Abstract
Numerous mutations in the mitochondrial genome are associated with maternally transmitted diseases and syndromes that affect muscle and other high energy-demand tissues. The mitochondrial genome encodes 13 polypeptides, 2 rRNAs and 22 interspersed tRNAs via long bidirectional polycistronic primary transcripts, requiring precise excision of the tRNAs. Despite making up only ~10% of the mitochondrial genome, tRNA genes harbor most of the pathogenesis-related mutations. tRNase Z endonucleolytically removes the pre-tRNA 3′ trailer. The flexible arm of tRNase Z recognizes and binds the elbow (including the T-loop) of pre-tRNA. Pathogenesis-related T-loop mutations in mitochondrial tRNAs could thus affect tRNA structure, reduce tRNase Z binding and 3′ processing, and consequently slow mitochondrial protein synthesis. Here we inspect the effects of pathogenesis-related mutations in the T-loops of mitochondrial tRNAs on pre-tRNA structure and tRNase Z processing. Increases in KM arising from 59A > G substitutions in mitochondrial tRNAGly and tRNAIle accompany changes in T-loop structure, suggesting impaired substrate binding to enzyme.
Keywords: Kinetics, T-loop mutations, mitochondrial tRNAs, tRNA structure, tRNase Z
Introduction
Numerous maternally transmitted diseases and syndromes affecting muscle (myopathies, including cardiomyopathies) and other high energy-demand tissues (e.g., defects in vision and hearing; a form of epilepsy) arise from mutations in the mitochondrial genome. The 16,569 bp mitochondrial genome encodes 13 polypeptides (essential components of respiratory transport chain complexes), two rRNAs and 22 tRNAs via long bidirectional polycistronic transcripts.1 Several thousand other proteins required for mitochondrial metabolism are nuclear encoded and transported from the cytoplasm. In contrast, few RNAs are known to be imported into human mitochondria.
A set of mitochondrially encoded tRNAs, one for each of 18 amino acids, two for tRNALeu [(UUR) and (CUN)] and two for tRNASer [(UCN) and (AGY)], is sufficient for translation of mitochondrial messages. Of over 220 mutations in the mitochondrial genome related to maternally transmitted diseases, more than 150 are located in tRNA genes (see ref. 2 for a compilation). Almost every tRNA harbors at least one pathogenesis-related mutation, suggesting that all 22 are required for efficient mitochondrial protein synthesis and for mitochondrial function (reviewed in ref. 3).
Molecular mechanisms by which pathogenesis arises from mutations in mitochondrial tRNAs remain largely unknown and the mutations don’t fall into obvious categories (for reviews, see refs. 3, 4). Neutral polymorphisms have been described in conserved regions and pathogenesis-related mutations in nonconserved ones, suggesting absence of a common theme.5 Thorough examination of all mutations may thus be required to understand their mechanisms; it will be useful to select a subset and look for correlations and patterns between the distributions of mutations in different tRNAs, positions in the tRNAs, and mitochondrial pathologies. For example, mutations in tRNASer(UCN) are often associated with non-syndromic deafness (reviewed in ref. 6), and the symptoms arising from mutations in tRNAIle (mainly ophthalmoplagias and cardiomyopathies) correlate with the reduction in aminoacylation efficiency.4
Mitochondrial tRNAs punctuate the mitochondrial genome and must be precisely excised for function of the mitochondrial mRNAs and rRNAs7,8 (reviewed in ref. 9). 5′ ends of human mitochondrial tRNAs are produced by mitochondrial RNase P, a protein-only enzyme consisting of three polypeptides.10 3′ ends are cut on the 3′ side of the discriminator (the last unpaired nucleotide following the acceptor stem) by tRNase Z, leaving a 3′-OH prepared for CCA addition.9 Since CCA at the 3′ end of mature tRNAs is not transcriptionally encoded, CCA-adding activity is essential11 and tRNase Z cleavage is central to tRNA maturation.
All tRNAs undergo post-transcriptional modification, and intron-containing tRNAs require splicing. Although important, these reactions were not investigated here. tRNAs engage in numerous additional interactions including the aminoacylation cycle, the Tu/Ts cycle (or the equivalent in eukaryotes and organelles) and other aspects of translation (e.g., decoding, peptidyl transfer, translocation). Ability of a mutant pre-tRNA to undergo tRNase Z reaction would thus not be an exclusive indicator of molecular deficiency, but tRNase Z reaction combined with tRNA secondary structure probing can effectively report the type and degree of harm caused by pathogenesis-related mutations.6,9,12-16
tRNase Z is encoded by two separate genes in some eukaryotes including humans17,18: a short form (tRNase ZS) and a long form (tRNase ZL) that may have arisen from tandem duplication of the short form followed by adaptation.18-20 Archaea and bacteria have only tRNase ZS. S. cerevisiae, C. elegans and D. melanogaster have only tRNase ZL. tRNase Z homologs in fungi and higher plants were recently thoroughly characterized using a bioinformatics approach.21-23 D. melanogaster tRNase Z demonstrably functions in vivo in both nuclear and mitochondrial pre-tRNA maturation.24,25 tRNase ZL is the better candidate for an essential function in human tRNA (including mitochondrial tRNA) maturation due to its ~2,000x higher reaction efficiency15 and dual localization26-28; tRNase ZS function is unknown.
The flexible arm (FA) of tRNase Z, a unique recognition and binding domain
Enzymes involved in general tRNA metabolism would not be expected to distinguish between tRNAs, despite the noncanonical structure of organellar tRNAs,29 but must distinguish tRNAs from other RNAs. Up to 25 y ago, tRNA end processing enzymes including RNase P, tRNase Z and CCA-adding enzyme were shown to utilize the same deleted substrate consisting of a half-tRNA minihelix (coaxially stacked acceptor stem and T arm30-32), suggested to be the primary recognition determinant for all three enzymes.
tRNase Z and CPSF-73 (the pre-mRNA 3′ end endonuclease33) are both members of the β-lactamase superfamily of metal-dependent hydrolases,34,35 and their metal-binding and active sites are virtually superimposable.33,36,37 The active site of CPSF-73 is covered by a large flap (the β-CASP region33) and a battalion of accessory proteins is required for cleavage, presumably to recognize the cleavage site, open the flap and activate the endonuclease.
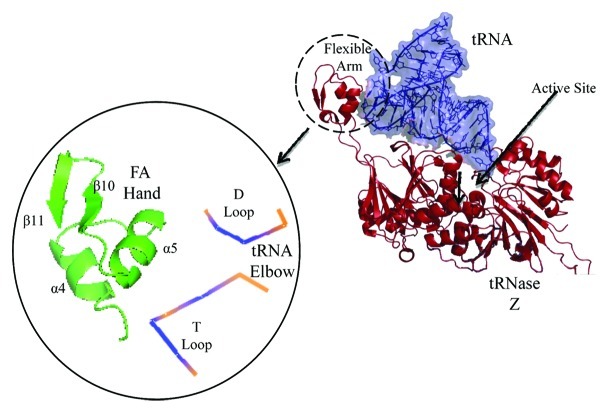
In contrast, tRNase Z has a flexible arm (FA) which recognizes the elbow that caps the coaxially stacked acceptor stem/T arm common to tRNAs,38 and requires no accessory proteins for tRNA binding or cleavage. The FA, located far from the active site, consists of a globular ααββ hand extruded from the body of the enzyme by a structured polypeptide stalk (Fig. 1, cf19,20,36,38-41). Deleting the FA hand causes close to a 100-fold increase in KM with little change in kcat,20 quantifying its substrate recognition/binding function.
Figure 1. The Flexible Arm (FA) of tRNase Z binds the elbow (D/T loops) of tRNA. The structure of tRNA complexed with B. subtilis tRNase Z was redrawn from Li et al., 2006 (PDB#2FK6). Inset: the FA hand and elbow of the tRNA.
A structure change in the T loop of tRNA could interfere with precursor binding to tRNase Z and impair tRNA maturation. Previous work31,42 suggests that structural determinants for tRNase Z activity are present in the T-loop, effects of substitutions in the D-loop being modulatory and less definitive. This investigation of mutant mitochondrial tRNAs was therefore limited to the T-loop. Thirteen pathogenesis-related mutations in the T-loops of 11 different mitochondrial tRNAs (compiled from Mitomap2) are presented in Table 1. To investigate effects on tRNase Z processing kinetics and tRNA structure arising from T loop substitutions, six pathogenesis-related T-loop substitutions were chosen for analysis (enclosed in ellipses in Table 1); secondary structures are presented in Figure 2 (adapted from Mamit,43 supported by references from 2). The in vitro effects of these mutations on tRNase Z processing, supported by changes in T-loop structure of the mitochondrial tRNAs, suggest a contributing molecular mechanism for mitochondrial pathology.
Table 1. Pathogenesis-related mutations in the T-loops of human mitochondrial tRNAs including their positions and related illnesses.
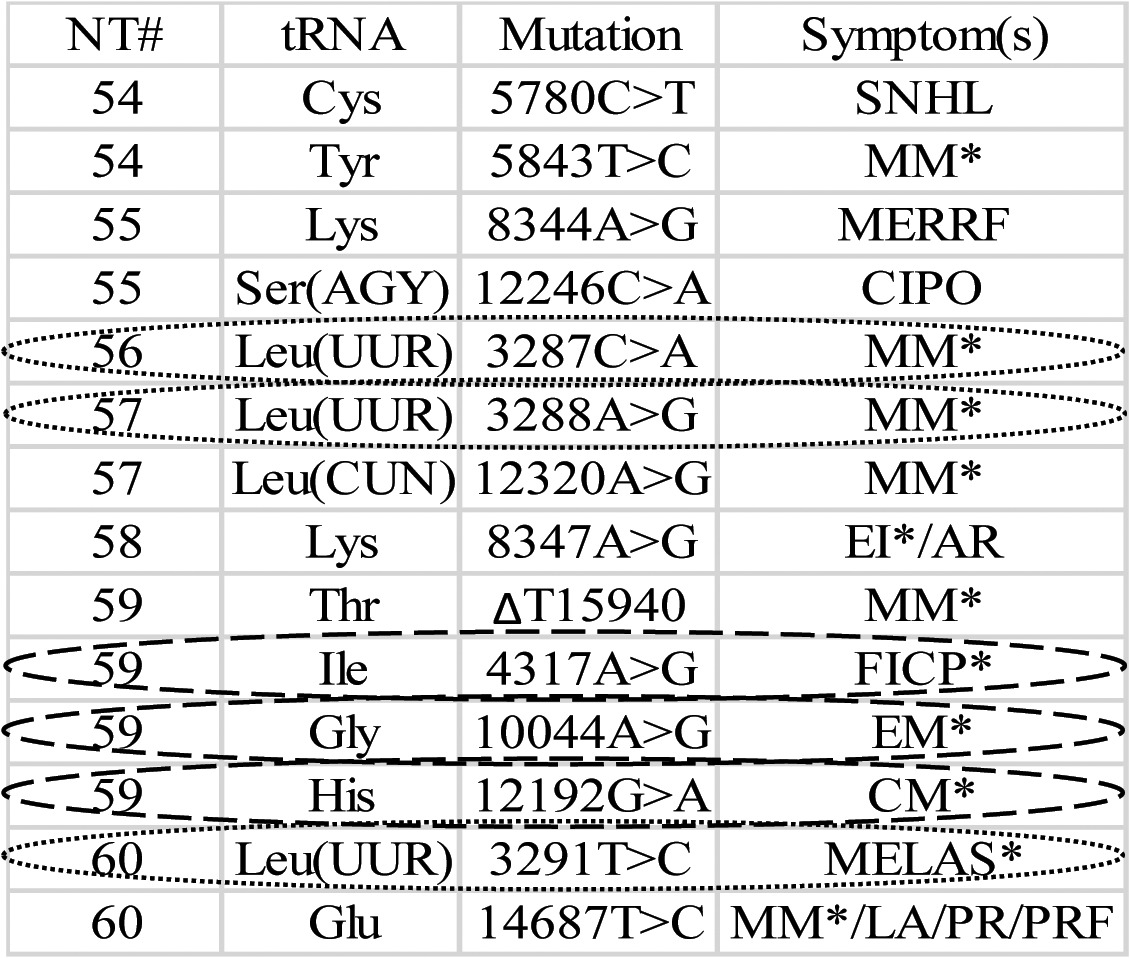
The table was assembled from Mamit and Mitomap. NT#: canonical nucleotide numbers for each tRNA. Mutation: nucleotide numbers based on the reference (Cambridge) sequence. Symptom abbreviations (from Mitomap): SNHL, Sensorineural Hearing Loss; MM, Mitochondrial Myopathy; MERRF, Myoclonic Epilepsy and Ragged Red muscle Fibers; CIPO, Chronic Intestinal Pseudo Obstruction with myopathy; EI, Exercise Intolerance; AR, Axenfeld-Rieger anomaly; FICP, Fatal Infantile Cardiomyopathy Plus a MELAS-associated cardiomyopathy; EM, Encephalomyopathy; CM, Cardiomyopathy; MELAS, Mitochondrial Encephalomyopathy, Lactis Acidose, and Stroke-like episodes; LA, Lactis Acidose; PR, Pigmentary Retinopathy; PRF, Progressive Respiratory Failure. Ellipses indicate specific mutations selected for further analysis (dotted: tRNALeu[UUR] nt 56, 57 and 60; dashed: nt 59 substitutions in tRNAIle, Gly and His).
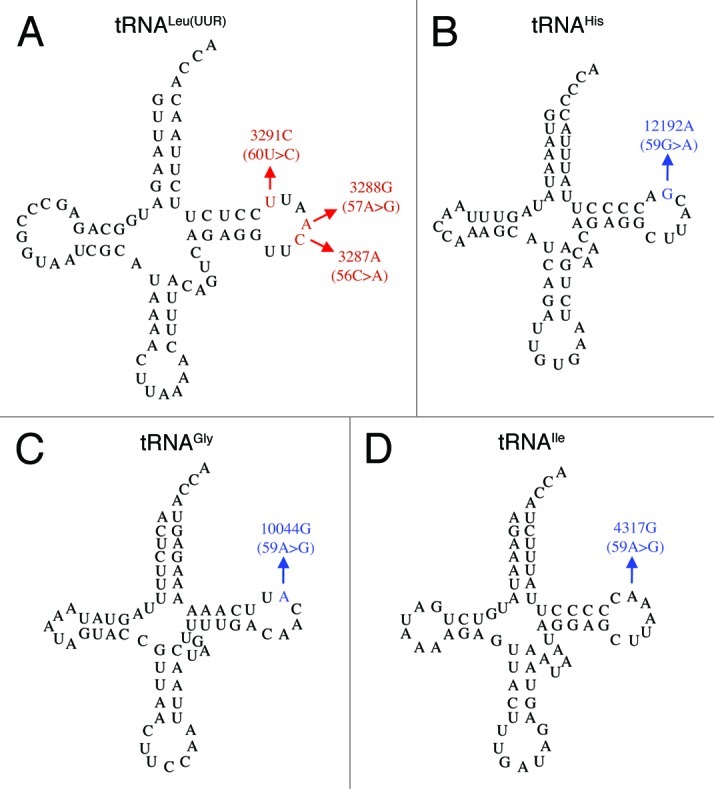
Figure 2. Secondary structures of selected human mitochondrial tRNAs with pathogenesis-related T-loop mutations (Reference Mamit with updates from Mitomap). The T-loop substitutions are assigned canonical nt #s with the mitochondrial reference sequence numbers in (). (A) tRNALeu(UUR) 56C > A (3287C > A), 57A > G (3288A > G), 60U > C (3291U > C); (B) tRNAIle 59A > G (4317A > G); (C) tRNAGly 59A > G (10044C > G); (D) tRNAHis 59G > A (12192G > A).
Results
Mitochondrial tRNAs with T loop substitutions selected for processing analysis
The rationale for analyzing the effects of pathogenesis-related T-loop substitutions in mitochondrial tRNAs is given in Introduction, a compilation of the substitutions is presented in Table 1 and tRNA secondary structures are shown in Figure 2. tRNALeu(UUR) has the conserved D and T loop sequences with the potential for canonical tertiary contacts. tRNAIle is next closest to canonical with the T loop sequence pyrimidine-pyrimidine-pyrimidine-purine (YYYR—) and the potential for tertiary pairing with the D-loop sequence AA corresponding to the canonical G18G19. Next is tRNAHis with the T-loop sequence YYYR—but no obvious D loop pairing partners and tRNAGly has the least canonical T-loop length and sequence and no obvious pairing potential with the D-loop apart from one U which could pair with a noncanonical A at T-loop position 55 or 56. Application of Leontis-Westhof pairing rules44 for non-Watson-Crick appositions is beyond the scope of this project.
Mitochondrial tRNA precursor 3′ end processing
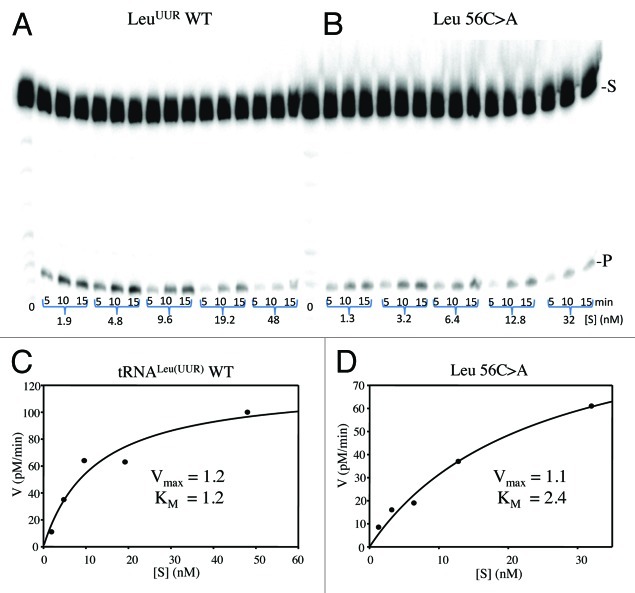
tRNase Z reaction kinetics was performed using wild type and variant precursors with a mature 5′ end and a natural sequence 3′ trailer. Processing data for wild type tRNALeu(UUR) and tRNALeu(UUR) with the 56C > A substitution are shown in Figure 3; the arrow in Figure 4C indicates the tRNase Z cleavage site. Figure 3C and D show the fit to Michaelis-Menten kinetics and the effects of the substitution on tRNase Z processing relative to wild type. The complete data set for tRNase Z processing is presented in Table 2. The greatest increases in KM relative to wild-type tRNAs, in tRNAIle 59A > G and tRNAGly 59A > G, consistent with impaired substrate binding, are highlighted.
Figure 3. Processing kinetics of tRNALeu(UUR) wild type and 56C > A mutant. (A, B) Electrophoresis gel. Designations at right: S-pre-tRNA substrate; P-tRNase Z product. (C, D) Data (from A, B) analyzed with ImageQuant were used to create Michalis-Menten kinetic plots using SigmaPlot.
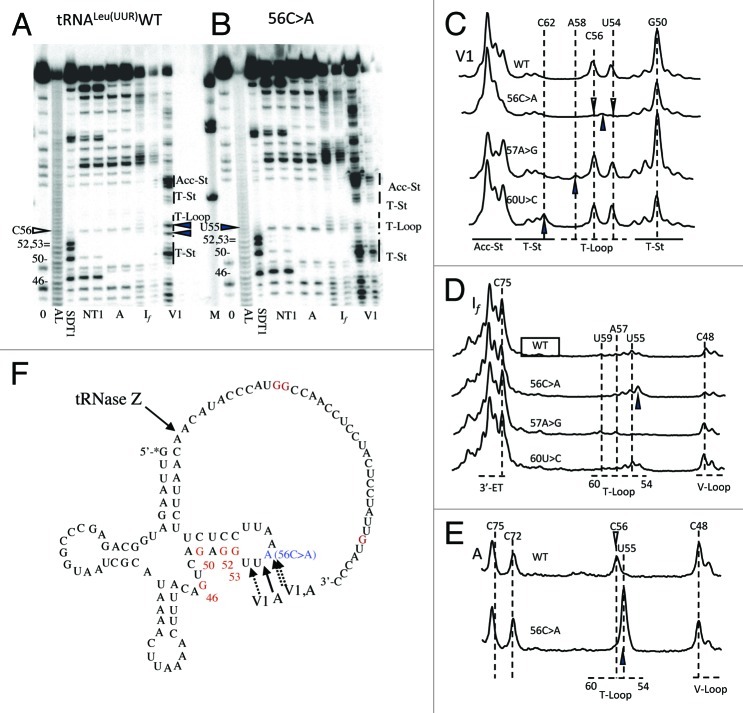
Figure 4. Structure probing of tRNALeu(UUR) WT and 56C > A, 57A > G, and 60U > C variants. (A, B) Electrophoresis gel with data for WT and 56C > A, respectively. Abbreviations: 0, no enzyme added; AL, Alkaline Ladder; SDT1, semi-denaturing T1; NT1, Native T1; A, RNase A; If, nuclease If; V1, nuclease V1. (C, D, E) Superimposed V1, If, and RNase A traces, respectively, allow structural comparison between WT and the variants. Open arrows pointing down indicate decreased sensitivity and solid arrows pointing up indicate increased sensitivity of the variants. (F) Secondary structure shown for the 56C > A variant. Arrow pointing toward the discriminator indicates the tRNase Z cleavage site. Ribonuclease sites differing from WT are shown on the secondary structure with dotted arrows indicating decrease in sensitivity, and solid arrows indicating increase in sensitivity.
Table 2. tRNase ZL processing kinetics with mitochondrially encoded wild-type tRNALeu(UUR), tRNAHis, tRNAGly, and tRNAIle and pathogenesis-related mutant T loop substrates.
| tRNA |
kcat (min−1) |
KM (nM) |
kcat/KM (X 108 M−1min−1) |
kcat Re WT |
KM Re WT |
kcat/KMre WTa |
|---|---|---|---|---|---|---|
| tRNALeu(UUR) | ||||||
| WT |
15.0 ± 1.9 |
24 ± 6 |
9.3 ± 2.9 |
N/A |
N/A |
N/A |
| 56C > A |
11.1 ± 1.8 |
32 ± 11 |
4.8 ± 1.2 |
0.81 ± 0.15 |
1.4 ± 0.3 |
0.52 ± 0.09 |
| 57A > G |
6.0 ± 0.7 |
17 ± 6 |
4.4 ± 1.1 |
0.47 ± 0.05 |
1.3 ± 0.3 |
0.42 ± 0.10 |
| 60U > C | 6.9 ± 1.0 | 17 ± 5 | 5.4 ± 1.7 | 0.53 ± 0.01 | 1.3 ± 0.1 | 0.40 ± 0.01 |
| tRNAIle | ||||||
|---|---|---|---|---|---|---|
| WT |
1.1 ± 0.3 |
48 ± 14 |
0.25 ± 0.06 |
N/A |
N/A |
N/A |
| 59A > G | 1.3 ± 0.3 | 169 ± 39 | 0.078 ± 0.01 | 1.5 ± 0.4 | 3.5 ± 1.0 | 0.46 ± 0.13 |
| tRNAGly | ||||||
|---|---|---|---|---|---|---|
| WT |
2.1 ± 0.2 |
29 ± 5 |
0.74 ± 0.04 |
N/A |
N/A |
N/A |
| 59A > G | 2.7 ± 1.0 | 103 ± 12 | 0.26 ± 0.07 | 1.2 ± 0.3 | 3.6 ± 0.1 | 0.35 ± 0.10 |
| tRNAHis | ||||||
|---|---|---|---|---|---|---|
| WT |
14.2 ± 3.8 |
18 ± 1 |
7.6 ± 2.3 |
N/A |
N/A |
N/A |
| 59G > A | 21.7 ± 1.3 | 33 ± 1 | 6.9 ± 1.9 | 1.6 ± 0.3 | 1.8 ± 0.4 | 0.87 ± 0.01 |
are WT (n-fold reduction re wild type) refers to the ratio of processing efficiencies: the mean kcat/KM (Mutant) / kcat/KM (WT) obtained in parallel kinetic experiments; N/A, Not Applicable; Averages of two or more Michaelis-Menten experiments with each variant are presented; wild-type and variant experiments were performed the same day. ± indicates standard error. kcat/KM re WT for the variants is the n-fold reduction relative to wild type. The two mutations with greatest increase in KM, tRNAIle 59A > G and tRNAGly 59A > G, are highlighted.
Wild type kinetics varies with tRNA substrate. KMs for four wild type tRNAs cover a range of less than a factor of 2.5 centered on 30 nM, in close agreement with previous results.14,16 kcat covers a range of more than an order of magnitude; tRNALeu(UUR) and tRNAHis have the highest kcat and tRNAGly and tRNAIle have the lowest. The only published kcat from this set of mitochondrial tRNAs, for tRNAIle 16, was a bit higher than results reported here.
The tRNALeu(UUR) 56C > A substitution causes a slight reduction in kcat accompanied by a slight increase in KM, producing a ~2x reduction in processing efficiency (kcat/KM) relative to wild type. tRNALeu(UUR)57A > G causes a slightly greater reduction in kcat, a slight increase in KM, and a ~2.4x reduction in processing efficiency. tRNALeu(UUR) 60U > C causes a ~2x reduction in kcat, a slight increase in KM, and a ~2.5x reduction in processing efficiency.
tRNAIle 59A > G causes a slight increase in kcat, a 3.5x increase in KM, and ~2x reduction in processing efficiency. tRNAGly 59A > G causes a slight increase in kcat, a 3–4x increase in KM, and a ~3x reduction in processing efficiency. tRNAHis 59G > A causes a slight increase in kcat, a slight increase in KM, and a slight reduction in processing efficiency relative to wild type.
Effects of the T-loop substitutions on tRNA precursor structure
Four nucleases were used to probe for structure changes caused by the substitutions. V1 and If display alternating patterns, generally consistent with the stem-loop cloverleaf structure of canonical tRNAs. The 3′ trailer is also structured, as previously reported.13,14 Wild-type tRNALeu(UUR) displays broad nuclease If sensitivity suggesting a floppy D-stem and anticodon arm, as previously noted.14
Wild type tRNALeu(UUR) shows pronounced V1 sensitivity at U54 and C56 (Fig. 4). The 56C > A substitution sharply reduces V1 susceptibility at these positions and increases susceptibility at U55. U54 becomes more susceptible to If cleavage, U55 becomes more susceptible to RNase A and A56 is less susceptible to RNase A than C56 in wild type (Fig. 4). tRNALeu(UUR) 57A > G shows increased V1 susceptibility at A58, as well as decreased sensitivity to If cleavage at U54, U55, and C56. tRNALeu(UUR) 60U > C shows increased V1 susceptibility at C62.
Increased V1 susceptibility of tRNALeu(UUR)57A > G at A58 (Fig. 4) shows that the T loop is more structured at this position. Decreased sensitivity to If at U54, U55 and C56 suggests tighter structure at the start of the T-loop. Combining the two observations suggests that the tRNALeu(UUR) 57A > G substitution starts a wave of increasing structure that spreads in both directions through the T-loop. tRNALeu(UUR)60U > C shows increased V1 susceptibility at C62 (Fig. 4C) which, as the second base in the T-stem, is expected to already be structured.
tRNAIle 59A > G displays decreased If sensitivity at C54 (Fig. 5), suggesting a more structured T-loop which could impair tRNA-tRNase Z binding, increasing KM. tRNAGly 59A > G displays decreased V1 sensitivity at C62 and C58 as well as decreased RNase A sensitivity at C58 (Fig. 6). A > G substitution at this position disrupts the CA step. RNase A cleaves after unstructured pyrimidines, thus there are eight possible dinucleotide sequences that could be cleaved by RNase A (C or U followed by any of the four nucleotides). The CA step is especially susceptible to RNase A (reviewed in 45). Four instances of changes involving a CA step were observed: tRNALeu(UUR) C56↓A becomes A56A in 56C > A; the C56↓A step becomes CG in tRNALeu(UUR) 57A > G; tRNAGly C↓A59 becomes CG in 59A > G; tRNAHis CG59 becomes C↓A in 59G > A. As expected, in the first three cases the RNase A susceptibility decreases and in the last one it increases.
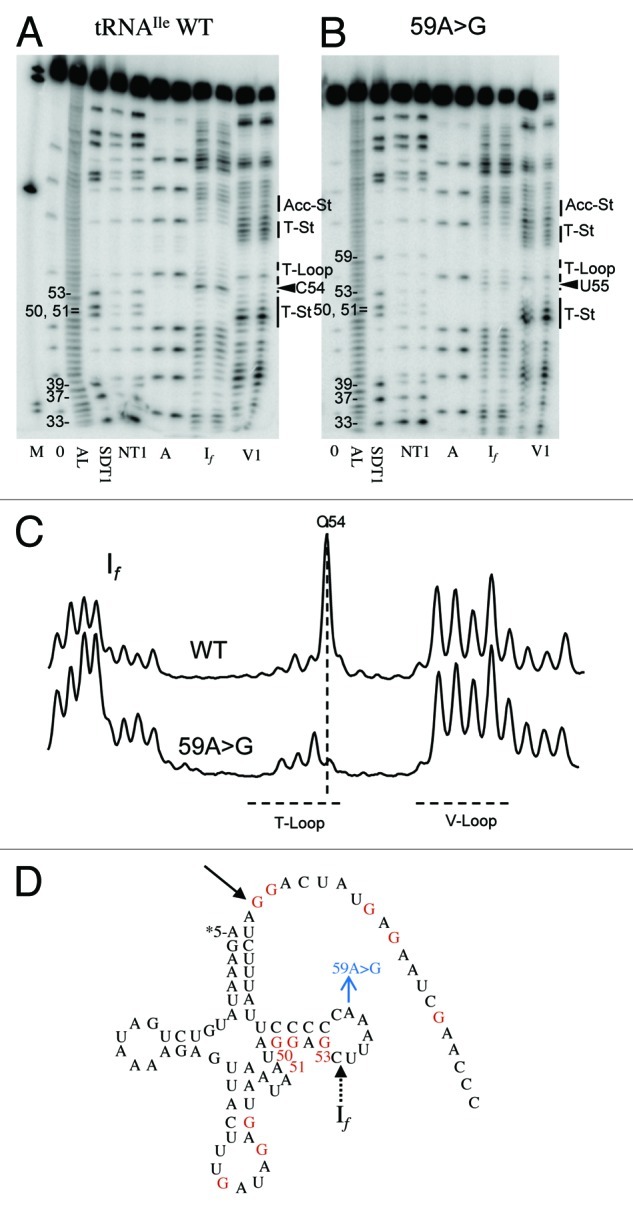
Figure 5. Structure probing of tRNAIle WT and 59A > G variant. Designations are the same as in Figure 4.
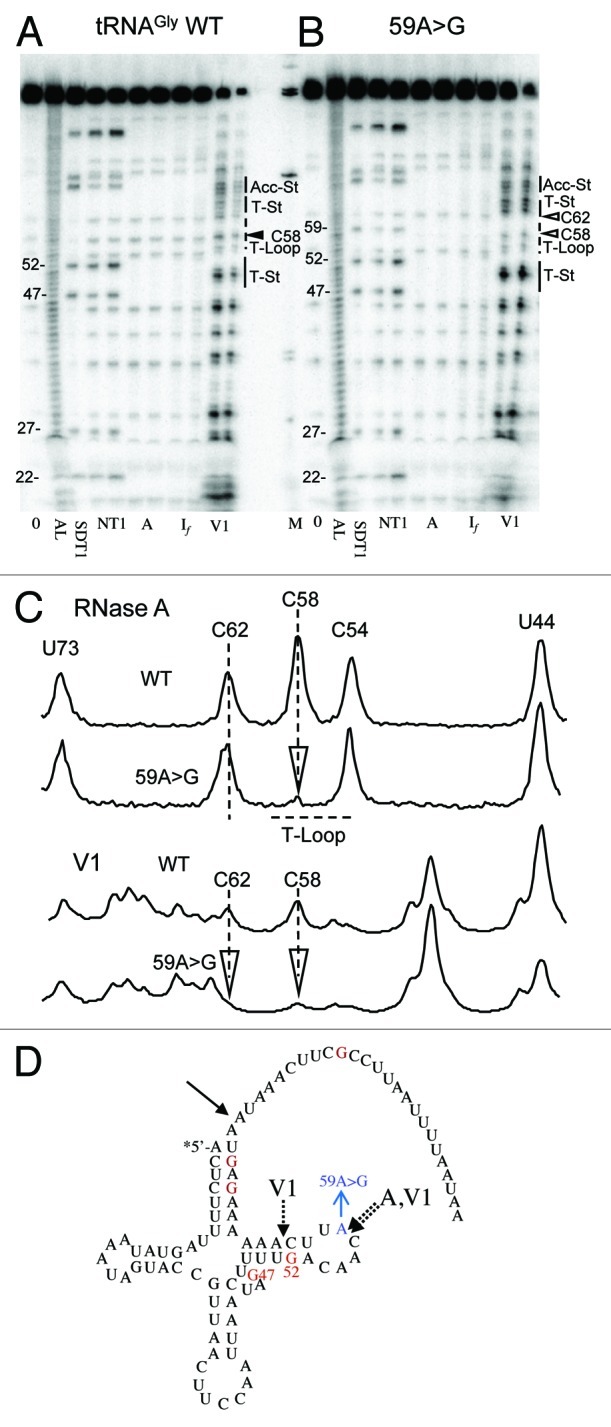
Figure 6. Structure probing of tRNAGly WT and 59A > G variant. Designations are the same as in Figure 4.
tRNAHis 59G > A displays decreased V1 susceptibility at G50 (Fig. 7) within the T-stem near the V-loop boundary, a structural disruption beyond the T loop. C58 becomes more susceptible to RNase A because 59G > A produces a C↓A59 step (see above). Position A58 was the most sensitive in a survey of nuclear encoded tRNAArg with D and T loop substitutions.42 These changes may impair the tRNA-tRNase Z interaction, as suggested by the increases in KM (Table 2).
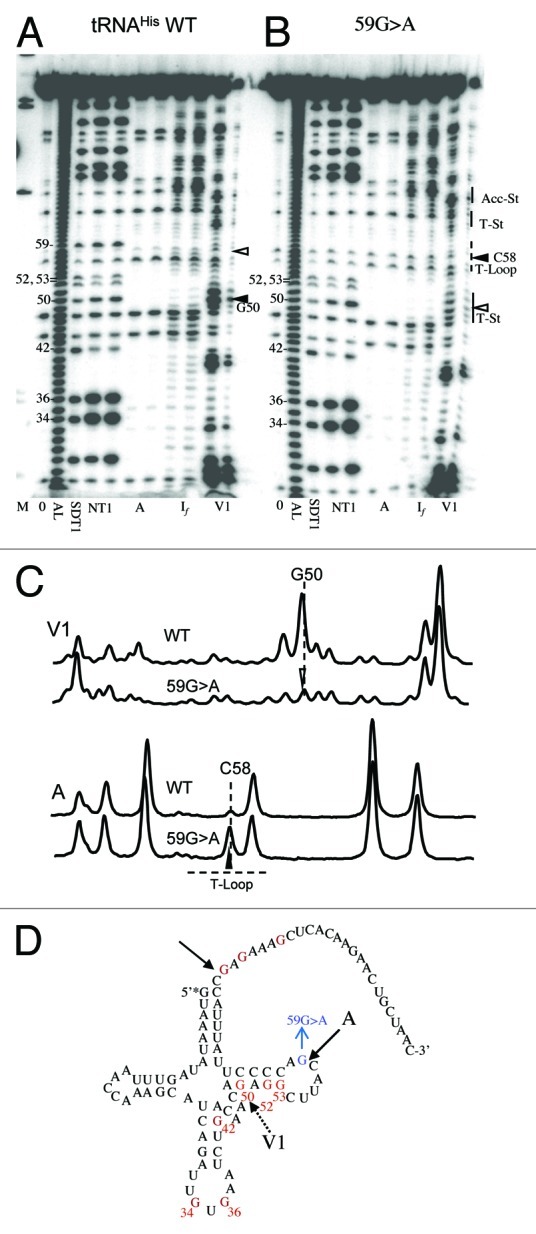
Figure 7. Structure probing of tRNAHis WT and 59G > A variant. Designations are the same as in Figure 4.
Discussion
All the pathogenesis related T-loop substitutions reduce tRNase Z processing efficiency (Table 2). Most notably, KM increases more than 3-fold with tRNAIle 59A > G and tRNAGly 59A > G, consistent with weaker binding between the flexible arm of tRNase ZL and the T-loops of these variant tRNAs, and compatible with the observed structural changes. T-loop substitutions often cause both kcat and KM to increase (Table 231,42), which could be explained if product release is the overall rate-limiting step in catalysis and the substitution reduces inhibition by product, as previously suggested.31,46
Most of the observed structure changes are within the T-loop. Since tRNALeu(UUR) has the canonical T-loop sequence and GG at the corresponding positions in the D-loop (nt 18, 19), compatible with D/T loop tertiary contacts, it was reasonable to look for corresponding structure changes in the D loop, which were not, however, observed (data not shown). Because the canonical T-loop is internally structured with a U-turn and a T54-A58 base pair across the loop, the observed structure changes could arise principally from local rearrangements. Additionally, three of the tRNAs analyzed have a structurally weak T-stem (a C/A mismatch in tRNAIle and tRNAHis and four out of five A/U pairs in tRNAGly. Structure changes triggered by T-loop substitutions could thus propagate laterally beyond the T-loop into the already weak T-stems.
All the mutations analyzed cause structure changes and reduce the catalytic efficiency of tRNase Z. Analysis of mitochondrial tRNAs with pathogenesis-related T-loop substitutions reveals a previously unnoticed richness of internal T-loop structure. The most consistently observed patterns are changes in susceptibility to RNase A at C↓A steps and association of structure changes with increases in KM in the 59A > G substitutions with tRNAIle and tRNAGly. Effects of the mutations on tRNase Z processing are mild, but the window of pathogenicity model (reviewed in ref. 9) argues that the most damaging mutations would seldom be observed in human patients due to lethality. If the effective concentration of tRNase Z is limiting in human mitochondria, the reduced processing efficiency could contribute to the pathomechanism of mutation. Observed structure changes could also affect other steps in tRNA maturation, aminoacylation or function of the tRNAs in the translation cycle.
Methods
Selection and preparation of tRNA precursors
Three pathogenesis-related T loop mutations are found in tRNALeu(UUR) [56C > A, associated with encephalopathy47; 57A > G, associated with mitochondrial myopathy48 and 60U > C, associated with Mitochondrial Encephalopathy, Lactic Acidosis, and Stroke-like episodes (MELAS)49; Fig. 2A] and three have a substitution at nucleotide 59 in different tRNAs (tRNAIle 59 A > G, associated with Fatal Infantile Cardiomyopathy plus a MELAS-associated cardiomyopathy [FICP47]; tRNAGly 59A > G—associated with Sudden Infant Death Syndrome [SIDS51], and tRNAHis 59G > A, associated with Maternal Inherited Cardiomyopathy49 and Optic Neuropathy50; Fig. 2B-D).
Three of the four T-loops are canonical in length. tRNAGly is short by one nucleotide; the numerical ambiguity thus caused may be resolved by numbering from the 5′ side (54-55-56) and from the 3′ side (60-59-58) with the missing nucleotide in the middle (57).
Ten mitochondrially encoded pre-tRNA genes (wild types and the pathogenesis-related variants of tRNALeu(UUR), tRNAIle, tRNAGly and tRNAHis; see Figure 2 for secondary structures) were constructed using long overlapping oligonucleotide primers (Sigma-Genosys). A natural sequence was included at the 3′ end of each precursor long enough to distinguish between the tRNase Z substrate and product using gel assays: 38 nt for tRNALeu(UUR), 25 nt for tRNAHis, and 20 nt for tRNAIle and tRNAGly, with SmaI runoff sites for tRNALeu(UUR) and tRNAIle, DraI for tRNAGly and PstI for tRNAHis (see C panels of Fig. 4–7 for the precursor sequences).
For T7 transcription and to use a cis-acting hammerhead ribozyme to cleave at +1 of the tRNAs,52 constructs begin with a T7 promoter followed by strong start (GGGAGA), a 5–7 nt hybridization box to target the hammerhead to +1, the hammerhead, tRNA gene, 3′ trailer and runoff site. A short universal forward primer consisting of the EcoRI subcloning site, T7 promoter and strong start and a short tRNA-specific reverse primer consisting of the BamHI subcloning site, runoff site and enough tRNA sequence to anneal specifically were used for primer extension/amplification with VENT DNA polymerase (New England Biolabs). Inserts were subcloned into the small high copy vector pHC624 and confirmed by sequencing (Genewiz).
Unlabeled transcripts were prepared from cloned runoff templates with T7 RNA polymerase accompanied by hammerhead self-cleavage as previously described15,54 with additional separate hammerhead reactions if necessary. tRNA precursors were gel purified, extracted by diffusion and recovered by ethanol precipitation. tRNA concentrations were determined by A260 using a conversion factor of 950,000 A260 M−1 for tRNALeu(UUR) and 875,000 A260 M−1 for tRNAs with shorter 3′ trailers. tRNA precursors were 5′ end-labeled with T4 polynucleotide kinase and [γ-32P]ATP for 30 min at 37°C, gel purified, visualized by phosphorimaging and recovered.
3′ Processing
Human tRNase ZL was baculovirus-expressed and affinity purified as described.15 Twenty-five microliter processing reactions were performed in a buffer containing 25 mM K-MOPS pH 6.75, 2 mM MgCl2 (3 mM CaCl2 for tRNALeu(UUR)), 1 mM dithiothreitol, 4 units/ml RNasin, and 100 μg/ml bovine serum albumin at 37°C. Five μl samples were taken after 5, 10, and 15 min of incubation, added to 2.5 μl formamide marker dye mix, and electrophoresed on denaturing 6% polyacrylamide gels. The gels were dried and exposed overnight using phosphor storage plates, scanned with a Typhoon imager (GE Life Sciences), and analyzed using ImageQuant software.
To determine processing efficiencies, reactions were performed at several different enzyme concentrations with a variant and simultaneously with the corresponding wild type pre-tRNA using a trace amount of labeled RNA and no added unlabeled substrate. Under these conditions, the % product/min of reaction (V/[S]) approximates the first order rate constant kcat/KM. Steady-state kinetic experiments were performed at a tRNase ZL concentration of 10 pM using 2, 5, 10, 20, and 50 nM unlabeled substrate with a constant (trace) concentration of labeled substrate for visualization. For tRNAIle 59A > G, both tRNase ZL concentration and the substrate range were 5X higher than for the other reactions. Michaelis-Menten plots (SigmaPlot) were used to determine kcat, KM, and processing efficiencies (kcat/KM; Fig. 3; Table 2) and the same parameters relative to wild type for each variant. Concentrations of tRNase Z and unlabeled tRNA were independently checked using fluorescently stained protein and RNA gels with appropriate standards and corrections were introduced for calculation of kinetic parameters. Experiments were repeated until acceptable standard errors were obtained for all kinetic parameters.
Structure Probing
Structure probing was performed as previously described (13). Under non-denaturing conditions, T1 cleaves after unstructured Gs, RNase A after unstructured pyrimidines, If in unstructured regions and V1 in stems and otherwise structured regions. Although not strictly confined to structured RNA regions,53 nuclease V1 is reliable enough to make useful comparisons between wild type and variant.
Reactions were terminated with 5 μl formamide marker dye mix and placed at -20°C. Concentrations of 32P-labeled tRNAs were sufficient for visualization on overnight exposure. Six microliter samples were loaded directly and electrophoresed on 6% and 8% denaturing urea-polyacrylamide gels. Imaging and analysis were performed as described above. A RNA refolding protocol did not affect results.
Acknowledgments
We are grateful to Rita Rai, Kyla-Gaye Pinnock, Sabina Paul and Christopher Wilson for technical assistance, and to Mel Silberklang for helpful discussions. Support from the National Institutes of Health (grants SC3GM08153 and R15CA120072) is gratefully acknowledged.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/19025
References
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.MITOMAP. A Human Mitochondrial Genome Database. http://www.mitomap.org, 2008. [DOI] [PMC free article] [PubMed]
- 3.Florentz C, Sohm B, Tryoen-Tóth P, Pütz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–75. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittenhagen LM, Kelley SO. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci. 2003;28:605–11. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Florentz C, Sissler M. Disease-related versus polymorphic mutations in human mitochondrial tRNAs. Where is the difference? EMBO Rep. 2001;2:481–6. doi: 10.1093/embo-reports/kve111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toompuu M, Levinger LL, Nadal A, Gomez J, Jacobs HT. The 7472insC mtDNA mutation impairs 5′ and 3′ processing of tRNA(Ser(UCN)) Biochem Biophys Res Commun. 2004;322:803–13. doi: 10.1016/j.bbrc.2004.07.181. [DOI] [PubMed] [Google Scholar]
- 7.Montoya J, Ojala D, Attardi G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–70. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 8.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–4. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 9.Levinger L, Mörl M, Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–41. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–74. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, et al. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265:16216–20. [PubMed] [Google Scholar]
- 12.Levinger L, Jacobs O, James M. In vitro 3′-end endonucleolytic processing defect in a human mitochondrial tRNA(Ser(UCN)) precursor with the U7445C substitution, which causes non-syndromic deafness. Nucleic Acids Res. 2001;29:4334–40. doi: 10.1093/nar/29.21.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levinger L, Giegé R, Florentz C. Pathology-related substitutions in human mitochondrial tRNA(Ile) reduce precursor 3′ end processing efficiency in vitro. Nucleic Acids Res. 2003;31:1904–12. doi: 10.1093/nar/gkg282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levinger L, Oestreich I, Florentz C, Mörl M. A pathogenesis-associated mutation in human mitochondrial tRNALeu(UUR) leads to reduced 3′-end processing and CCA addition. J Mol Biol. 2004;337:535–44. doi: 10.1016/j.jmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Zareen N, Levinger L. Naturally occurring mutations in human mitochondrial pre-tRNASer(UCN) can affect the transfer ribonuclease Z cleavage site, processing kinetics, and substrate secondary structure. J Biol Chem. 2006;281:3926–35. doi: 10.1074/jbc.M509822200. [DOI] [PubMed] [Google Scholar]
- 16.Schaller A, Desetty R, Hahn D, Jackson CB, Nuoffer JM, Gallati S, et al. Impairment of mitochondrial tRNAIle processing by a novel mutation associated with chronic progressive external ophthalmoplegia. Mitochondrion. 2011;11:488–96. doi: 10.1016/j.mito.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffer S, Rösch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 2002;21:2769–77. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tavtigian SV, Simard J, Teng DHF, Abtin V, Baumgard M, Beck A, et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–80. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 19.Schilling O, Späth B, Kostelecky B, Marchfelder A, Meyer-Klaucke W, Vogel A. Exosite modules guide substrate recognition in the ZiPD/ElaC protein family. J Biol Chem. 2005;280:17857–62. doi: 10.1074/jbc.M500591200. [DOI] [PubMed] [Google Scholar]
- 20.Levinger L, Hopkinson A, Desetty R, Wilson C. Effect of changes in the flexible arm on tRNase Z processing kinetics. J Biol Chem. 2009;284:15685–91. doi: 10.1074/jbc.M900745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao W, Yu H, Li S, Huang Y. Identification and analysis of candidate fungal tRNA 3′-end processing endonucleases tRNase Zs, homologs of the putative prostate cancer susceptibility protein ELAC2. BMC Evol Biol. 2010;10:272. doi: 10.1186/1471-2148-10-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan X, Yang J, Li J, Yu H, Dai H, Liu J, et al. The fission yeast Schizosaccharomyces pombe has two distinct tRNase Z(L)s encoded by two different genes and differentially targeted to the nucleus and mitochondria. Biochem J. 2011;435:103–11. doi: 10.1042/BJ20101619. [DOI] [PubMed] [Google Scholar]
- 23.Fan L, Wang Z, Liu J, Guo W, Yan J, Huang Y. A survey of green plant tRNA 3′-end processing enzyme tRNase Zs, homologs of the candidate prostate cancer susceptibility protein ELAC2. BMC Evol Biol. 2011;11:219. doi: 10.1186/1471-2148-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 2004;32:255–62. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie X, Dubrovskaya VA, Dubrovsky EB. RNAi knockdown of dRNaseZ, the Drosophila homolog of ELAC2, impairs growth of mitotic and endoreplicating tissues. Insect Biochem Mol Biol. 2011;41:167–77. doi: 10.1016/j.ibmb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Mineri R, Pavelka N, Fernandez-Vizarra E, Ricciardi-Castagnoli P, Zeviani M, Tiranti V. How do human cells react to the absence of mitochondrial DNA? PLoS One. 2009;4:e5713. doi: 10.1371/journal.pone.0005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossmanith W. Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS One. 2011;6:e19152. doi: 10.1371/journal.pone.0019152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–26. doi: 10.4161/rna.8.4.15393. [DOI] [PubMed] [Google Scholar]
- 29.Helm M, Brulé H, Friede D, Giegé R, Pütz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–79. doi: 10.1017/S1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClain WH, Guerrier-Takada C, Altman S. Model substrates for an RNA enzyme. Science. 1987;238:527–30. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- 31.Levinger L, Bourne R, Kolla S, Cylin E, Russell K, Wang X, et al. Matrices of paired substitutions show the effects of tRNA D/T loop sequence on Drosophila RNase P and 3′-tRNase processing. J Biol Chem. 1998;273:1015–25. doi: 10.1074/jbc.273.2.1015. [DOI] [PubMed] [Google Scholar]
- 32.Shi PY, Weiner AM, Maizels N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA. 1998;4:276–84. [PMC free article] [PubMed] [Google Scholar]
- 33.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–6. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucleic Acids Res. 2002;30:3592–601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominski Z. Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 36.Li de la Sierra-Gallay I, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–61. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- 37.Karkashon S, Hopkinson A, Levinger L. tRNase Z catalysis and conserved residues on the carboxy side of the His cluster. Biochemistry. 2007;46:9380–7. doi: 10.1021/bi700578v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li de la Sierra-Gallay I, Mathy N, Pellegrini O, Condon C. Structure of the ubiquitous 3′ processing enzyme RNase Z bound to transfer RNA. Nat Struct Mol Biol. 2006;13:376–7. doi: 10.1038/nsmb1066. [DOI] [PubMed] [Google Scholar]
- 39.Ishii R, Minagawa A, Takaku H, Takagi M, Nashimoto M, Yokoyama S. Crystal structure of the tRNA 3′ processing endoribonuclease tRNase Z from Thermotoga maritima. J Biol Chem. 2005;280:14138–44. doi: 10.1074/jbc.M500355200. [DOI] [PubMed] [Google Scholar]
- 40.Kostelecky B, Pohl E, Vogel A, Schilling O, Meyer-Klaucke W. The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins. J Bacteriol. 2006;188:1607–14. doi: 10.1128/JB.188.4.1607-1614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishii R, Minagawa A, Takaku H, Takagi M, Nashimoto M, Yokoyama S. The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:637–41. doi: 10.1107/S1744309107033623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopkinson A, Levinger L. Effects of conserved D/T loop substitutions in the pre-tRNA substrate on tRNase Z catalysis. RNA Biol. 2008;5:104–11. doi: 10.4161/rna.5.2.6086. [DOI] [PubMed] [Google Scholar]
- 43.Pütz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA. 2007;13:1184–90. doi: 10.1261/rna.588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–66. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 46.Beebe JA, Fierke CA. A kinetic mechanism for cleavage of precursor tRNA(Asp) catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry. 1994;33:10294–304. doi: 10.1021/bi00200a009. [DOI] [PubMed] [Google Scholar]
- 47.Anitori R, Manning K, Quan F, Weleber RG, Buist NR, Shoubridge EA, et al. Contrasting phenotypes in three patients with novel mutations in mitochondrial tRNA genes. Mol Genet Metab. 2005;84:176–88. doi: 10.1016/j.ymgme.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Hadjigeorgiou GM, Kim SH, Fischbeck KH, Andreu AL, Berry GT, Bingham P, et al. A new mitochondrial DNA mutation (A3288G) in the tRNA(Leu(UUR)) gene associated with familial myopathy. J Neurol Sci. 1999;164:153–7. doi: 10.1016/S0022-510X(99)00062-3. [DOI] [PubMed] [Google Scholar]
- 49.Goto Y, Tsugane K, Tanabe Y, Nonaka I, Horai S. A new point mutation at nucleotide pair 3291 of the mitochondrial tRNA(Leu(UUR)) gene in a patient with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) Biochem Biophys Res Commun. 1994;202:1624–30. doi: 10.1006/bbrc.1994.2119. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka M, Obayashi T, Yoneda M, Kovalenko SA, Sugiyama S, Ozawa T. Mitochondrial DNA mutations in cardiomyopathy: combination of replacements yielding cysteine residues and tRNA mutations. Muscle Nerve. 1995;3:S165–9. doi: 10.1002/mus.880181432. [DOI] [PubMed] [Google Scholar]
- 51.Santorelli FM, Schlessel JS, Slonim AE, DiMauro S. Novel mutation in the mitochondrial DNA tRNA glycine gene associated with sudden unexpected death. Pediatr Neurol. 1996;15:145–9. doi: 10.1016/0887-8994(96)00163-4. [DOI] [PubMed] [Google Scholar]
- 52.Shin WS, Tanaka M, Suzuki J, Hemmi C, Toyo-oka T. A novel homoplasmic mutation in mtDNA with a single evolutionary origin as a risk factor for cardiomyopathy. Am J Hum Genet. 2000;67:1617–20. doi: 10.1086/316896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mimaki M, Ikota A, Sato A, Komaki H, Akanuma J, Nonaka I, et al. A double mutation (G11778A and G12192A) in mitochondrial DNA associated with Leber’s hereditary optic neuropathy and cardiomyopathy. J Hum Genet. 2003;48:47–50. doi: 10.1007/s100380300005. [DOI] [PubMed] [Google Scholar]
- 54.Fechter PJ, Rudinger J, Giegé R, Théobald-Dietrich A. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 1998;436:99–103. doi: 10.1016/S0014-5793(98)01096-5. [DOI] [PubMed] [Google Scholar]
- 55.Maizels N, Weiner AM, Yue D, Shi PY. New evidence for the genomic tag hypothesis: archaeal CCA-adding enzymes and tDNA substrates. Biol Bull. 1999;196:331–3, discussion 333-4. doi: 10.2307/1542963. [DOI] [PubMed] [Google Scholar]