Abstract
Agrobacterium species are capable of interkingdom gene transfer between bacteria and plants. The genome of Agrobacterium tumefaciens consists of a circular and a linear chromosome, the At-plasmid and the Ti-plasmid, which harbors bacterial virulence genes required for tumor formation in plants. Little is known about promoter sequences and the small RNA (sRNA) repertoire of this and other α-proteobacteria. We used a differential RNA sequencing (dRNA-seq) approach to map transcriptional start sites of 388 annotated genes and operons. In addition, a total number of 228 sRNAs was revealed from all four Agrobacterium replicons. Twenty-two of these were confirmed by independent RNA gel blot analysis and several sRNAs were differentially expressed in response to growth media, growth phase, temperature or pH. One sRNA from the Ti-plasmid was massively induced under virulence conditions. The presence of 76 cis-antisense sRNAs, two of them on the reverse strand of virulence genes, suggests considerable antisense transcription in Agrobacterium. The information gained from this study provides a valuable reservoir for an in-depth understanding of sRNA-mediated regulation of the complex physiology and infection process of Agrobacterium.
Keywords: RNA-seq, deep sequencing, plant-microbe interaction, regulatory RNA, small RNA
Agrobacterium tumefaciens, is a broad-host range plant pathogen, which induces the so-called crown gall disease. Its genome comprises four different replicons: a circular chromosome, a linear chromosome, the At-plasmid and the tumor inducing (Ti)-plasmid.1 The latter includes a segment of DNA (T-DNA) which is excised, transported via a type IV secretion system, and integrated into the plant chromosome. Expression of the T-DNA-encoded genes results in tumor formation by the synthesis of plant hormones and in production of untypical amino acids, either opines or nopalines, which are used by Agrobacterium as nutrients.2
To induce virulence (vir)-gene expression in the bacterium, the VirAG two-component system is needed. It is stimulated by diverse plant signals, including acetosyringone.3 The membrane-bound sensor kinase VirA phosphorylates the response regulator VirG, which activates vir-gene transcription by binding to specific 12-bp DNA sequences, called vir-boxes.4-7 Expression of annotated genes after stimulation with acetosyringone has been analyzed in considerable detail.8-10 In contrast, little is known about small regulatory RNAs (sRNAs) in A. tumefaciens in general and under virulence conditions in particular. We attempted to close this gap in the present study.
The importance of sRNAs in regulation of bacterial gene expression is now widely appreciated.11,12 Most sRNAs, usually ranging from 50 to 500 nucleotides (nt) in length, act through base pairing with target mRNAs and modulate their translation and/or stability. They are often encoded in intergenic regions (IGRs) between two annotated genes and regulate mRNAs encoded elsewhere on the chromosome. These “trans-encoded sRNAs” usually show only limited complementarity with their targets.12 In contrast, “cis-encoded antisense sRNAs” are partially or entirely expressed from the reverse strand of a protein-encoding DNA and thus share extended regions of full complementarity with their target.13 Several sRNAs that act as key players in bacterial virulence have been reported.14,15 sRNAs are also known to control a variety of cellular processes, e.g., transcription, oligopeptide transport, plasmid replication or cell devision, just to name a few.16-19 Moreover, there is growing evidence that sRNAs regulate multiple, functionally related target genes and thereby control large posttranscriptional regulons.20
Both, trans- and cis-encoded sRNAs have been identified in a great variety of organisms by a number of different strategies. Biocomputational predictions revealed a number of bacterial sRNAs.21 Alternatively, numerous sRNAs can be identified by experimental approaches, like cDNA cloning of small-sized RNA species or by tiling microarrays.22 Most recently, high-throughput sequencing of cDNA (RNA-seq) has revolutionized transcriptome analysis and led to the discovery of a vast number of sRNAs in many organisms, including several microbial pathogens.23-29 Another benefit of RNA-seq approaches is its potential to identify 5′ transcriptional start sites (TSS) with high resolution in an annotation-independent manner.30,31
Although genome-wide profiling for sRNAs has become very popular, our knowledge on the sRNA inventory of α-proteobacteria is still restricted. Tiling array-based transcriptome studies have been reported for Caulobacter crescentus and Rhizobium etli.32,33 Deep sequencing uncovered several photooxidative stress induced sRNAs in Rhodobacter sphaeroides.34 A recent 454-sequencing study with Sinorhizobium meliloti revealed that about 3% of all genes encode trans-encoded sRNAs and about 2% cis-antisense transcripts in this plant symbiont.28 To the best of our knowledge, the sRNA inventory of a plant pathogen has not been reported yet. The only experimentally characterized A. tumefaciens sRNAs are the Ti-plasmid encoded RepE RNA, which regulates Ti-plasmid replication, and two recently described, homologous trans-acting sRNAs, AbcR1 and AbcR2.18,35 AbcR1 plays an important role in controlling the expression of an ABC transporter involved in γ-aminobutyric acid (GABA) and proline uptake.
Both latter sRNAs were derived from a biocomputational prediction which was restricted to the circular chromosome of A. tumefaciens. To detect sRNAs experimentally on a genome-wide scale, we used a previously established differential RNA-sequencing (dRNA-seq) approach, which allows for a discrimination of primary transcripts and processed RNAs.31 The successful identification of 228 sRNA candidates on the four replicons underlines the power of this method and the potential of sRNA-mediated gene regulation in A. tumefaciens.
Results
Sequencing of cDNA libraries from normally grown and virulence-induced Agrobacterium cells
To analyze the RNA population of A. tumefaciens, cDNA libraries of total RNA were analyzed by dRNA-seq.31 To examine transcriptional differences under virulence and non-virulence conditions, we grew cultures in minimal medium (pH 5.5) in the absence (-Vir) or presence (+Vir) of the vir-gene inducer acetosyringone. After RNA isolation, two different cDNA libraries were constructed for each of the two conditions. One was generated from the original, untreated total RNA, and the other following treatment with terminator 5′-phosphate-dependent exonuclease (TEX) that degrades processed RNAs with a 5′P but not primary transcripts, which carry a 5′PPP and thus leads to a relative enrichment of primary transcripts. The resulting four libraries [-Vir, -Vir (+TEX) and +Vir, +Vir (+TEX)] were then subjected to 454-pyrosequencing.
This strategy yielded 422,204 cDNA sequences. 348,998 cDNA reads being longer than 18 nt were mapped to the four A. tumefaciens replicons (Table 1). Correlating with replicon sizes, more cDNAs were mapped to the circular and linear chromosomes than to the plasmids. In the RNA preparations from cells harvested under virulence conditions (+Vir libraries), the number of cDNA reads from the Ti-plasmid significantly increased, suggesting successful virulence-gene induction.
Table 1. cDNA transcripts on all four A. tumefaciens replicons.
| Library | Total BLAST hits | Circular chrom. | Linear chrom. | At-plasmid | Ti-plasmid |
|---|---|---|---|---|---|
| -Vir |
46733 |
21436 |
22835 |
1687 |
775 |
| -Vir (+TEX) |
117641 |
54550 |
57112 |
4426 |
1553 |
| +Vir |
50458 |
19314 |
26101 |
1499 |
3544 |
| +Vir (+TEX) | 93251 | 41194 | 44935 | 2674 | 4448 |
All sequenced cDNAs longer than 18 nucleotoides (nt) were blasted. Total BLAST hits are listed and separated on the A. tumefaciens replicons.
Annotation of transcriptional start sites
The terminator exonuclease treatment enriches for primary transcripts characterized by a 5′ tri-phosphate end, leading to a characteristic cDNA enrichment pattern (Fig. 1AB).31 TSS were retrieved from regions upstream of annotated genes. At least five reads starting at exactly the same nucleotide position were required to define a TSS (see precise mapping criteria in the material and methods section). Overall, 356 potential TSS were identified upstream of annotated ORFs (Table S1), including the previously published TSS for flaA, flaB, and ccrM.36,37 Exemplarily, screenshots of the known TSS of flaA, coding for a flagella associated protein (Fig. 1A) and the newly identified TSS of ftsH, encoding a membrane-anchored protease (Fig. 1B) are demonstrated in Figure 1.38 32 additional TSS belong to tRNA transcripts and 5S rRNA (Table S2). Start sites that were mapped for tRNA transcripts slightly differed from the corresponding NCBI annotations, most probably because mature tRNAs rather than the authentic unprocessed pre-tRNA transcripts had been annotated. 239 of all 5′ ends were located on the circular, 89 on the linear, 20 on the At- and 8 on the Ti-plasmid. Distances between the identified TSS and start codons of protein-coding genes ranged from 0 to 544 nt with an average distance of 87 nt (Fig. 1C). 32 mRNAs possessed 5′-untranslated regions (5′UTRs) of less than 10 nt, and can thus be classified as leaderless mRNAs. For six genes (indicated by asterisks in Table S1), a TSS within the coding sequence hinted at an incorrectly annotated start codon.
Figure 1.A. tumefaciens TSS revealed by dRNA-seq. (A) and (B) Enrichment of primary 5′ ends by TEX treatment (in –Vir libraries). Screenshots of (A) flaA and (B) ftsH are shown. The distance between the TSS and the AUG start codon is indicated. (C) Histogram representing the 5′UTR length distribution. (D) A. tumefaciens consensus promoter sequence. Sequences 40 nt upstream and 10 nt downstream of all identified TSS were extracted and aligned. Conserved sequences were visualized with the WebLogo 3.0 program. Numbering of bases is given relative to the TSS.
Upstream 40 nt of each identified TSS were extracted in order to compile putative promoter sequences. The deduced Agrobacterium-specific -35 and -10 regions (CTTGNN and -10: TATNNT, respectively; Figure 1D) deviated from typical E. coli housekeeping promoters (-35: TTGACA and -10: TATAAT) at several positions. In addition, a conserved motif (CAT) from the -1 to +2 position was observed.
Validation of the dRNA-seq Data Set
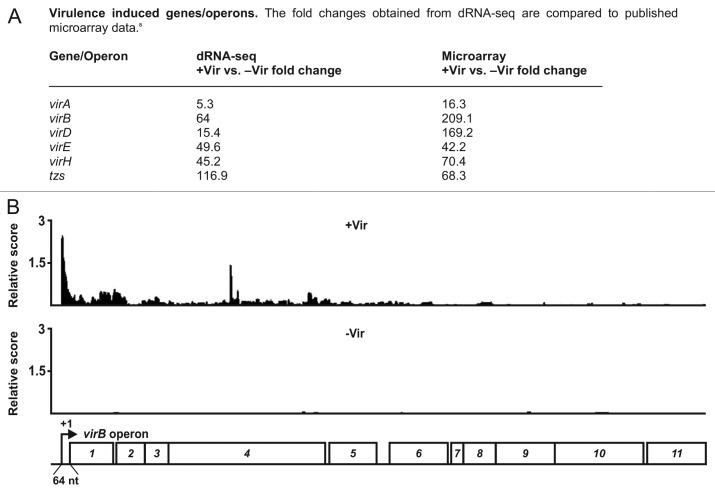
To evaluate the dRNA-seq results, we compared them with a recently published microarray study conducted with cells under identical virulence and non-virulence conditions.8 Seventy-one genes were found to be upregulated by a factor of at least two in the array study (Table S3). 45 of these also showed increased expression in the dRNA-seq data, indicating that dRNA-seq can provide reliable quantitative estimates of transcript levels in A. tumefaciens. A consistent set of 22 genes was strongly virulence-induced in both experimental setups. The classical virulence-induced genes or operons virA, virB, virD, virE, virH and tzs were included in this group (Fig. 2A). Among the most strongly induced genes in both approaches was virB1, the first gene of an extended operon of 11 genes, coding for the major components of the T4SS (Fig. 2B).
Figure 2. Virulence-induced genes/operons. (A) List of six classical virulence-induced genes. The fold changes obtained from dRNA-seq are compared with previously published microarray data.8 (B) Screenshot of the virB virulence induction. cDNA reads of +Vir (top) and -Vir (bottom) libraries are shown. The virB operon and the distance of the mapped TSS to the virB1 AUG start codon are indicated.
Identification of sRNA Transcripts
For the discovery of sRNAs in A. tumefaciens, cDNA reads along the whole genome were visualized by calculating graph files and displaying them in the Integrated Genome Browser (IGB, Affymetrix). We manually analyzed the 454-sequencing results for transcripts derived from intergenic regions or for reads complementary to protein-coding genes. Transcripts were listed as putative sRNA if they were represented by five or more cDNA reads in at least one library. This procedure resulted in a total of 228 candidate sRNAs (Table S4), including cis-encoded antisense sRNAs that overlap partially or completely with annotated ORFs. sRNAs were identified on all four A. tumefaciens replicons, namely 129 on the circular chromosome, 59 on the linear chromosome, 20 on the At- and 20 on the Ti-plasmid, respectively (Fig. 3). Interestingly, more than half of the sRNAs found on the two plasmids were cis-antisense sRNAs, whereas the majority of chromosomally encoded sRNAs were trans-encoded.

Figure 3.Distribution of sRNAs on all four A. tumefaciens replicons. The diagram depicts all identified sRNAs on the circular chromosome, the linear chromosome, the At- and the Ti-plasmid. Numbers of trans- and cis-encoded transcripts are given.
As expected, transcripts of four housekeeping sRNAs, namely the 6S RNA, the signal recognition particle (SRP) RNA 4.5S, RNase P, and tmRNA were identified. The RNase P transcript, which is encoded on the circular chromosome, was found to be located some nucleotides further upstream from its annotated site. The three recently published sRNAs RepE, AbcR1 and AbcR2 were also present in the data set.18,35
Expression profiling of 22 sRNAs
We used RNA gel blot analysis to confirm selected sRNAs by an independent method and to monitor their expression under various conditions (Table 2). Twenty-two of 28 candidates were detected (Figs. 4, 5, 6 and S1). They were named according to their position on the genome (e.g., C1, C2 etc. on the circular chromosome; L1, L2 etc. on the linear chromosome; see Table S4). Homologs of several of the validated chromosomal sRNAs (C2, C5, C6 and L5) have previously been reported in Rhizobium species (see Table S4 for references). All other sRNAs are initially described in this study. Blast searches revealed that C7, C10, L2, L3, L6, At2, Ti1 and Ti4 are unique to Agrobacterium, whereas sequences homologous to C1, C3, C4, C8, C9, L1, L4, At1, Ti2 and Ti3 can be found in at least one other α-proteobacterium (data not shown).
Table 2. Validated sRNAs.
| sRNA |
Position |
Flanking Genes |
Antisense Gene |
Size dRNA-seq |
Size Northern |
|---|---|---|---|---|---|
| circular chromosome | |||||
| C1 |
109476 – 109593 |
- |
atu0105 |
117 |
100, 70 |
| C2 |
112675 – 112536 |
atu0109 / atu8081 |
- |
139, 98 |
140, 95 |
| C3 |
317175 – 317330 |
atu0323 / atu0324 |
- |
155 |
170, 70 |
| C4 |
998103 – 998034 |
atu1000 / atu1001 |
- |
69 |
50 |
| C5 |
1275442 – 1275296 |
- |
atu1287 |
146 |
140 |
| C6 |
1288585 – 1288698 |
atu1298 / atu1299 |
- |
113 |
100 |
| C7 |
1345809 – 1345652 |
atu1350 / atu1351 |
- |
157, 103 |
160, 105 |
| C8 |
1995974 – 1995854 |
atu2036 / atu2038 |
- |
120 |
120 |
| C9 |
2087200 – 2087380 |
atu2122 / atu2123 |
- |
180 |
180 |
| C10 | 2667194 – 2667282 | atu2683 / atu2684 | - | 88 | 85 |
| linear chromosome | |||||
|---|---|---|---|---|---|
| L1 |
1179739 – 1179913 |
- |
atu4061 |
174 |
170, 130 |
| L2 |
1563974 – 1564060 |
atu4421 / atu4422 |
- |
86 |
85 |
| L3 |
1758411 – 1758628 |
- |
atu4611 |
217 |
140 |
| L4 |
1830948 – 1831214 |
- |
atu4669 |
266 |
270 |
| L5 |
1831445 – 1831606 |
atu4670 / atu4671 |
- |
161, 114 |
160, 115 |
| L6 | 1966571 – 1966697 | atu4791 / atu4792 | - | 126 | 130 |
| At-plasmid | |||||
|---|---|---|---|---|---|
| At1 |
55274 – 55161 |
atu5055 / atu5056 |
- |
113 |
115 |
| At2 | 363304 – 363131 | - | atu5372 | 173 | 170 |
| Ti-plasmid | |||||
|---|---|---|---|---|---|
| Ti1 |
148374 – 148279 |
- |
atu6129 |
95 |
110, 75 |
| Ti2 |
173670 – 173783 |
atu6154 / atu6155 |
- |
113 |
113 |
| Ti3 |
190702 – 190613 |
- |
atu6175 |
89 |
120 |
| Ti4 | 194399 – 194505 | - | atu6979 | 106 | 100 |
sRNAs are named after their location on the genome. Position on the genome, flanking genes or antisense transcript and sizes estimated from dRNA-seq data and from RNA gel blots are listed.
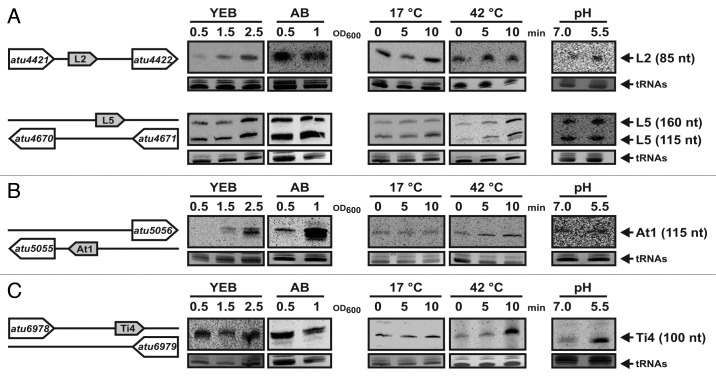
Figure 4. Differential expression of 10 sRNAs from the circular chromosome. Schematic diagrams of the genomic context of each sRNA are given on the left. Northern hybridizations were performed with RNA from A. tumefaciens from various growth conditions, i.e., different media, growth-phases, temperatures and pH-value, as indicated. Eight µg of total RNA were separated on a 10% polyacrylamide gel containing 7 M urea and detected by RNA gel blot analysis using RNA probes. Sizes of the sRNAs are given on the right. Ethidiumbromide-stained tRNAs were used as loading controls.
Figure 5.Differential expression of sRNAs from the linear chromosome and both A. tumefaciens plasmids. Validation of (A) two sRNAs located on the linear chromosome, (B) one on the At-plasmid and (C) one on the Ti-plasmid. Conditions were as described in Figure 4.

Figure 6. The virulence-induced transcript Ti2. (A) Genomic location of Ti2. (B) Schematic drawings of Ti2-dRNA-seq results. From top to bottom, diagrams from +Vir (+TEX), +Vir, -Vir (+TEX) and -Vir libraries are shown. (C) Predicted secondary structure of Ti2 calculated by the mfold program.39 (D) RNA gel blot analysis of Ti2 in the A. tumefaciens WT and VirA- and VirG deletion mutants. The three strains were grown in AB medium in the absence (-Vir) or the presence (+Vir) of the vir-gene inducer acetosyringone. The Ti2 size is given on the left. Ethidiumbromide-stained tRNAs were used as loading controls.
As bacterial sRNAs are often involved in stress adaptation, their expression profiles were recorded under different growth conditions, i.e., different cultivation media, growth phases, temperatures and pH values (Figs. 4 and 5). Seven sRNAs were hardly detectable and thus were only tested under normal and virulence conditions (Fig. S1). Generally, plasmid-encoded sRNAs and cis-antisense sRNAs were more difficult to detect than chromosome-encoded or trans-acting candidates.
Expression of the C1, C4, C5 and C7 RNAs was barely affected by the growth conditions (Fig. 4). Four sRNAs displayed a growth-phase dependent expression profile. C6, C10, L2 and At1 were strongest expressed at high optical densities, whereas C9 was most abundant in early stages of growth. Consistent with the remarkably high number of sequence reads (2,470), the C10 RNA showed the strongest expression in the Northern analysis (Figs. Four and 5). The expression of other sRNAs responded to temperature (L5 and Ti4) and pH alterations (C2, C3, C8 and Ti4), suggesting a biological role under these conditions.
In most cases, the transcript lengths estimated from Northern hybridizations were consistent with the lengths determined in the dRNA-seq approach (Table 2). However, eight sRNAs (C1, C2, C3, C7, L1, L4, L5 and Ti1) showed more than one Northern signal, suggesting that these sRNAs are processed into two or three smaller products. In several cases (C1, C2, C7 and L5) such processing events were already apparent in the sequencing data set (data not shown).
A total of nine cis-antisense transcripts were validated by RNA gel blot analysis. A fraction of them (C5, L4, Ti4) overlap only partly with the open reading frame encoded on the opposite strand. For instance, Ti4 reaches 48 nt into the 3′end of the coding sequence of the virulence-induced gene virC2. Other antisense sRNAs (C1, L1, L3, AT2, Ti1 and Ti3) show complete or nearly complete complementarity to an open reading frame on the other DNA strand. Interestingly, the virulence-gene virB9 (atu6175) and the conjugal transfer gene traB (atu6129) have an sRNA with perfect complementarity encoded on the reverse strand (Ti3 and Ti1, respectively; Fig. S1).
The virulence-induced sRNA Ti2
The Ti2 RNA, which is encoded in the intergenic region of virF-atu6155, was almost exclusively expressed under virulence conditions (Fig. 6). With 1,772 cDNA reads, the 113 nt transcript was the most abundant transcript from the Ti-plasmid. Ti2 is located upstream of the atu6155-virK-operon and is preceded by a vir-box sequence (Fig. 6A). The predicted secondary structure of Ti2 is given in Figure 6C. RNA gel blot experiments confirmed the dramatic induction in the presence of the virulence-gene inductor acetosyringone (Fig. 6D). Ti2 was not detectable at all under any other growth condition (data not shown).
Induction of virulence-genes in A. tumefaciens is mediated by the VirA/VirG two-component system, which is activated by several plant signals. To check whether expression of Ti2 is dependent on these two regulatory proteins, Northern analysis was performed with RNA from virA and virG deletion mutants. Since acetosyringone-induced Ti2 expression was lost in these backgrounds, Ti2 is a likely target of the VirA/VirG system (Fig. 6D).
Discussion
TSS mapping in A. tumefaciens
Within only a few years, RNA-seq technologies have outperformed classical transcriptome analyses by microarrays, in particular when it comes to the precise mapping of 5′ends of transcripts. In the present study, we used 454-pyrosequencing to deduce an A. tumefaciens consensus housekeeping promoter from the start sites of a total of 388 primary transcripts. Knowledge on α-proteobacterial promoter sequences is valuable because it is commonly observed that genes from Agrobacterium and Rhizobium species are poorly expressed in E. coli when transcribed from their authentic promoters although their -10 and -35 regions often resemble E. coli-type sigma 70 promoters. The Agrobacterium consensus promoter extracted from our dRNA-seq data (5′-CTTGNN-N18-TATNNT) deviates slightly from the promoter (5′-CTTGAC-N17-CTATAT) compiled from nearly 100 known and predicted promoters in S. meliloti and other rhizobia.40 Notably, an additional conserved box with a CAT motif was identified around the TSS. Our data will thus aid in the prediction of promoters of other Agrobacterium genes of interest.
A. tumefaciens genes and proteins prevalent under virulence conditions have been identified by several global studies using microarray or proteomics studies, respectively.8-10 Consistent with these findings, we confirmed that nearly all virulence-induced genes were located on the Ti-plasmid. Our approach revealed the virulence-induced short transcript Ti2 that had never been detected in the previous studies. Virulence induction in A. tumefaciens depends on the VirA/VirG two component system.3 Using a vir-box consensus sequence published by Cho and Winans, we searched for typical VirG binding sites upstream newly annotated TSS from virulence-associated genes.9 A vir-box with only one mismatch from the given consensus sequence was found upstream of virF, virB1 and the newly identified sRNA candidate Ti2. Among the most strongly induced transcripts were Ti2 and virB1. The corresponding vir-boxes are located 61.5 nt (Ti2) and 63.5 nt (virB1) upstream of the TSS (counted from the vir-box center). For virF, which was only slightly induced under virulence conditions, a vir-box-TSS distance of 53.5 nt was observed. As proposed previously, both the sequence and the position of the vir-box relative to the TSS seem to determine the induction rate of virulence-genes in A. tumefaciens.6
sRNA discovery: prediction vs. experiment
Prior to a wave of studies taking advantage of RNA-seq approaches, sRNAs have primarily been discovered through bioinformatics predictions and subsequent experimental validation.21 In a recently published study from our lab, we used a comparative bioinformatics approach to find sRNAs on the circular chromosome of A. tumefaciens.35 All IGRs longer than 50 nt on the circular chromosome were compared with different rhizobial genomes. Regions with high sequence conservation were collected as candidates for sRNA-encoding genes. A total of 231 sRNAs were predicted, four of which were validated by RNA gel blot analysis. 129 sRNAs were identified on the circular chromosome by deep sequencing. The overlapping sRNA candidates pool consists of only 26 sRNAs (Fig. 7). This may have several reasons. (1) It is quite possible that some of the computer-predicted sRNAs are not expressed at all. In fact, 9 out of 13 of the predicted sRNAs could not be identified by RNA gel blot hybridization.35 (2) It is also possible that suitable conditions, under which some of the in silico predicted candidates are expressed, have not been analyzed yet. It is noteworthy that many sRNAs in the present study and in other studies are differentially expressed under various stress conditions and might thus have escaped detection under the two conditions used for our pyrosequencing approach.28,32,41-43 (3) Since the computer-based search was restricted to IGRs, it will have missed all cis-antisense sRNAs. (iv) Comparative bioinformatics is limited to the discovery of sRNAs that are conserved in closely related species. Only experimental approaches, like RNA-seq, will be able to discover unique sRNAs with a specific function in an organism of interest.
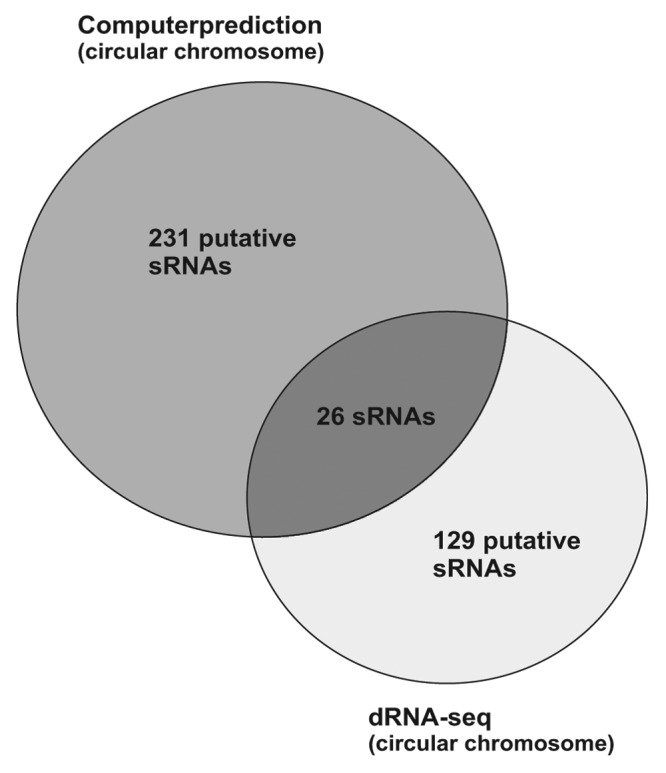
Figure 7. Comparison of sRNA identification by computer prediction and dRNA-seq. Venn diagram comparing the results for the circular chromosome of A. tumefaciens. Twenty-six sRNAs were found by both methods.
In this context, findings on the sRNome of Listeria monocytogenes are noteworthy because most exhaustive sRNA searches using bioinformatics, tiling arrays and deep sequencing techniques have been performed in this organism.27,44-46 Despite substantially overlapping sets of sRNAs, each of the four studies reported up to 71 unique sRNAs.45 Such findings lend value to each of the possible strategies used for sRNA identification.
sRNAs on all four A. tumefaciens replicons
56% of the 228 identified sRNAs were located on the circular chromosome, 26% on the linear chromosome, 9% on the At-plasmid, and 9% on the Ti-plasmid. sRNAs being located on mega-plasmids have been described earlier in several bacteria, including S. meliloti, R. etli or Chlamydia trachomatis, indicating that non-essential plasmid-encoded functions can be prone to sRNA-mediated regulation.26,28,33 Except for sRNAs involved in the control of plasmid replication, the function of all of these sRNAs is currently unknown.18,47
Homologous sRNAs of some identified candidates exist in related bacteria (Table S4), suggesting they might have conserved functions. Homologs of the 270-nt L4 sequence were found in all other rhizobia and many additional α-proteobacteria. This transcript always codes for a small protein of unknown function. A closer inspection of A. tumefaciens L4 revealed a possible AUG start codon preceded by an AG-rich sequence, which may serve as Shine-Dalgarno sequence. Thus, L4 might code for an unannotated peptide rather than a small RNA. Small proteins routinely missed by automated genome annotation and classical proteomic studies have recently received a lot of attention.28,48-50 Another interesting class of short transcripts serves dual functions as regulatory RNA and as mRNA, e.g., RNA III from Staphylococcus aureus, the SgrS RNA from E. coli or SR1 from Bacillus subtilis.51-53
For some of the identified sRNAs more than one transcript was observed, suggesting posttranscriptional processing of a primary transcript. Many enterobacterial sRNAs are activated or inactivated by processing events.20,54-56 In many bacterial species, RNaseE is responsible for sRNA processing. ArcZ for instance is processed by RNaseE into three transcript variants in E. coli and Salmonella.20,54 Since an RNaseE homolog is present in A. tumefaciens, it might be involved in sRNA processing.
In most cases, cis-antisense sRNAs modulate expression of the overlapping sense gene on transcriptional or posttranscriptional level, allowing us to speculate about some putative sRNA targets.57 The sRNA L3 is located antisense to tnp, encoding a transposase. Additionally, we found a second antisense sRNA overlapping with another transposase gene (antisense sRNA to atu4601, Table S4). Thus, transposon mobility might be regulated by an RNA-antisense mechanism in A. tumefaciens, as previously reported for E. coli.58 Likewise, a number of other studies revealed sRNAs in opposite orientation to transposase transcripts.24,28,59
Other cis sRNAs identified and verified in this study are encoded antisense to exoX (exopolysaccharide production repressor protein), traB (conjugal transfer protein), virC2 (excision of the T-DNA) and virB9 (component of the typeIV secretion system). As the latter two putative antisense sRNA targets encode important virulence factors, it is tempting to speculate that the corresponding sRNAs (Ti1 and Ti4) play regulatory roles in the A. tumefaciens plant infection process.
One of the most appealing sRNA revealed in this study is Ti2. This transcript was highly induced under virulence conditions in dependence of the VirA/VirG two component system. The distance between the 3′ end of Ti2 and the downstream operon atu6155-virK is only 93 nt. Microarrays demonstrated induction of the atu6155-virK transcript.8,9 However, the function of both genes remains unclear. VirK might be involved in the wide host range of Agrobacterium, although an A. tumefaciens virK deletion strain was not defective in tumor formation.7,60,61 As apparent from the dRNA-seq data, atu6155 is not transcribed from a separate TSS but—together with virK—derives from the promoter upstream of Ti2. The remarkable differential accumulation of Ti2 and the downstream mRNA raises the question whether elongation of most of the transcripts initiated at the VirAG-promoter is prematurely terminated at the 3′end of Ti2. Alternatively, the Ti2 RNA could be a processing product from the 5′UTR of atu6155. The ubiquitous 6S RNA and the Vibrio MicX sRNA are two examples processed from longer transcripts with their neighboring genes.62,63 Small RNA species derived from processing of 3′ or 5′ UTRs were reported for E. coli.64 Cis-regulatory elements (including riboswitches), which can act in trans as non-coding RNA have been described in several organisms.31,59,65
We wondered whether the massively induced Ti2 RNA plays a role in the virulence of A. tumefaciens. Like a virK mutant, deletion of Ti2 did not significantly affect tumorigenesis of Kalanchoe daigremontiana leaves (data not shown).61 Assuming that the massive induction of Ti2 should have a physiological function we are going to do proteomics and/or transcriptomics studies to identify possible targets of Ti2. Similar studies will be performed with other selected sRNA mutants.
Some recent studies hypothesized a secretion of sRNAs into host cells akin to effector protein translocation.12,14,15 It certainly is an attractive possibility that A. tumefaciens directly transfers sRNAs or—along with the T-DNA—sRNA genes into the host during plant transformation, which might mimic plant microRNA precursors. A recent study demonstrated that the plant microRNA pathway is essential for Agrobacterium disease development.66 Additionally, it is known that microRNAs are involved in defense mechanisms and in plant-microbe interactions including the response to A. tumefaciens.67,68
The large number of Agrobacterium sRNAs, some of which are differentially expressed, together with our recent finding that a single sRNA like AbcR1 can address several targets35 promises many interesting findings for the numerous sRNAs in this plant pathogen.
Material and Methods
Bacterial growth conditions and stress experiments
A. tumefaciens C58 was cultivated in YEB complex medium or in AB minimal medium at 30°C. For heat and cold shock conditions, cells were grown in YEB medium to an OD600 of 0.5 and subsequently shifted to pre-warmed or pre-cooled flasks to either 42°C (heat shock) or 17°C (cold shock). Cells for RNA isolation were collected before the temperature shift and after 5 and 10 min. pH-stress was performed in AB minimal medium adjusted to different pH values (5.5 and 7). After 5 h of growth (OD600 of 0.2), cells were harvested for RNA preparation. To induce virulence-gene expression, A. tumefaciens cells were pre-cultivated in AB minimal medium to an OD600 of 0.2 at 30°C before addition of acetosyringone (Sigma-Aldrich, Munich, Germany) to a final concentration of 0.1 mM. Cells were further incubated for 16 h at 23°C. Control cultures (non-induced) were treated with solvent (DMSO and H2O) only. Virulence induction in samples used for dRNA-seq was confirmed by protein gel blotting with a VirB9-probe using standard protocols.69
RNA preparation
Cells (10 ml) were harvested by centrifugation. After washing in ice cold AE-buffer (20 mM Na acetate, pH 5.5), pellets were immediately frozen in liquid nitrogen. Total RNA of cultured bacteria was isolated using the hot acid phenol method.70 Precipitated RNA used for 454-sequencing was treated with 100 U of DNase I (Roche, Mannheim, Germany) for 30 min at 37°C in a reaction volume of 100 µl. SUPERase InTM (100 U, Ambion, Huntingdon, UK) was added in the reaction to inhibit RNase activity. RNA was checked for integrity by agarose gel electrophoresis. The absence of genomic DNA was controlled by PCR.
cDNA library construction and sequence analysis
cDNA libraries were prepared at Vertis Biotechnology AG (Germany) and analyzed on a Roche FLX sequencer as previously described.23 Primary transcripts of total RNA were enriched by performing a selective degradation of RNAs containing a 5′mono-phosphate (5′P) using a 5′P-dependent terminator exonuclease (Epicenter).31 For each library [-Vir, -Vir (+TEX), +Vir, +Vir (+TEX)], graphs representing the number of mapped reads per nucleotides were calculated and visualized using the Integrated Genome Browser software from Affymetrix for each of the four A. tumefaciens replicons.
TSS mapping
Transcriptional start sites (TSS) were identified similar to previously published methods.31 Briefly, sequence reads were mapped to the A. tumefaciens reference genome (NC_003062.2, NC_003063.2, NC_003064.2, NC_003065.3) using Bowtie.1,71,72 Combined mapping data from the samples that were enriched for primary transcripts were then used to map TSS according to the following criteria with custom-made Perl scripts: Coverage at the TSS site was ≥ 5 reads and at least 5-fold the number of reads than the previous position in 5′ direction (saw-tooth pattern at the TSS). Coverage at the TSS had to be higher in the samples enriched for primary transcripts than in those not enriched, and coverage at the 18 nucleotide positions downstream of the TSS had to be at least 0.8-fold that of the TSS to prevent spurious annotation. TSS in front of annotated features were listed.
Comparison of dRNA-seq Data to Published Microarray Results
For a quantitative analysis of gene expression, Bowtie mapping data (see previous section) were used to determine the number of reads for each annotated CDS in the A. tumefaciens reference genome using custom-made Perl scripts based on BioPerl packages.73 Quantitative analysis of read counts was performed with the Bioconductor package DESeq that allows the analysis of experimental data even without replicates (www.bioconductor.org).74 Comparison of expression ratios (+Vir/-Vir) from this study (dRNA-seq) with previously published microarray data was performed with custom-made Perl scripts.8
Northern analysis
For the detection of small RNAs, 8–10 µg total RNA was separated on 10% polyacrylamide gels containing 7 M urea and subsequently transferred onto nylon membranes by semi-dry electroblotting (Biometra, Göttingen, Germany). Hybridizations were performed at 37–48°C over night using digoxygenine (DIG)-labeled DNA- or RNA probes (primers used for probe generation are listed in table S5), which were produced according to the instruction manual (Roche, Mannheim, Germany). After hybridization, membranes were first washed for 5 min at room temperature with solution 1 (5 x SSC, 0.1% SDS) followed by incubation with solution 2 (1 x SSC, 0.1% SDS) and 3 (0.5 x SSC, 0.1% SDS). Detection was performed by exposing the blot to a luminescence detector, using chemiluminescence substrate (CDP-Star; Roche Molecular Biochemicals, Mannheim, Germany).
Supplementary Material
Acknowledgments
We thank Rosemarie Gurski, Anna-Maria Stock and Marion Pesch for excellent technical assistance and Philip Möller and Christiane Fritz for providing the A. tumefaciens virA and virG deletion strains. We thank Fritz Thümmler (Vertis Biotechnology AG) for cDNA library construction, Richard Reinhardt (Max-Planck Institute of Molecular Genetics, Berlin, Germany) for 454-pyrosequencing. CMS is grateful to Jörg Vogel for continuous support. MN would like to thank Ulrich Kück and the Protein Research Department (Ruhr University Bochum) for support. This work was financed by a grant from the German Research Foundation (DFG priority program SPP 1258: Sensory and regulatory RNAs in Prokaryotes) to FN and a fellowship from the RUB Research School to IW.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/17212
References
- 1.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–23. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 2.Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;29:1021–32. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stachel SE, Nester EW. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986;5:1445–54. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 2002;3:3013–8. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazour GJ, Das A. Characterization of the VirG binding site of Agrobacterium tumefaciens. Nucleic Acids Res. 1990;18:6909–13. doi: 10.1093/nar/18.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin SG, Roitsch T, Christie PJ, Nester EW. The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J Bacteriol. 1990;172:531–7. doi: 10.1128/jb.172.2.531-537.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–95. doi: 10.1128/JB.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klüsener S, Hacker S, Tsai YL, Bandow JE, Gust R, Lai EM, et al. Proteomic and transcriptomic characterization of a virulence-deficient phosphatidylcholine-negative Agrobacterium tumefaciens mutant. Mol Genet Genomics. 2010;283:575–89. doi: 10.1007/s00438-010-0542-7. [DOI] [PubMed] [Google Scholar]
- 9.Cho H, Winans SC. VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc Natl Acad Sci USA. 2005;102:14843–8. doi: 10.1073/pnas.0503458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand A, Uppalapati SR, Ryu CM, Allen SN, Kang L, Tang Y, et al. Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 2008;146:703–15. doi: 10.1104/pp.107.111302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narberhaus F, Vogel J. Regulatory RNAs in prokaryotes: here, there and everywhere. Mol Microbiol. 2009;74:261–9. doi: 10.1111/j.1365-2958.2009.06869.x. [DOI] [PubMed] [Google Scholar]
- 12.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georg J, Hess WR. cis-Antisense RNA, Another Level of Gene Regulation in Bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–27. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: regulators of bacterial virulence. Nat Rev Microbiol. 2010;8:857–66. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 16.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–23. doi: 10.1016/S0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 17.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–68. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 18.Chai Y, Winans SC. A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol Microbiol. 2005;56:1574–85. doi: 10.1111/j.1365-2958.2005.04636.x. [DOI] [PubMed] [Google Scholar]
- 19.Bouché F, Bouché JP. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol. 1989;3:991–4. doi: 10.1111/j.1365-2958.1989.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 20.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–58. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 21.Backofen R, Hess WR. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010;7:33–42. doi: 10.4161/rna.7.1.10655. [DOI] [PubMed] [Google Scholar]
- 22.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536–46. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jäger D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA. Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci USA. 2009;106:21878–82. doi: 10.1073/pnas.0909051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–77. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, et al. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics. 2009;10:641. doi: 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Janicke S, et al. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010;11:245. doi: 10.1186/1471-2164-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–9. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Vliet AH. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol Lett. 2010;302:1–7. doi: 10.1111/j.1574-6968.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- 31.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 32.Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol. 2008;68:600–14. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vercruysse M, Fauvart M, Cloots L, Engelen K, Thijs IM, Marchal K, et al. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics. 2010;11:53. doi: 10.1186/1471-2164-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol. 2009;74:1497–512. doi: 10.1111/j.1365-2958.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- 35.Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol. 2011;80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
- 36.Chesnokova O, Coutinho JB, Khan IH, Mikhail MS, Kado CI. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–90. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 37.Kahng LS, Shapiro L. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J Bacteriol. 2001;183:3065–75. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narberhaus F, Obrist M, Führer F, Langklotz S. Degradation of cytoplasmic substrates by FtsH, a membrane-anchored protease with many talents. Res Microbiol. 2009;160:652–9. doi: 10.1016/j.resmic.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLellan SR, MacLean AM, Finan TM. Promoter prediction in the rhizobia. Microbiology. 2006;152:1751–63. doi: 10.1099/mic.0.28743-0. [DOI] [PubMed] [Google Scholar]
- 41.Valverde C, Livny J, Schlüter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics. 2008;9:416. doi: 10.1186/1471-2164-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, et al. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–27. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wassarman KM. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–4. doi: 10.1016/S0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 44.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–6. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 45.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011;39:4235–48. doi: 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livny J, Waldor MK. Mining regulatory 5′UTRs from cDNA deep sequencing datasets. Nucleic Acids Res. 2010;38:1504–14. doi: 10.1093/nar/gkp1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkova-Canova T, Soberon NE, Ramirez-Romero MA, Cevallos MA. Two discrete elements are required for the replication of a repABC plasmid: an antisense RNA and a stem-loop structure. Mol Microbiol. 2004;54:1431–44. doi: 10.1111/j.1365-2958.2004.04366.x. [DOI] [PubMed] [Google Scholar]
- 48.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–51. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnleitner E, Sorger-Domenigg T, Madej MJ, Findeiss S, Hackermuller J, Huttenhofer A, et al. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology. 2008;154:3175–87. doi: 10.1099/mic.0.2008/019703-0. [DOI] [PubMed] [Google Scholar]
- 50.Hemm MR, Paul BJ, Miranda-Rios J, Zhang A, Soltanzad N, Storz G. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol. 2010;192:46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–75. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA. 2007;104:20454–9. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gimpel M, Heidrich N, Mader U, Krugel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- 54.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Görke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–80. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–9. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Ma C, Simons RW. The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J. 1990;9:1267–74. doi: 10.1002/j.1460-2075.1990.tb08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–36. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hattori Y, Iwata K, Suzuki K, Uraji M, Ohta N, Katoh A, et al. Sequence characterization of the vir region of a nopaline type Ti plasmid, pTi-SAKURA. Genes Genet Syst. 2001;76:121–30. doi: 10.1266/ggs.76.121. [DOI] [PubMed] [Google Scholar]
- 61.Kalogeraki VS, Winans SC. Wound-released chemical signals may elicit multiple responses from an Agrobacterium tumefaciens strain containing an octopine-type Ti plasmid. J Bacteriol. 1998;180:5660–7. doi: 10.1128/jb.180.21.5660-5667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr Opin Microbiol. 2007;10:164–8. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65:373–85. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–50. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–9. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 66.Dunoyer P, Himber C, Voinnet O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet. 2006;38:258–63. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- 67.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010;48:225–46. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pruss GJ, Nester EW, Vance V. Infiltration with Agrobacterium tumefaciens induces host defense and development-dependent responses in the infiltrated zone. Mol Plant Microbe Interact. 2008;21:1528–38. doi: 10.1094/MPMI-21-12-1528. [DOI] [PubMed] [Google Scholar]
- 69.Wessel M, Klüsener S, Godeke J, Fritz C, Hacker S, Narberhaus F. Virulence of Agrobacterium tumefaciens requires phosphatidylcholine in the bacterial membrane. Mol Microbiol. 2006;62:906–15. doi: 10.1111/j.1365-2958.2006.05425.x. [DOI] [PubMed] [Google Scholar]
- 70.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–10. [PubMed] [Google Scholar]
- 71.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294:2323–8. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 72.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–8. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.