Summary
Sterol Regulatory Element-Binding Proteins (SREBPs) activate genes involved in the synthesis and trafficking of cholesterol and other lipids, and therefore are critical for maintaining lipid homeostasis. Aberrant SREBP activity, however, can result in excess stored fat and contribute to obesity, fatty liver disease and insulin resistance, hallmarks of metabolic syndrome. Our studies identify a conserved regulatory circuit in which SREBP-1 controls production of the methyl donor S-adenosylmethionine (SAMe). Methylation is critical for synthesis of phosphatidylcholine (PC), a major membrane component, and we find that blocking SAMe or PC synthesis in C. elegans, mouse liver and human cells causes elevated SREBP-1-dependent transcription and lipid droplet accumulation. Distinct from negative regulation of SREBP-2 by cholesterol, our data suggest a mechanism where maturation of nuclear, transcriptionally active SREBP-1 is controlled by levels of PC. Thus, nutritional or genetic conditions limiting SAMe or PC production may activate SREBP-1, contributing to human metabolic disorders.
Introduction
Disorders of metabolism, such as obesity, insulin resistance and nonalcoholic fatty liver disease (NAFLD) are associated with metabolic syndrome, and represent risk factors for more serious diseases such as type II diabetes and cardiovascular disease. However, molecular mechanisms causing dysfunction in these complex diseases remain unclear. The Sterol Regulatory Element-Binding Protein family (SREBP-1a, -1c, SREBP-2) respond to nutrient levels and regulate transcription of genes required for many aspects of lipid metabolism (Osborne and Espenshade, 2009). SREBP-2 preferentially controls expression of many cholesterogenic genes, whereas SREBP-1 isoforms primarily regulate fatty acid and phospholipid biosynthesis genes. In accord with distinct gene regulatory functions, activities of the isoforms appear to be controlled by different regulatory cues. The cholesterol-mediated negative feedback regulation of SREBP-2 is particularly well understood (Brown and Goldstein, 1997). SREBP-2 is produced as an inactive precursor stored with the cholesterol-sensing INSIG/SCAP chaperone complex in the endoplasmic reticulum (ER). Depletion of cholesterol promotes release from INSIG and SCAP-mediated transit of SREBP-2 to the Golgi, followed by proteolytic maturation and nuclear translocation of the transcriptionally active portion to stimulate cholesterogenic gene expression. SREBP-1a and -1c are produced through alternative promoter usage and splicing of the SREBF1 gene (Horton et al., 2002). While SREBP-1 isoforms are also synthesized as ER-targeted precursors, SREBP-1c is not controlled by cholesterol, but rather responds to feeding cues and to insulin signaling (Browning and Horton, 2004). In Drosophila, palmitate and phosphatidylethanolamine may block processing of the fly SREBP-1 ortholog dSREBP (Dobrosotskaya et al., 2002; Seegmiller et al., 2002) or affect expression of dSREBP target genes in the heart (Lim et al., 2011). However, specific mechanisms linking metabolites from SREBP-1-dependent transcriptional pathways to its maturation as a transcription factor remain unclear.
Invertebrate systems such as Caenorhabditis elegans are particularly attractive for determining links between genetics, diet, and metabolism. The C. elegans intestine has both digestive and endocrine functions and may model aspects of both hepatic and adipose lipogenesis (Ashrafi et al., 2003). Importantly, the single ortholog of SREBP in C. elegans, SBP-1, appears highly conserved, regulating lipid storage similarly to mammalian SREBP-1 (Horton et al., 2002; McKay et al., 2003). For example, both mammalian SREBP-1 and SBP-1 activate stearoyl CoA desaturase genes (SCD and fat-6, fat-7, respectively), requiring the ARC105/MED15/MDT-15 subunit of the Mediator transcriptional co-activator (Yang et al., 2006). We also recently found that both C. elegans SBP-1 and mammalian SREBP can be downregulated by the NAD+-dependent sirtuin SIR-2.1/SIRT1 during fasting (Walker et al., 2010). Thus, the C. elegans system is a powerful tool to elucidate conserved gene regulatory mechanisms by SREBP orthologs.
Employing C. elegans and mammalian models, we have uncovered a conserved set of SBP-1/SREBP-1 target genes in the one-carbon cycle (1CC), a pathway involving folate-methionine metabolism and manufacture of the predominant methyl donor, S-adenosylmethionine (SAMe). Because SAMe is required for the methylation-dependent synthesis of phosphatidylcholine (PC), a key membrane component, regulation of the 1CC by SBP-1/SREBP-1 is consistent with its function in maintaining phospholipid homeostasis (Hagen et al., 2010). Importantly, we have found that SBP-1/SREBP-1-dependent gene expression and lipogenesis increases when cellular methylation capacity or phospholipid biosynthesis is limited. Decreased levels of SAMe or PC have been linked to multiple models of hepatosteatosis in rodents and humans, however mechanisms underlying the strongly elevated lipogenesis have been unclear (Mato, 2008; Vance et al., 2007; Vance and Vance, 2004; Zeisel, 2008). Our observations indicate that an important regulatory mechanism for SREBP-1 activation may operate when methylation capacity or PC biogenesis is diminished and suggests this pathway may contribute to lipid accumulation in fatty liver disease.
Results
SREBPs regulate 1CC genes in C. elegans and in mammalian cells
The SREBP family of transcription factors regulates genes involved in biosynthesis and trafficking of cholesterol and other lipids in mammals (Osborne and Espenshade, 2009). Employing the nematode C. elegans to elucidate conserved functions associated with SREBP regulation in metazoans, we have carried out genome-wide gene expression analysis on worms depleted of the single SREBP ortholog SBP-1. As expected, the DNA microarray studies showed that expression of many genes important for fatty acid, TAG and phospholipid production are dependent on SBP-1 (Figure 1A and S1A). Intriguingly, our analysis also found enrichment of genes predicted to function in the 1-carbon cycle (1CC) (Figure 1A–C). The 1CC coordinates folate and methionine metabolism with production of the methyl donor S-adenosylmethionine (SAMe) (Mato and Lu, 2005). SAMe is critical for most cellular methylation reactions, including methylation-dependent synthesis of a large number of cellular metabolites and phospholipids (Mato, 2008). Because of the central importance of the 1CC to metabolic homeostasis, we examined regulation of the 1CC by SBP-1/SREBP in more detail. Analysis of individual 1CC genes identified in DNA microarray studies, as well as closely related genes, by quantitative RT-PCR (qRT-PCR) after sbp-1 RNAi confirmed that a broad array of 1CC genes depend on SBP-1 for full expression (Figure 1C).
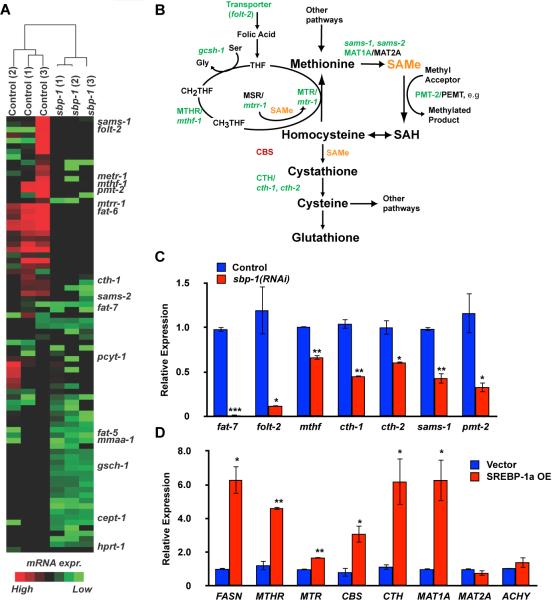
Figure 1. Co-regulation one-carbon cycle and fatty acid biosynthesis genes by SBP-1/SREBP-1.
A. Unbiased hierarchical clustering of genes more than two-fold downregulated in sbp-1(RNAi) nematodes and with metabolic regulation KEGG terms shows that one-carbon cycle (1CC) genes are decreased similarly to known SBP-1-responsive genes in lipid metabolism. B. Schematic diagram of the one-carbon cycle (1CC). SBP-1-dependent genes in C. elegans are shown in green; genes responsive to mammalian SREBP-1 are shown in red. C. Quantitative RT-PCR (qRT-PCR) from control or sbp-1(RNAi) nematodes reveals that multiple 1CC genes require SBP-1 for expression. D. qRT-PCR from 293T cells overexpressing SREBP-1a shows that expression of multiple 1CC genes increases with elevated SREBP-1 activity. Error bars show standard deviation. Statistical relevance (p value) shown by <0.05, *; <0.01, **; <0.005, ***.
Because 1CC genes had not been identified in searches for mammalian SREBP target genes, we also examined their regulation in human cells. We found that overexpression of SREBP-1a in human embryonic kidney 293T cells resulted in upregulation of multiple 1CC genes (Figure 1D). Several of these, such as CTH and MAT1A, are orthologs of genes identified in our C. elegans studies, whereas others, such as CBS, appeared specific to regulation in mammalian cells. We also found that expression of MAT1A specifically depended on SREBPs in human cells, whereas MAT2A did not (Figure S1B). This suggests that SREBP regulation of 1CC genes is conserved among metazoans and that metabolic flux through this pathway may be controlled by SREBP orthologs.
Increased SBP-1-dependent lipogenesis and gene expression after sams-1 depletion in C. elegans
To better understand the link between methyl donor homeostasis and regulation of these genes by SBP-1, we examined phenotypes of animals depleted for the SAMe synthase-encoding gene sams-1. The sams-1 gene was initially identified in an RNAi screen for extended C. elegans lifespan (Hansen, 1995). Surprisingly, we find that sams-1(RNAi) and sams-1(lof) nematodes also exhibit large refractile droplets within the intestine and body cavity that stained with Sudan Black (Figure 2A–C), suggesting lipid accumulation was increased. Accordingly, we found that TAGs in sams-1(RNAi) nematodes were significantly elevated when compared to controls (Table 1). Although C. elegans harbor 4 additional sams genes, RNAi of sams-1 resulted in an approximately 65% decrease in SAMe levels with similar decreases in S-adenosylhomocysteine (SAH), the product of SAMe-dependent methyltransferase reactions (Figure 2D). This suggests that sams-1 is required for the majority of SAMe production. The lipid accumulation seen after sams-1 RNAi led us to hypothesize that low methylation capacity may feedback activate SBP-1 and promote increased lipogenesis, as is the case with low cholesterol for mammalian SREBP-2 regulation.
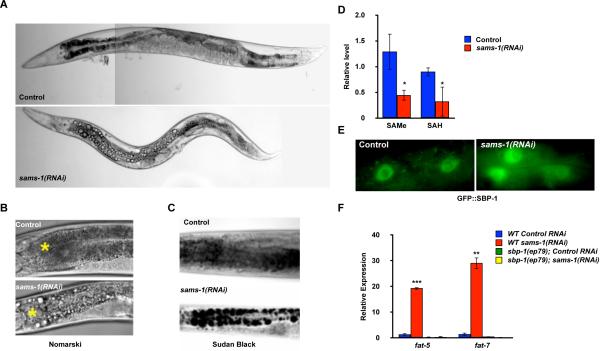
Figure 2. In C. elegans, SBP-1-dependent lipogenesis and gene expression are increased after sams-1(RNAi).
A. RNAi knockdown of sams-1 revealed large refractile droplets in the intestine and body cavity by Nomarski optics. B. Enlarged view of droplets by Nomarski optics. The first sets of intestinal cells are shown, the position of the pharynx is marked with a yellow star. C. Staining with Sudan Black shows that droplets in sams-1(RNAi) nematodes contain lipids. D. SAMe and SAH levels are significantly decreased after sams-1(RNAi). Error bars represent standard error between triplicate independent experiments. E. Nuclear accumulation of a GFP∷SBP-1 fusion protein is increased in sams-1(RNAi) intestinal cells. F. qRT-PCR comparing levels of fat-5 and fat-7 in wild-type or a hypomorphic sbp-1 allele (ep79) demonstrated that FA desaturase upregulation in sams-1(RNAi) animals depends on sbp-1. Statistical relevance (p value) shown by <0.05, *; <0.01, **, <0.005, ***.
Table 1.
Increased triacylglycerols in sams-1, pcyt-1 orcept-1(RNAi) nematodes
| %TAG | SEM | pvalue | |
|---|---|---|---|
| Control | 42.8 | 2.6 | |
| sams-1 (RNAi) | 64.5 | 4.1 | 0.0024 |
| pcyt-1 (RNAi) | 51.4 | 1.6 | 0.0401 |
| cept-1(RNAi) | 50.8 | 1.6 | 0.0801 |
Quantitative analysis demonstrates increased storage of neutral lipids after RNAi knockdown of genes necessary for PC production in nematodes when comparing percent triacylglycerol (TAG) to total lipids. SEM is standard error of the mean.
To determine if decreased methylation capacity could affect nuclear SBP-1 levels and elevated lipogenesis, we examined cellular localization of a GFP∷SBP-1 fusion protein and SBP-1-dependent transcription in sams-1(RNAi) nematodes. Indeed, GFP∷SBP-1 showed increased nuclear accumulation after sams-1 RNAi (Figure 2E), suggesting that levels of transcriptionally active SBP-1 are increased. Concomitantly, expression of multiple SBP-1-dependent genes, including the palmitoyl-CoA desaturase fat-5 and the stearoyl-CoA desaturases fat-6 and fat-7 were increased in both sams-1(RNAi) and sams-1(lof) animals (Figure 2F, S2A). Similar levels of sbp-1 mRNA were present in control or sams-1(lof) and sams-1(RNAi) nematodes, showing that regulation of SBP-1 in response to SAMe depletion is likely to be post-transcriptional (Figure S2A and data not shown). To determine if SBP-1 was necessary for increased lipogenic gene expression after sams-1 RNAi, we examined sams-1 knockdown phenotypes in nematodes expressing a hypomorphic sbp-1 allele (sbp-1(ep79)). Elevated expression of fat-5, fat-6 and fat-7, or the characteristic lipid droplets, were not observed when both sams-1 and sbp-1 function was limited (Figure 2F, S2B), suggesting that SBP-1 is essential for increased lipogenesis after sams-1 depletion.
Decreased phosphatidylcholine production in C. elegans is linked to elevated SBP-1-dependent gene expression and lipogenesis after sams-1 RNAi
We next examined whether depletion of sams-1 caused specific metabolic changes that might be linked to increases in SBP-1 activity. Intriguingly, methylation-dependent phosphatidylcholine (PC) biogenesis intermediates were altered in metabolomic analysis of sams-1-depleted nematodes (Figure 3B,C). PC is produced by two alternative pathways. It can be synthesized from choline (Kennedy pathway), or by methylation of phosphatidylethanolamine (PE) by PEMT in mammals, or of phosphoethanolamine by PMT-1 and PMT-2 orthologs in C. elegans and plants (Figure 3A) (Brendza et al., 2007; Palavalli et al., 2006; Vance and Vance, 2004). We found significantly lower levels of the product of the PMT-1 methyltransferase, phosphocholine (Figure 3B) and also observed that PC levels were dramatically lower after sams-1 RNAi, whereas PE was not significantly changed (Figure 3C). Thus, marked alterations in phospholipid metabolism occur along with increased lipogenesis in sams-1(RNAi) nematodes. Similar to our observation in sams-1-deficient nematodes, accumulation of large lipid droplets has been associated with low PC levels, or changes in PC/PE ratios in Drosophila and mammalian models (Guo et al., 2008; Jacobs et al., 2008; Li et al., 2006). We therefore wished to determine whether this increased lipid droplet formation in might be linked mechanistically to increased SBP-1/SREBP-1 activity.
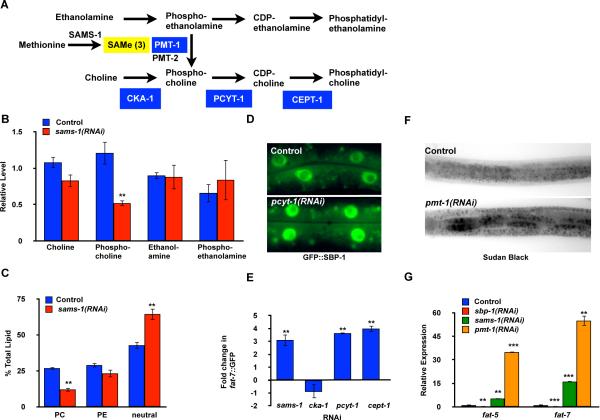
Figure 3. Increased lipogenesis and SBP-1-dependent gene expression after sams-1 RNAi in C. elegans is linked to limited phosphatidylcholine (PC) production.
A. Schematic diagram of links between methyl production and phospholipid biosynthesis in nematodes. Enzymes whose functions are disrupted by RNAi in subsequent panels are shown in blue boxes. B. Analysis of metabolites in sams-1(RNAi) nematodes shows that PC precursors downstream of methylation steps are decreased (phosphocholine), whereas metabolites upstream (choline, ethanolamine, phosphoethanolamine) are unchanged or slightly increased. Error bars represent standard error between quadruplicate independent experiments. C. Quantitative analysis show that PC levels are diminished after sams-1(RNAi). Error bars represent standard error between quadruplicate independent experiments. D. GFP∷SBP-1 accumulates in intestinal nuclei after pcyt-1(RNAi). E. Quantitative measurement of pfat-7∷GFP intensity in C. elegans populations by a COPAS biosorter shows that RNAi knockdown of PC biosynthesis genes downstream of methylation steps activate this SBP-1-dependent reporter at similar levels to sams-1 RNAi. Blocking methylation-dependent PC production by interference with PMT-1 produces similar phenotypes to sams-1(RNAi), such as increased lipid droplet formation (F) and overexpression of SBP-1 transcriptional targets, fat-5 and fat-7 (G). Error bars show standard deviation, statistical relevance (p value) shown by <0.05, *; <0.01, **, <0.005, ***.
Production of PC from either phosphoethanolamine or PE requires three sequential SAMe-dependent methylation steps, however, synthesis from choline is methylation-independent (Vance and Vance, 2004). To determine whether rescuing PC production would reverse SBP-1-dependent lipogenic events in sams-1(RNAi) nematodes, we provided excess dietary choline. Strikingly, dietary choline supplementation completely rescued accumulation of lipid droplets corrected overexpression of the SBP-1 transcriptional targets fat-5, fat-6 and fat-7 in sams-1(RNAi) worms (Figure S3A,B and data not shown). These results support our hypothesis that decreased levels of PC in sams-1(RNAi) worms may be directly linked to the increased SBP-1-dependent lipogenesis.
Defective PC production results in increased SBP-1-dependent gene expression and lipogenesis in C. elegans
While decreased methylation capacity prevents adequate PC synthesis, lowering levels of SAMe could also affect a broad array of methyltransferases. We next asked if PC biosynthesis was mechanistically associated with SBP-1-dependent lipogenesis and lipid droplet formation in C. elegans. Indeed, RNAi knockdown of pcyt-1 or cept-1, encoding the final enzymes in the PC biosynthesis pathway (Figure 3A), resulted in decreased PC levels, elevated nuclear localization of GFP∷SBP-1 and increased stored lipids, similar to what we observed in sams-1(RNAi) nematodes (Figure 3D, Table 1, Table S1, data not shown). Furthermore, in an independent, unbiased screen (V. Rottiers and A.M. Näär, unpublished observations), we found that RNAi of genes involved in SAMe production or genes encoding enzymes acting downstream of the methylation step in PC biogenesis resulted in strong upregulation of a GFP reporter fused to the promoter of the SBP-1 target gene fat-7 (pfat-7∷GFP) (Figure 3E). By contrast, RNAi of cka-1, encoding a choline kinase upstream of the methylation-dependent PC synthesis step, did not increase pfat-7∷GFP expression (Figure 3E). We also examined lipid accumulation and expression of SBP-1 target genes after knockdown of pmt-1, encoding phosphoethanolamine methyltransferase, a key consumer of SAMe during PC production in C. elegans. Reduction of pmt-1 expression causes a late larval arrest and defects in PC production that can be rescued by dietary choline supplementation (Brendza et al., 2007). We also found that pmt-1 knockdown increased lipid droplet formation and caused a marked elevation in expression of the SBP-1-responsive genes fat-5 and fat-7 (Figure 3F,G). Limitation of SAMe production could cause pleiotropic phenotypes by reducing activity of various distinct methyltransferases. However, the similar effects on SBP-1-dependent gene expression and lipogenesis after sams-1 or pmt-1 depletion suggests that reduced production of PC is directly affecting SBP-1 activity.
Activation of ER stress response in sams-1(RNAi) animals is not linked to elevated SBP-1-dependent lipogenic gene expression
Because the decrease in PC levels tightly correlates with increases in SBP-1 activity, we hypothesized that lowered PC in intracellular membranes could alter proteolytic processing and maturation of SREBP-1 orthologs. ER stress may be sufficient to initiate SREBP processing in mammals, independent of the cholesterol feedback cycle (Ferre and Foufelle, 2010). Therefore, we asked if sams-1 depletion initiated ER stress responses and whether this activation was sufficient to upregulate SBP-1-dependent gene expression. In C. elegans, the BiP/GRP78 ortholog hsp-4 is induced by ER stressors such as the glycosylation inhibitor tunicamycin and depends on the activity of the ER stress effector XBP-1 (Calfon et al., 2002). RNAi of sams-1 in a hsp-4∷GFP reporter line shows a strong xbp-1-dependent increase in GFP expression (Figure S3C), suggesting that ER stress responses are robustly activated. To determine if ER stress response was functionally important for the elevated SBP-1 activity in response to loss of sams-1, we knocked down the ER stress effectors atf-6 or ire-1 (Shen et al., 2005) in sams-1(lof) nematodes and found that SBP-1 target gene upregulation and lipid droplets accumulation were not altered (Figure S3D, data not shown). Next we asked if cellular responses to ER stress could activate SBP-1 in wild-type animals and found that SBP-1 target gene expression was not affected by doses of tunicamycin causing mild or moderate levels of lethality in control cultures (Figure S3E, data not shown). While our results point to a significant activation of ER stress responses when sams-1 is knocked down, ER stress does not appear to cause increases in SBP-1 gene expression and lipogenesis.
Increase in mammalian SREBP-1 nuclear localization and lipid droplet size after blocking PC biogenesis in human hepatoma cells
Our C. elegans results suggest that SBP-1 activity increases in sams-1(RNAi) nematodes as methylation capacity is diminished, and that mechanisms underlying this phenomenon are directly related to decreased PC levels. In order to determine if mammalian SREBP-1 might also be regulated by PC depletion, we used short interfering (si) RNA technology to knockdown genes necessary for PC production in human cells. As in invertebrates, PC production in mammals may proceed through methylation-dependent or -independent pathways, although in mammals PE is methylated to generate PC (Figure 4A) (Vance and Vance, 2004). We found that blocking PC production through siRNA-mediated depletion of enzymes in the methylation-dependent steps (MAT1A, PEMT), or enzymes in the methylation-independent CDP-choline pathway (CTα, CEPT) in HepG2 human hepatoma cells resulted in increased accumulation of large lipid droplets, akin to our observations in C. elegans (Figure 4B,D,F, S4E, data not shown). This data is in accord with the development of fatty liver in Mat1a- (Lu et al., 2001), Ctα–, and Pemt-deficient mice on a high-fat diet (Jacobs et al., 2008; Jacobs et al., 2010; Watkins et al., 2003), and with accumulation of lipid droplets in cell culture models which limit PC production (Guo et al., 2008; Jacobs et al., 2008; Testerink et al., 2009). Our results reveal that along with increases in lipid droplet size, HepG2 cells depleted of CTα, CEPT, MAT1A or PEMT showed SREBP-1 concentrated in the nucleus in immunofluorescence assays (Figure 4C,E, S4B,C), whereas controls exhibited stronger cytoplasmic staining where the inactive SREBP-1 precursor is stored. Furthermore, qRT-PCR of CTα knockdown cells showed increased expression of SREBP-1 transcriptional targets SCD1 and MAT1A, but no increases in the expression of SREBF1 gene encoding the SREBP-1 protein (Figure 4F), suggesting that elevation of SREBP-1 activity is likely post-transcriptional. As in our C. elegans studies, these experiments suggest that limitation of PC biosynthesis in mammalian cells promotes elevated levels of nuclear SREBP-1, resulting in increased SREBP-1-dependent gene expression, and in elevated lipogenesis and lipid droplet formation.
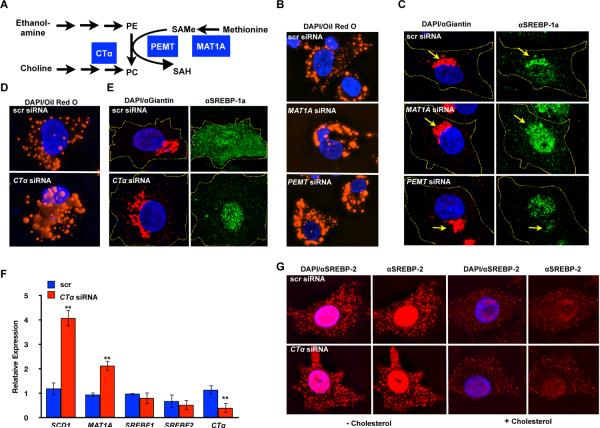
Figure 4. Increase in lipid droplet formation and SREBP-1a nuclear localization in human hepatoma cells in response to SAMe/PC depletion.
A. Schematic diagram of links between mammalian methyl production and phospholipid biosynthesis. Enzymes whose functions are disrupted by siRNA in subsequent panels are shown in blue boxes. siRNA knockdown of PEMT, MAT1A (B) or CTα (D) causes increased accumulation of large lipid droplets by Oil Red O staining in HepG2 cells in lipid-depleted serum. Immunostaining with an antibody against SREBP-1a shows increased nuclear accumulation after PEMT, MAT1A (C) or CTα (E) knockdown. Cells were co-stained with antibodies recognizing the Golgi marker Giantin. Yellow lines show outline of cells. F. Analysis of gene expression in CTα knockdown HepG2 cells by qRTPCR shows increases in SREBP-1-dependent genes such as SCD1 and MAT1A. G. Antibodies directed against SREBP-2 were used to stain HepG2 cells in lipid-depleted serum in the presence or absence of 10 μg/ml cholesterol in control or CTα siRNA-treated cells.
In contrast with potent effects of PC limitation on SREBP-1 nuclear accumulation, nuclear levels of SREBP-2, which regulates genes for cholesterol biosynthesis (Osborne and Espenshade, 2009), were already high in control cells and did not increase when PC synthesis was blocked by siRNA of CTα, CEPT, MAT1A or PEMT (Figure 4G, S4D, data not shown). Because strong nuclear accumulation of SREBP-2 is likely due to low levels of cholesterol in our culture conditions, which could mask additional effects from decreased PC, we also compared SREBP-1 and SREBP-2 nuclear accumulation in the presence of cholesterol. We found that SREBP-1 was preferentially nuclear in CTα and CEPT knockdown cells regardless of cholesterol levels (Figure 4E, S4B,C). However, nuclear levels of SREBP-2 were high in low cholesterol and then decreased when cholesterol was added, whether in control, CTα or CEPT knockdown cells (Figure 4G, S4D). This suggests that under these culture conditions, blocking PC synthesis has a greater effect on SREBP-1 nuclear accumulation, gene expression and lipogenesis, and that SREBP-2 responds preferentially to cholesterol levels.
Decreased PC biosynthesis in murine liver promotes SREBP-1 proteolytic maturation and target gene expression
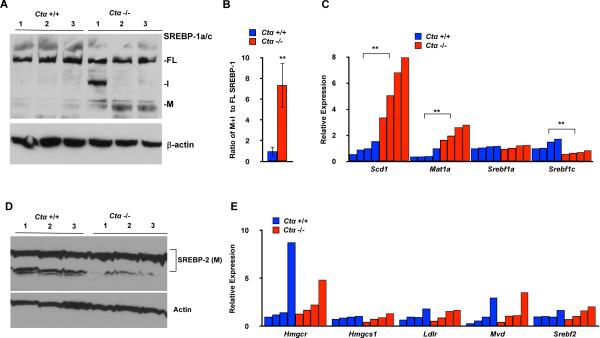
To determine whether inhibition of PC biosynthesis in a mammalian in vivo model also altered SREBP-1 processing and target gene expression, we examined livers from mice deficient in PC biosynthesis due to a liver-specific deletion in Ctα, encoding the rate-limiting enzyme in PC biosynthesis from choline (Vance and Vance, 2004) and the ortholog to C. elegans pcyt-1. Livers from Ctα−/− mice on a high-fat diet exhibit decreased PC and increased TAG compared to controls, but have normal cholesterol levels (Jacobs et al., 2004). To determine whether SREBP function was altered, we examined SREBP-1 and SREBP-2 in extracts from Ctα−/− livers by immunoblotting and found higher levels of mature, processed SREBP-1 (Figure 5A,B), suggesting that lowering PC is sufficient to affect mammalian SREBP-1a/c proteolytic maturation. In accord with our findings in C. elegans, expression of the Δ9 fatty acid desaturase and key lipogenic SREBP-1 target gene Scd1 (orthologous to fat-6 and fat-7) was increased in livers from Ctα−/− mice (Figure 5C). Previous studies of Ctα−/− mice did not uncover changes in Scd1 expression, probably because of the use of fasted mice (Jacobs et al., 2004), a condition known to strongly inhibit SREBP target gene expression (Horton et al., 1998; Walker et al., 2010). As previously reported (Jacobs et al., 2005), expression of the sams-1 ortholog Mat1a is also upregulated in these livers. However, we found that levels of mature SREBP-2 and expression of SREBP-2 target genes were not increased in Ctα−/− livers, and that there were no increases in the expression of the Srebf1a/c and Srebf2 genes themselves (Figure 5C–E). This in vivo study suggests that, as in our cell culture studies, SREBP-2 activity may not be significantly affected in response to altered PC levels whereas processed, mature SREBP-1 is increased, along with SREBP-1-dependent transcription and concomitant lipogenesis.
Figure 5. Increase in SREBP-1 processing and target gene expression in Ctα knockout mouse livers.
A. Immunoblotting of extracts from mice with a liver-specific knockout of Ctα (Jacobs et al., 2004) limiting the CDP-choline pathway shows increased SREBP-1a/c processing (Jacobs et al., 2004). Fl is full-length SREBP-1a/c precursor; I, intermediate form; M, mature, transcriptionally active form. B. Analysis of the immunoblot by densitometry shows increases in proteolytic products of SREBP-1 in livers from Ctα knockout mice. C. Analysis of gene expression by qRT-PCR from Ctα knockout mice show increased expression of Scd1, an SREBP-1 target, as well as Mat1a, ortholog of sams-1. Bars represent individual mice, error bars show standard deviation; statistical relevance (p value<0.01) shown by (**). Analysis of gene expression by qRT-PCR from the liver-specific Ctα knockout mice (Jacobs et al., 2004) show no statistically significant changes in levels of mature, processed SREBP-2 (D) or target gene expression (E). Bars represent individual mice, error bars show standard deviation; statistical relevance (p value<0.01) shown by (**).
Inhibition of PC biosynthesis affects localization of SREBP-activating proteases and parallels inactivation of ARF-GTPases regulating COPI Golgi-ER retrograde transport
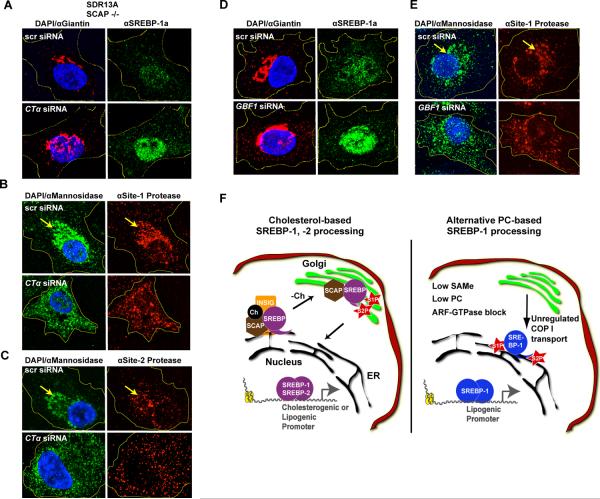
Our results indicate that lipogenic SREBPs (SBP-1/SREBP-1) exhibit increased activity when PC synthesis is limited. This regulation occurs in the presence of cholesterol, suggesting that it represents an independent mechanism to regulate SREBP-1. Therefore, we examined the dependence of this regulatory circuit on SCAP, the cholesterol-binding chaperone essential for the classical feedback pathway controlling SREBP-2 processing and maturation (Brown and Goldstein, 1997). RNAi-mediated depletion of Ctα in SRD13A cells, in which SCAP is mutated and inactivated (Hua et al., 1996) resulted in robust nuclear accumulation of SREBP-1 (Figure 6A, S5A), suggesting that the mechanism activating SREBP-1 in response to PC depletion is indeed distinct from the classical pathway involving SCAP-dependent transfer of SREBPs from the ER to the Golgi when cholesterol is limiting.
Figure 6. Relocalization of SREBP-activating proteases in mammalian cells when PC production is blocked.
A. SREBP-1 accumulates in the nucleus after CTα siRNA treatment in SRD13A cells, which lack a functional SCAP. Yellow lines show cell outlines. Co-immunostaining of HepG2 cells with the Golgi marker α-mannosidase and antibodies to S1P (B) or S2P (C) shows a strong shift away from an organized Golgi structure and disorganization of SREBP-activating protease staining after CTα knockdown. Yellow arrow marks Golgi body in control cells. Interference with ARF-GTPase signaling by GBF1 siRNA treatment of HepG2 cells increases SREBP-1 nuclear accumulation (D) and disrupts Golgi-specific partitioning of S1P (E.) F. Schematic model for relocalization of SREBP-activating proteases, resulting in activation of SREBP-1 and transit to the nucleus upon decreases in SAMe or PC levels or blocks in ARF-GTPase cycles.
Because PC is a major membrane component, and SREBP-1 activation occurs within the confines of the ER and Golgi membranes, we hypothesized that changes in these membranes could lead to increased SREBP-1 activity. The balance of PC/PE ratio affects membrane fluidity and curvature, and depletion of PC can alter protein trafficking and lipid accumulation (Hagen et al., 2010; Testerink et al., 2009). The antibiotic brefeldin A (BFA) inactivates the ARF-GTPase cycle critical for regulation of COPI, or retrograde protein transport from the Golgi apparatus to the ER (Donaldson and Lippincott-Schwartz, 2000). BFA has also been reported to disrupt localization of the Golgi-resident SREBP-activating Site-1 and Site-2 proteases (S1P, S2P), causing ectopic proteolytic cleavage of SREBP-2 in the ER (DeBose-Boyd et al., 1999). We asked whether decreased PC levels could similarly to affect S1P and S2P localization to induce SREBP-1a processing and nuclear accumulation. Interestingly, when CTα or CEPT were depleted in HepG2 cells, S1P and S2P lost Golgi-specific localization and appeared instead in a diffuse cytoplasmic pattern consistent with KDEL ER-marker staining (Figures 6B,C, S5B–D, data not shown). Indeed, localization of two additional Golgi-specific proteins, Giantin and α–mannosidase was also disrupted (Figures 4C,E, 6B,C, S5B,C), suggesting the Golgi may be broadly affected by changes in PC levels.
Recent studies have found that the ARF-activating protein/GEF GBF1 affects S2P localization in the unfolded protein response (Citterio et al., 2008). To gain further insights into ER-Golgi trafficking of SREBP-activating proteases, we used siRNA to deplete GBF1 and found increases in SREBP-1 nuclear accumulation and redistribution of S1 and S2 proteases to an ER-like pattern, similar to the effects of PC depletion (Figures 6D,E, and S5E,F). ARF signaling may also be inhibited by increases in membrane curvature (Bigay et al., 2003), which is predicted to occur when membrane PC levels drop (Lev, 2006). Thus, SREBP-1 nuclear accumulation could occur if PC levels drop and membrane curvature inhibits ARF-GTPase signaling, allowing relocalization of S1P and S2P to the ER and proteolytic activation of SREBP-1 (Figure 6F). Limitations in choline or methionine metabolism affecting PC synthesis could activate SREBP-1 through this mechanism, promoting the lipid accumulation in fatty liver disease.
Discussion
Studies by a number of groups have revealed a complex regulatory relationship between SREBPs and phospholipids. SREBP-1a has been implicated as a regulator phospholipid biogenesis genes in mammals (Hagen et al., 2010), and we have found that multiple PC biosynthesis genes are dependent on SBP-1 in C. elegans (Figure 1A,1B, Figure S1A). In a Drosophila cell culture model, Dobrosotskaya et al. showed that palmitate and precursors to PE are potent inhibitors of dSREBP processing (Dobrosotskaya et al., 2002). However, genes or metabolites leading directly to PC biosynthesis were not examined. The differences in our studies may reflect a distinct SREBP-regulatory circuit in Drosophila, or perhaps that specific combinations of membrane phospholipids may differently affect SREBP processing. Nevertheless, a recent study showed that low PE levels in the Drosophila heart affect cardiac lipid accumulation and expression of dSREBP target genes (Lim et al., 2011), strengthening links between phospholipids and regulation of SREBP-1 orthologs.
Phosphatidylcholine, 1CC metabolism and links to hepatic lipogenesis
Lipid accumulation in the liver (hepatosteatosis) is an early step in nonalcoholic fatty liver disease (NAFLD) (Browning and Horton, 2004) and appears to predispose to subsequent, more severe pathological changes (Browning and Horton, 2004; Li et al., 2006). In metabolic syndrome, hepatic lipid accumulation may be driven by SREBP-1c in response to high insulin levels (Browning and Horton, 2004; Ferre and Foufelle, 2010), but may also occurs when dietary or genetic lesions alter 1C metabolism, SAMe production or decrease PC levels (Larter and Yeh, 2008; Mato, 2008). PEMT polymorphisms, and a reduced PC/PE ratio, have also been associated with NAFLD in humans (Li et al., 2006; Song et al., 2005). When PC biosynthesis is limited in mice, hepatosteatosis is not concurrent with insulin resistance (Ctα−/−) (Jacobs et al., 2004) and Pemt−/− mice in fact exhibit increased insulin sensitivity (Jacobs et al., 2010), suggesting that the excess lipid production when PC is limiting is not linked to SREBP-1c upregulation as a function of insulin resistance. Unlike insulin- or oxysterol-dependent effects on SREBP-1c function (Osborne and Espenshade, 2009), the PC-linked increases are not accompanied by upregulation of the SREBF1 transcript. This indicates that lower levels of PC resulting from diminished methylation capacity act by promoting SREBP-1 processing and may contribute to steatosis in early stages of fatty liver diseases.. Indeed, increased hepatic lipids are associated with diverse conditions such as intravenous (parenteral) feeding (Buchman et al., 1995), cystic fibrosis-associated liver disease (Colombo et al., 1998), or excess alcohol consumption (Kharbanda, 2009), in which dietary availability, absorption or metabolism of choline and/or other 1CC metabolites could be affected. Choline supplementation can reverse hepatic lipid accumulation associated with total parenteral feeding (Buchman et al., 1995) and improve abnormal 1CC metabolite plasma levels in cystic fibrosis patients (Innis et al., 2007). Our results provide a mechanistic rationale for further exploring choline and SAMe supplementation as treatments for fatty liver diseases in humans.
Activation of SBP-1/SREBP-1 in response to low PC: an alternative to cholesterol-based regulatory mechanisms
Activation of SREBPs involve the ER, where full-length SREBP precursors are stored, and the Golgi, housing the SREBP-activating proteases, S1P and S2P. Our data suggest that changes in PC levels may alter membrane function leading to SREBP-1 activation. ER stress has previously been linked to SREBP activation (Hotamisligil, 2010), and recently to PC/PE ratios in leptin-deficient, ob/ob mice (Fu et al., 2011). The ER-stress inducer ATF-6 is regulated similarly to SREBPs (Ye et al., 2000), thus ATF-6 could also be activated if S1P and S2P lose Golgi-specific localization. However, our data suggest that ER stress responses may occur in parallel to SBP-1 activation, because stimulating the ER stress response was not sufficient to increase SBP-1 activity (Figure S3E and data not shown). Therefore, we conclude that SBP-1 activation mechanisms in response to low PC are likely independent of ER stress responses.
Elegant studies have shown that Golgi and ER separation is crucial for SREBP regulation (Bartz et al., 2008; DeBose-Boyd et al., 1999). In these studies, cells were treated with brefeldin A (BFA), which disrupts Golgi function by inhibiting ARF-GTPase cycles essential for protein sorting in COPI-dependent Golgi-to-ER retrograde transport (Mansour et al., 1999; Peyroche et al., 1999). When cells are treated with BFA, S1 and S2 proteases collect in the ER (Bartz et al., 2008; DeBose-Boyd et al., 1999), activating SREBP-2. We have found that availability of methyl donors or PC also alters localization of S1P and S2P (Figure 6B,C, S5B,C) and increases in mature, nuclear SREBP-1 (Figure 4C,E, S4B,C). Intriguingly, lowering membrane PC content and BFA treatment are both predicted to inhibit ARF-GTPase cycles, although by different mechanisms. BFA binds ARF-GDP, preventing formation of active GTP-bound ARF (Donaldson and Lippincott-Schwartz, 2000). Membranes with low PC/PE ratios exhibit increased curvature (Lev, 2006), activating the ARF-GTPase repressor ARF-GAP (Bigay et al., 2003). In support of this model, S2P moved from Golgi to the ER when an ARF-activating factor, GBF1, was depleted (Citterio et al., 2008). Importantly, similar to depletion of PC synthesis enzymes (Figure 6D), we have found that depletion of GBF1 also causes SREBP-1 nuclear accumulation, showing that alterations in ARF-GTPase cycles are sufficient to activate SREBP-1. Taken together, this data suggests a simple mechanism for increasing SREBP-1 processing when membrane PC is limiting; increased curvature of the membrane may affect ARF signaling, deregulate COPI transport and shift the distribution of S1P or S2P towards the ER, where they cleave and activate SREBP-1. Intriguingly, we found that SREBP-2 was not affected by this PC-based activation mechanism (Figures 4G, S4D), although it is activated by BFA (DeBose-Boyd et al., 1999) suggesting the possibility that SREBP-1 and SREBP-2 precursors may respond differently to these cues.
To conclude, we propose a model where SBP-1/SREBP-1 is part of a conserved feedback loop responding to PC levels, to regulate expression of 1CC biogenesis genes and ensure adequate SAMe levels for PC production. Activation of other SBP-1/SREBP-1 targets in fatty acid biosynthesis pathways may also occur concomitantly, driving TAG production when PC levels are chronically low. Thus, abrogated PC feedback could be a major contributor to SREBP-1-dependent lipogenesis when methyl group metabolism is compromised by diet or genetics, and predispose to hepatic lipid accumulation and fatty liver disease.
Experimental Procedures
The Supplemental Information includes Extended Experimental Procedures. C. elegans strains, RNAi protocols, dietary rescue, and COPAS biosorting procedures are presented. Also included are cell culture conditions and antibodies.
C. elegans: whole genome analysis
Synchronized larvae from control and sbp-1 RNAi cultures were harvested and RNA was isolated. cDNA was produced and hybridized to Affymetrix C. elegans arrays. Responsive genes were defined as having a greater than two-fold change.
C. elegans: metabolite analysis
Replicates of synchronized L4/young adult C. elegans cultures from control or sams-1 RNAi treatment were harvested, frozen in liquid N2 and analyzed by Metabolon (Research Triangle Park, NC). TAG, PC and PE were measured as in (Watts and Browse, 2000), see Supplemental Information for more detail.
Cell culture and siRNA
For analysis of gene expression in HEK293T cells (cultured as in Walker, 2010), SREBP-1a was transfected by Lipofectamine 2000 (Life Technologies), and cells were harvested after 48 hours. For gene knockdown in HepG2 cells, siRNA Smartpools (Thermo-Fisher Scientific) were transfected by Amaxa Nucleofector (Lonza) and cells were placed in 1% Lipid Depleted Serum, then harvested after 24 hours.
Immunohistochemistry or staining with lipid soluble dyes
Antibody staining of SREBPs in cultured cells and C. elegans Sudan Black staining were performed as in (Walker et al., 2010). S1P and S2P antibody staining were performed as in (Bartz et al., 2008). For Oil Red O staining, cells were transfected with siRNA oligonucleotides by Amaxa Nucleofector and placed in 1% lipid depleted serum. After 24 hours, cells were fixed, stained with Oil Red O for 10 minutes and visualized. Microscopy for immunostaining and Oil Red O staining was performed on an Olympus ix81. Montages of images deconvolved by Nearest Neighbor algorithms were further processed by Adobe Photoshop.
Animal handling, liver extract preparation
Male liver-specific Ctα knockout and control mice (12 weeks old) were given free access to the high-fat diet (Bioserve, #F3282) for 3 days. Liver samples were collected from fed animals and either processed for immunoblotting with antibodies to SREBP or into Tri-reagent (Sigma) for RNA isolation. All procedures were approved by the University of Alberta's Institutional Animal Care Committee in accordance with guidelines of the Canadian Council on Animal Care.
mRNA isolation and qPCR
mRNA extracted by Tri-reagent (Sigma). cDNA was made from total RNA by Transcriptor cDNA synthesis kit (Roche). qPCR was run on a Roche 480 light cycler. Duplicate samples from at least three biologically independent experiments were analyzed. Statistical significance was determined by calculating standard deviation and Students T Test. For mRNA isolation from mouse liver, after homogenization in Tri-Reagent RNA was further purified by RNAeasy columns (Qiagen) before proceeding to cDNA synthesis and qPCR as above.
Supplementary Material
Acknowledgments
We thank T. Keith Blackwell for advice on examining ER stress in C. elegans as well as members of the Näär lab, Drs. Nick Dyson, Johnathan Whetstine, Andrew Gladden and Tom Rapoport for helpful discussions. We express our gratitude to Drs. Stefan Taubert and Raul Mostoslavsky for critical reading of the manuscript. We thank Dr. Michael Brown for the SRD13A, SCAP-deficient cells and Dr. Joachim Seemann for advice on S1P staining. We thank the Caenorhabditis Genetics Center for strains. Funding for A.K.W. was provided by the Claflin Distinguished Scholar Award and R01DK084352. Funding for A.M.N. was provided by The Paul F. Glenn Laboratories for the Biological Mechanisms of Aging at Harvard Medical School and the following grants from NIH: R01DK078332 and R01GM071449. Funding for J.L.W. was provided by NIH grant R01DK074114. Funding for Research in D.V.'s lab was from the Canadian Institutes of Health Research (MOP5182).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information Supplemental information includes 5 figures, 1 table, associated legends and Extended Experimental Procedures.
Bibliography
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Bartz R, Sun LP, Bisel B, Wei JH, Seemann J. Spatial separation of Golgi and ER during mitosis protects SREBP from unregulated activation. EMBO J. 2008;27:948–955. doi: 10.1038/emboj.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- Brendza KM, Haakenson W, Cahoon RE, Hicks LM, Palavalli LH, Chiapelli BJ, McLaird M, McCarter JP, Williams DJ, Hresko MC, et al. Phosphoethanolamine N-methyltransferase (PMT-1) catalyses the first reaction of a new pathway for phosphocholine biosynthesis in Caenorhabditis elegans. Biochemical Journal. 2007;404:439–448. doi: 10.1042/BJ20061815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AL, Dubin MD, Moukarzel AA, Jenden DJ, Roch M, Rice KM, Gornbein J, Ament ME. Choline deficiency: a cause of hepatic steatosis during parenteral nutrition that can be reversed with intravenous choline supplementation. Hepatology. 1995;22:1399–1403. [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Citterio C, Vichi A, Pacheco-Rodriguez G, Aponte AM, Moss J, Vaughan M. Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1. Proc Natl Acad Sci U S A. 2008;105:2877–2882. doi: 10.1073/pnas.0712224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo C, Battezzati PM, Strazzabosco M, Podda M. Liver and biliary problems in cystic fibrosis. Semin Liver Dis. 1998;18:227–235. doi: 10.1055/s-2007-1007159. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Lippincott-Schwartz J. Sorting and signaling at the Golgi complex. Cell. 2000;101:693–696. doi: 10.1016/s0092-8674(00)80881-8. [DOI] [PubMed] [Google Scholar]
- Ferre P, Foufelle F. Hepatic steatosis: a role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes Metab. 2010;12(Suppl 2):83–92. doi: 10.1111/j.1463-1326.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen RM, Rodriguez-Cuenca S, Vidal-Puig A. An allostatic control of membrane lipid composition by SREBP1. FEBS Lett. 2010;584:2689–2698. doi: 10.1016/j.febslet.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New Genes tied to Endocrine, Metabolic and Dietary Regulation of Lifespan from a Caenorhabditis elegans RNAi Screen. PLoS Genetics. 1995;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clini Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Innis SM, Davidson AG, Melynk S, James SJ. Choline-related supplements improve abnormal plasma methionine-homocysteine metabolites and glutathione status in children with cystic fibrosis. Am J Clin Nutr. 2007;85:702–708. doi: 10.1093/ajcn/85.3.702. [DOI] [PubMed] [Google Scholar]
- Jacobs RL, Devlin C, Tabas I, Vance DE. Targeted deletion of hepatic CTP:phosphocholine cytidylyltransferase alpha in mice decreases plasma high density and very low density lipoproteins. J Biol Chem. 2004;279:47402–47410. doi: 10.1074/jbc.M404027200. [DOI] [PubMed] [Google Scholar]
- Jacobs RL, Lingrell S, Zhao Y, Francis GA, Vance DE. Hepatic CTP:phosphocholine cytidylyltransferase-alpha is a critical predictor of plasma high density lipoprotein and very low density lipoprotein. J Biol Chem. 2008;283:2147–2155. doi: 10.1074/jbc.M706628200. [DOI] [PubMed] [Google Scholar]
- Jacobs RL, Stead LM, Devlin C, Tabas I, Brosnan ME, Brosnan JT, Vance DE. Physiological regulation of phospholipid methylation alters plasma homocysteine in mice. J Biol Chem. 2005;280:28299–28305. doi: 10.1074/jbc.M501971200. [DOI] [PubMed] [Google Scholar]
- Jacobs RL, Zhao Y, Koonen DP, Sletten T, Su B, Lingrell S, Cao G, Peake DA, Kuo MS, Proctor SD, et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J Biol Chem. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda KK. Alcoholic liver disease and methionine metabolism. Semin Liver Dis. 2009;29:155–165. doi: 10.1055/s-0029-1214371. [DOI] [PubMed] [Google Scholar]
- Larter CZ, Yeh MM. Animal models of NASH: getting both pathology and metabolic context right. J Gastroenterol Hepatol. 2008;23:1635–1648. doi: 10.1111/j.1440-1746.2008.05543.x. [DOI] [PubMed] [Google Scholar]
- Lev S. Lipid homoeostasis and Golgi secretory function. Biochem Soc Trans. 2006;34:363–366. doi: 10.1042/BST0340363. [DOI] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metabolism. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lim HY, Wang W, Wessells RJ, Ocorr K, Bodmer R. Phospholipid homeostasis regulates lipid metabolism and cardiac function through SREBP signaling in Drosophila. Genes Dev. 2011;25:189–200. doi: 10.1101/gad.1992411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SJ, Skaug J, Zhao XH, Giordano J, Scherer SW, Melancon P. p200 ARF-GEP1: a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci U S A. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato JM, Martinez-Chantar ML, Lu SC. Methionine Metabolism and Liver Disease. Ann Rev of Nutr. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM. C elegans: a model for exploring the genetics of fat storage. Developmental Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palavalli LH, Brendza KM, Haakenson W, Cahoon RE, McLaird M, Hicks LM, McCarter JP, Williams DJ, Hresko MC, Jez JM. Defining the role of phosphomethylethanolamine N-methyltransferase from Caenorhabditis elegans in phosphocholine biosynthesis by biochemical and kinetic analysis. Biochemistry. 2006;45:6056–6065. doi: 10.1021/bi060199d. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Developmental Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Song J, da Costa KA, Fischer LM, Kohlmeier M, Kwock L, Wang S, Zeisel SH. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD) FASEB J. 2005;19:1266–1271. doi: 10.1096/fj.04-3580com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink N, van der Sanden MH, Houweling M, Helms JB, Vaandrager AB. Depletion of phosphatidylcholine affects endoplasmic reticulum morphology and protein traffic at the Golgi complex. J Lipid Res. 2009;50:2182–2192. doi: 10.1194/jlr.M800660-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance DE, Li Z, Jacobs RL. Hepatic phosphatidylethanolamine N-methyltransferase, unexpected roles in animal biochemistry and physiology. J Biol Chem. 2007;282:33237–33241. doi: 10.1074/jbc.R700028200. [DOI] [PubMed] [Google Scholar]
- Vance JE, Vance DE. Phospholipid biosynthesis in mammalian cells. Biochemistry & Cell Biology. 2004;82:113–128. doi: 10.1139/o03-073. [DOI] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SM, Zhu X, Zeisel SH. Phosphatidylethanolamine-N-methyltransferase activity and dietary choline regulate liver-plasma lipid flux and essential fatty acid metabolism in mice. J Nutr. 2003;133:3386–3391. doi: 10.1093/jn/133.11.3386. [DOI] [PubMed] [Google Scholar]
- Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochemical & Biophysical Research Communications. 2000;272:263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Molecular Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Zeisel SH. Genetic polymorphisms in methyl-group metabolism and epigenetics: lessons from humans and mouse models. Brain Res. 2008;1237:5–11. doi: 10.1016/j.brainres.2008.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.