 1, and ε follows a standard normal distribution. We draw x from a mean zero normal population with the covariance Σ4 = (σij)p×p with entries σii = 1, for i = 1, · · ·, p, σi4 = σ4i = ρ1/2 for i ≠ 4, and σij = ρ, for i ≠ j, i ≠ 4 and j ≠ 4. That is, all predictors except for X4 are equally correlated with correlation coefficient ρ, while X4 has correlation ρ1/2 with all other p − 1 predictors. By design X4 is independent of Y, so that our method cannot pick it up except by chance, whereas X4 is indeed an active predictor when ρ ≠ 0. We also vary the value of ρ to be 0, 0.1, 0.5 and 0.9, with a larger ρ yielding a higher collinearity.
We compare both the non-iterative and the iterative versions of our screening method. For the iterative procedure, we choose M = 2 iterations with p(1) = [N/2] and N = [n/log(n)]. This simple choice performs very well in this example. Table 8 reports the proportion criterion
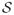 , where the iterative procedure dramatically improves over its non-iterative counterpart.
, where the iterative procedure dramatically improves over its non-iterative counterpart.
Table 8.
The selection criterion
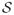 for Example 4 – proportion that all the truly active predictors are correctly identified out of 1000 replications. ISIRS denotes the iterative version of the proposed SIRS method.
for Example 4 – proportion that all the truly active predictors are correctly identified out of 1000 replications. ISIRS denotes the iterative version of the proposed SIRS method.
| Method | ρ = 0 | ρ = 0.1 | ρ = 0.5 | ρ = 0.9 |
|---|---|---|---|---|
| ISIRS | 0.925 | 1.000 | 1.000 | 0.940 |
| SIRS | 1.000 | 0.005 | 0.000 | 0.000 |
3.5 A Real Data Analysis
As an illustration, we apply the proposed screening method to the analysis of microarray diffuse large-B-cell lymphoma (DLBCL) data of Rosenwald et al. (2002). Given that DLBCL is the most common type of lymphoma in adults and has only about 35 to 40 percent survival rate after the standard chemotherapy, there has been continuous interest to understand the genetic factors that influence the survival outcome. The outcome in the study was the survival time of n = 240 DLBCL patients after chemotherapy. Measurements of p = 7,399 genes obtained from cDNA microarrays for each individual patient were the predictors. Given such a large number of predictors and small sample size, feature screening seems a necessary initial step as a prelude to any other sophisticated statistical modeling that does not cope well with such high dimensionality.
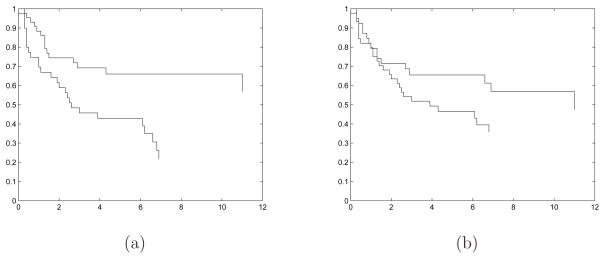
All predictors are standardized to have mean zero and variance one. We form the bivariate response consisting of the observed survival time and the censoring indicator. We use a data split of Li and Luan (2005) and Lu and Li (2008), which divides the data into a training set with n1 = 160 patients and a testing set with 80 patients. We apply the proposed screening method to the training data. Among 200 trials of the thresholding rule (2.11), 196 times the hard thresholding rule dominates. Therefore, we choose [n1/log(n1)] = 31 genes in our final set. This result seems to agree with the analysis of this same data set in the literature: only a small number of genes are relevant, and according to our simulations, the hard thresholding is more dominant in this scenario. Based on those selected genes, we fit a Cox proportional hazards model. We evaluate the prediction performance of this model following the approach of Li and Luan (2005) and Lu and Li (2008). That is, we apply the screening approach and fit a Cox model for the training data. We then compute the risk scores for the testing data and divide it to a low-risk group and a high-risk group, where the cutoff value is determined by the median of the estimated scores from the training set. Figure 1(a) shows the Kaplan-Meier estimate of survival curves for the two risk groups of patients in the testing data. The two curves are well separated, with the log-rank test yielding a p-value equal to 0.0025, indicating a good prediction of the fitted model.
Figure 1.
The Kaplan-Meier estimate of survival curves for the two risk groups in the testing data. (a) is based on the proposed feature screening, and (b) is based on the univariate Cox model screening.
Both Li and Luan (2005) and Lu and Li (2008) used a univariate Cox model to screen the predictors. Applying their screening approach, while retaining as many as 31 genes, yields a subset of genes among which 12 overlap with the ones identified by our method. As a simple comparison, we also fit a Cox model based on the genes selected by their marginal screening method, and evaluate its prediction performance. Figure 1(b) is constructed in the same fashion as Figure 1(a) except that the genes are selected by the univariate Cox model. The figure shows that the two curves are less well separated, with the p-value of the log-rank test equal to 0.1489, suggesting an inferior predictive performance compared to our method.
We remark that, without any information about the appropriate model form for this data set, our model-free screening result seems more reliable compared to a model-based procedure. We also note that choosing the Cox model after screening only serves as a simple illustration in this example. More refined model building and selection could be employed after feature screening, while the model-free nature of our screening method grants full flexibility in subsequent modeling.
Acknowledgments
The authors are grateful to Dr Yichao Wu for sharing the ideas through personal communication about the iterative screening approach presented in this paper. The authors thank the Editor, the AE and reviewers for their suggestions, which have helped greatly improve the paper. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF or NIDA.
Biographies
Li-Ping Zhu is Associate Professor, School of Statistics and Management, Shanghai University of Finance and Economics. luz15@psu.edu. His research was supported by National Natural Science Foundation of China grant 11071077 and National Institute on Drug Abuse (NIDA) grant R21-DA024260
Lexin Li is Associate Professor, Department of Statistics, North Carolina State University, Raleigh, NC 27695-8203. li@stat.ncsu.edu. His research was supported by NSF grant DMS 0706919
Runze Li is the corresponding author and Professor, Department of Statistics and The Methodology Center, The Pennsylvania State University, University Park, PA 16802-2111. rli@stat.psu.edu. His research was supported by NSF grant DMS 0348869, National Natural Science Foundation of China grant 11028103, and National Institute on Drug Abuse (NIDA) grant P50-DA10075
Li-Xing Zhu is Chair Professor of Statistics, Department of Mathematics, Hong Kong Baptist University. lzhu@hkbu.edu.hk. His research was supported by Research Grants Council of Hong Kong grant HKBU2034/09P
Appendix: proofs
Proof of Theorem 1
Without loss of generality, we assume that the basis matrix β = (β1, · · ·, βK) satisfies , where IK is a K × K identity matrix. In this case, the linearity condition (2.7) is simplified as . For ease of presentation, we denote the matrix vvT by v2 for a vector v.
Consider the left hand side of (2.8). Because x is independent of Y given βTx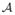 and Ỹ is an independent copy of Y, it follows that x is independent of Y and Ỹ given βTx
and Ỹ is an independent copy of Y, it follows that x is independent of Y and Ỹ given βTx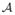 . This, together with the simplified linearity condition and the law of iterated expectations, yields that
. This, together with the simplified linearity condition and the law of iterated expectations, yields that
| (A.1) |
Then one can obtain that
| (A.2) |
where the first equality follows from (C2). Then it is straightforward to verify that
| (A.3) |
Here the second inequality follows because , and the third inequality holds due to the fact that λmax(CTBC) ≤ λmax(B)λmax(CTC) for any matrix B ≥ 0. After some algebra, we have
| (A.4) |
Then Condition (C1), together with (A.2), (A.3) and (A.4), entails (2.8).
Proof of Corollary 1
It follows from the definition in (2.4) that ωk = 0 is equivalent to E {Xk1(Y < y) = 0 for any y ∈ Ψy. Because Y relates to x only through linear combinations βTx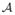 , it follows that there exists some y ∈ Ψy such that E{βTx
, it follows that there exists some y ∈ Ψy such that E{βTx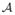 1(Y < y)} ≠ 0. Consequently, (A.1) implies that E{Xk1(Y < y)} = 0 if and only if cov(βTx
1(Y < y)} ≠ 0. Consequently, (A.1) implies that E{Xk1(Y < y)} = 0 if and only if cov(βTx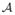 , Xk) = 0, which completes of proof of Corollary 1.
, Xk) = 0, which completes of proof of Corollary 1.
Proof of Theorem 2
To enhance readability, we divide the proof into two main steps.
Step 1
We first show that, under condition (C3),
| (A.5) |
Note that ω̂k can be expressed as follows:
Thus, ω̂k is a standard U-statistic. With Markov’s inequality, we can obtain that, for any 0 < t < s0k*, where k* = [n/3],
Through 5.1.6 of Serfling (1980), the U-statistic ω̂k can be represented as an average of averages of independent and identically distributed random variables; that is, , where each w(X1k, Y1; · · ·, Xnk, Yn) is an average of k* = [n/3] independent and identically distributed random variables, and denotes summation over n! permutations i1, · · ·, in of (1, · · ·, n). We denote that ψh(s) = E[exp {sh(Xjk, Yj; Xik, Yi; Xlk, Yl)}] for 0 < s < s0. Since the exponential function is convex, it follows by Jensen’s inequality that
Combining the above two results, we obtain that
| (A.6) |
where s = t/k*. Note that E {h(Xjk, Yj; Xik, Yi; Xlk, Yl)} = ωk, and with Taylor expansion, exp {sY} = 1 + sY + s2Z/2 for any generic random variable Y, where 0 < Z < Y2 exp {s1Y}, and s1 is a constant between 0 and s. It follows that
By invoking Condition (C3), it follows that there exists a constant C (independent of n and p) such that ; that is,
Recall that 0 < s = t/k* < s0. For a sufficiently small s, which can be achieved by selecting a sufficiently small t, we have that exp(−sε) = 1 − εs + O(s2) and therefore,
| (A.7) |
Combining the results (A.6) and (A.7), we show that, for any ε > 0, there exists a sufficiently small sε such that . Here we use the notation sε to emphasize s depending on ε. Similarly, we can prove that . Therefore,
| (A.8) |
This completes the proof of Step 1.
Step 2
We next show that
| (A.9) |
Recall the assumption that . Thus,
| (A.10) |
Proof of Theorem 3
Denote p* = p − |
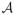 |. For a fixed r ∈ ℕ, the event that |
|. For a fixed r ∈ ℕ, the event that |
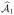 ∩
∩
 | ≥ r means there are at least r elements in {ω̂k : k ∈
| ≥ r means there are at least r elements in {ω̂k : k ∈
 } greater than all values of {ω̂k : k = p + 1, · · ·, p + d}. Because the auxiliary variables z and the inactive predictors x
} greater than all values of {ω̂k : k = p + 1, · · ·, p + d}. Because the auxiliary variables z and the inactive predictors x are equally likely to be recruited given Y, it follows that
are equally likely to be recruited given Y, it follows that
The result of Theorem 3 follows.
References
- Breiman L. Better subset regression using the nonnegative garrote. Technometrics. 1995:37, 373–384. [Google Scholar]
- Candes E, Tao T. The Dantzig selector: Statistical estimation when p is much larger than n (with discussion) Annals of Statistics. 2007:35, 2313–2404. [Google Scholar]
- Carroll RJ, Fan J, Gijbels I, Wand MP. Generalized partially linear single-index models. Journal of the American Statistical Association. 1997;92:477–489. [Google Scholar]
- Choi NH, Shedden K, Sun Y, Zhu J. Technical report. University of Michigan; 2009. Penalized regression methods for ranking multiple genes by their strength of unique association with a quantitative trait. [Google Scholar]
- Cox DR. Regression models and life tables. Journal of the Royal Statistical Society Series B. 1972;34:187–220. [Google Scholar]
- Donoho DL. High-dimensional data: The curse and blessings of dimensionality. American Mathematical Society Conference Mathematical Challenges of 21st Century 2000 [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle property. Journal of the American Statistical Association. 2001;96:1348–1360. [Google Scholar]
- Fan J, Li R. Statistical challenges with high dimensionality: Feature selection in knowledge discovery. In: Sanz-Sole M, Soria J, Varona JL, Verdera J, editors. Proceedings of the International Congress of Mathematicians. III. Freiburg European Mathematical Society; Zurich: 2006. pp. 595–622. [Google Scholar]
- Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space (with discussion) Journal of the Royal Statistical Society, Series B. 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Lv J. A selective overview of variable selection in high dimensional feature space. Statistica Sinica. 2010:20, 101–148. [PMC free article] [PubMed] [Google Scholar]
- Fan J, Samworth R, Wu Y. Ultrahigh dimensional feature selection: Beyond the linear model. Journal of Machine Learning Research. 2009;10:1829–1853. [PMC free article] [PubMed] [Google Scholar]
- Fan J, Song R. Sure independence screening in generalized linear models with NP-dimensionality. The Annals of Statistics. 2010;38:3567–3604. [Google Scholar]
- Fang KT, Kotz S, Ng KW. Symmetric Multivariate and Related Distributions . Chapman & Hall; London: 1989. [Google Scholar]
- Hall P, Li KC. On almost linearity of low dimensional projection from high dimensional data. Annals of Statistics. 1993:21, 867–889. [Google Scholar]
- Härdle W, Hall P, Ichimura H. Optimal smoothing in single-index models. Annals of Statistics. 1993;21:157–178. [Google Scholar]
- Härdle W, Liang H, Gao JT. Partially Linear Models. Springer Phisica-Verlag; Germany: 2000. [Google Scholar]
- Li KC. Sliced inverse regression for dimension reduction (with discussion) Journal of the American Statistical Association. 1991;86:316–342. [Google Scholar]
- Li L, Li H. Dimension reduction methods for microarrays with application to censored survival data. Bioinformatics. 2004;20:3406–3412. doi: 10.1093/bioinformatics/bth415. [DOI] [PubMed] [Google Scholar]
- Li H, Luan Y. Boosting proportional hazards models using smoothing spline, with application to high-dimensional microarray data. Bioinformatics. 2005:21, 2403–2409. doi: 10.1093/bioinformatics/bti324. [DOI] [PubMed] [Google Scholar]
- Lu W, Li L. Boosting methods for nonlinear transformation models with censored survival data. Biostatistics. 2008;9:658–667. doi: 10.1093/biostatistics/kxn005. [DOI] [PubMed] [Google Scholar]
- Luo X, Stefanski LA, Boos DD. Tuning variable selection procedure by adding noise. Technometrics. 2006:48, 165–175. [Google Scholar]
- Pettitt AN. Inference for the linear model using a likelihood based on ranks. Journal of Royal Statistical Society, Series B. 1982;44:234–243. [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Hermelink HK, Smeland EB, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England Journal of Medicine. 2002:346, 1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Serfling RJ. Approximation Theorems of Mathematical Statistics. New York: John Wiley & Sons Inc; 1980. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via lasso. Journal of the Royal Statistical Society, Series B. 1996;58:267–288. [Google Scholar]
- Wang H. Forward regression for ultra-high dimensional variable screening. Journal of the American Statistical Association. 2009;104:1512–1524. [Google Scholar]
- Wu Y, Boos DD, Stefanski LA. Controlling variable selection by the addition of pseudo variables. Journal of the American Statistical Association. 2007;102:235–243. [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society, Series B. 2006;68:49–67. [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]