Abstract
Nuclear RNA decay factors are involved in many different pathways including rRNA processing, snRNA and snoRNA biogenesis, pre-mRNA processing, and the rapid decay of cryptic intergenic transcripts. In contrast to its yeast counterpart, the mammalian nuclear decay machinery is largely uncharacterized. Here we report interactions of several putative components of the human nuclear RNA decay machinery, including the TRAMP complex protein Mtr4 and the nuclear exosome constituents PM/Scl-100 and PM/Scl-75, with components of the U4/U6.U5 tri-snRNP complex required for pre-mRNA splicing. The tri-snRNP component Prp31 interacts indirectly with Mtr4 and PM/Scl-100 in a manner that is dependent on the phosphorylation sites in the middle of the protein, while Prp3 and Prp4 interact with the nuclear decay complex independent of Prp31. Together our results suggest recruitment of the nuclear decay machinery to the spliceosome to ensure production of properly spliced mRNA.
Keywords: Mtr4, PM/Scl-100, Prp31, nuclear RNA decay, spliceosome, tri-snRNP
Introduction
Nuclear machinery not only synthesizes and processes several different types of coding and non-coding RNA molecules, but also ensures production of functionally competent RNAs by selective degradation of aberrant species. More specifically, the nuclear RNA degradation apparatus is known to participate in the proper processing of snRNAs, snoRNAs,1 and rRNAs,2,3 as well as in the decay of cryptic transcripts.4 Moreover, it plays crucial roles in gene silencing5 and in the quality control of tRNAs6 and mRNAs.7-14
Two macromolecular complexes, the Trf4/Air2/Mtr4 Polyadenylation (TRAMP) complex and the nuclear exosome, contribute both to these processing events and to nuclear RNA surveillance.10,15-19 Many non-coding RNAs are generated by endonucleolytic cleavage, followed by 3′-polyadenylation and processing of the polyadenylated precursor.1 The polyadenylation (TRAMP) complex that marks RNAs for degradation or processing consists of a noncanonical poly(A) polymerase (Trf4/Trf5), a Zn knuckle-containing RNA binding protein (Air1/Air2), and an RNA helicase Mtr4.20,21 The nuclear exosome that is responsible for degradation or processing contains a nuclear-specific subunit Rrp6 (PM/Scl-100 in humans) in addition to the nine-subunit exosome core.22-24 While Rrp6 is exclusively localized in the nucleus in Saccharomyces cerevisiae, the human homolog PM/Scl-100 is predominantly nuclear with a minor fraction present in the cytoplasm.25,26 Mutations in yeast factors associated with RNA splicing (Prp2), with splicing and export (Sub2), with transcription (THO), and with polyadenylation (Rna14, Rna15) have revealed a critical role for the nuclear exosome-specific subunit Rrp6 in counteracting the generation of aberrant messages produced in these mutant cells.7,8,11,14
Communication between all these nuclear events—transcription, splicing, cleavage, polyadenylation, and nuclear RNA degradation—is pivotal to the formation of functional eukaryotic RNAs.27 The complex steps of splicing must be strictly orchestrated in order to avoid creating aberrant messages. Adding an extra layer of complexity is alternative splicing in higher eukaryotes, which plays a regulatory role in generating mRNAs in a tissue-specific manner and can be important in disease (for review see ref. 28).
Although there are relatively few genes that undergo splicing in S. cerevisiae, a connection between the splicing and the nuclear decay machineries has been established. Bousquet-Antonelli et al. showed that levels of pre-mRNA remained low after disrupting splicing, either with a temperature-sensitive allele of Prp2 (a splicing factor required for the first transesterification step) or with a 3′-splice site mutation in the intron.8 However, inhibition of degradation by the nuclear exosome resulted in accumulation of both pre-mRNAs and spliced RNAs, suggesting a competition between splicing and degradation of pre-mRNAs.
In vertebrates, communication between splicing and the nuclear degradation machinery is likewise expected, but no such interactions have been reported. The TRAMP complex is conserved in T. brucei,29,30 D. melanogaster,31 S. cerevisiae and S. pombe, as is the exosome, including the nuclear-specific subunit Rrp6(PM/Scl-100).10,25,32 Yet, both the TRAMP complex and the exosome are poorly characterized in mammalian cells, and the current literature reports only a few investigations of the Trf4-mediated TRAMP complex and its connection with the Rrp6-mediated exosome complex.2,33,34 Two human proteins, Trf4-1 (pols) and Trf4-2 (papd5), share 37% identity and 56% similarity with the yTrf4 protein. Recently, Shcherbik et al. identified a role of papd5 in degradation of transcripts generated by RNA polymerase I.34 However, the composition of the mammalian complexes that include these proteins is yet to be reported.
We set out to characterize the human homolog of the yeast TRAMP complex, which may play a role in degradation of aberrant RNAs in the nucleus of mammalian cells. Here we report interactions of the nuclear decay factors Mtr4 and PM/Scl-100 (and -75) with tri-snRNP-associated proteins in HeLa and HEK293 cells. Our results suggest a possible mechanism for recruitment of the nuclear decay machinery to splicing complexes.
Results
Mammalian homologs of TRAMP complex components Trf4-1 and Mtr4 associate with the tri-snRNP protein Prp31
To isolate factors associated with human Trf4 proteins, a FLAG-tagged Trf4-1 was expressed in HEK293 cells, followed by extract preparation and immunoprecipitation with anti-FLAG antibody in the presence of ribonuclease (RNase) A. Immunoprecipitated proteins were eluted using FLAG peptide and were subjected to LC-MS/MS (Table S1). Surprisingly, among many proteins that may be non-specific background (for example, Gemin4 in Fig. 1A), we identified a tri-snRNP-specific splicing factor, Prp31, whose enrichment was verified by western blot analysis of the immunoprecipitate (IP) (Fig. 1A). Prp31 is part of the U4/U6 complex and plays an important role in the stability and integrity of the spliceosome.35-38 It is also linked to autosomal dominant retinitis pigmentosa, a genetic eye condition that progressively leads to incurable blindness.39
Figure 1. Human Trf4-1 and Mtr4 interact with the tri-snRNP component Prp31. (A) western blot analysis of proteins immunoprecipitated from nuclear extract of HEK293T cells expressing FLAG-tagged Trf4-1 using anti-FLAG agarose beads; the blot was probed with anti-Prp31 antibody (rabbit). (B) Anti-Prp31 antibody (Abnova, mouse polyclonal) was used for immunoprecipitation from HeLa nuclear extract after RNaseA treatment. The IP was run on SDS-PAGE, transferred and probed with anti-Mtr4 antibody (top panel) and rabbit anti-Prp31 (Lührmann laboratory) (bottom panel). (C) Immunoprecipitation was performed from 50 μl nuclear extract using rabbit anti-Prp31 or anti-Mtr4 (Novus Biologicals, NB100–1575) antibody after treatment with RNase A (100 ng/μl). Alkaline phosphatase (5U, FastAP, Fermentas) was added to the washed IP sample according to the manufacturer’s protocol, followed by further washing to remove disengaged proteins. The IPs were western blotted with anti-Mtr4 or rabbit anti-Prp31 antibody. Lanes 3–5, which were probed with anti-Prp31, show a shorter exposure of the same blot as in lanes 1–2 and 6–9.
Finding a splicing factor associated with a putative nuclear RNA decay factor suggests potential regulation of RNA processing. Since we discovered the presence of a tri-snRNP protein Prp31 in the Trf4-1 coimmunoprecipitate, we sought to establish whether this interaction serves to recruit the nuclear decay machinery to the spliceosome. Therefore, we asked if other members of the nuclear RNA decay complex (Mtr4) or of the exosome machinery (PM/Scl-100) also associate with Prp31. Immunoprecipitation of Prp31 using an anti-Prp31 antibody (mouse polyclonal, Abnova) immunoprecipitated a small fraction of Prp31 from the nuclear extract and resulted in modest but clear coimmunoprecipitation of Mtr4 with RNaseA treatment (Fig. 1B). In contrast, a rabbit polyclonal antibody35 raised against the C-terminus (aa484–497) of Prp31 failed to coimmunoprecipitate Mtr4 (Fig. 1C, lanes 4 and 5; for details see below). A reverse IP of Mtr4 from nuclear extract using an anti-Mtr4 antibody in the presence of RNaseA, followed by probing for Prp31 revealed low intensity bands of slightly slower mobility than the bulk of Prp31, which partially overlaps with antibody heavy chain (Fig. 1C, lanes 7–9). These slower migrating bands could reflect posttranslationally modified Prp31, which carries several putative phosphorylation sites (www.phosphosite.org/proteinAction.do?id=8981&showAllSites=true) (Fig. 3). Recently Schneider et al. thoroughly characterized some of these phosphorylation sites on Prp31.37 Treatment of the extract with alkaline phosphatase prior to immunoprecipitation with anti-Mtr4 modestly reduced the intensity of the upper band (25% decrease) in the Prp31 blot (Fig. 1C, lane 8).
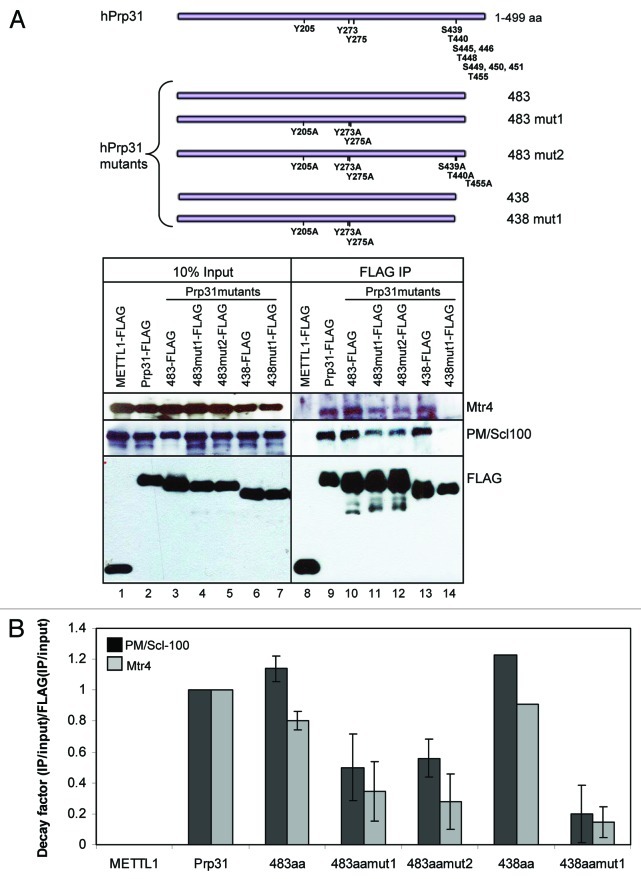
Figure 3. Interaction between Prp31 and Mtr4 is dependent on the phosphorylation sites of Prp31. (A, top) FLAG-tagged mutants of hPrp31, which include two different deletions at the C-terminus (aa1–483 and aa1–438), as well as the indicated amino acid substitutions at specific positions. (A, bottom) Binding of Prp31 to Mtr4 depends on putative phosphorylation sites in the middle of Prp31. Each Prp31 mutant was individually expressed in HEK293 cells followed by lysate preparation in RIPA buffer followed by sonication. FLAG-tagged Prp31 mutants and associated proteins were selected on anti-FLAG M2 agarose beads followed by western blotting to identify the interacting partners indicated on the right. A longer exposure did not reveal significant binding of Mtr4 and PM/Scl-100 in lane 14 (data not shown). (B) Relative binding of PM/Scl-100 and Mtr4 to each FLAG-tagged Prp31 protein is graphed. The western blot was analyzed using Image J; bound fractions of Prp31, PM/Scl-100, and Mtr4 proteins were normalized to their respective inputs, and binding data were plotted for the normalized PM/Scl-100 and Mtr4 relative to the normalized FLAG proteins in the IP lanes. Results are presented as an average of two experiments except for the 438aa Prp31 determination, which was from one experiment.
Prp31 interacts with both Mtr4 and PM/Scl-100 and the interaction depends on the phosphorylation sites of Prp31
Since the major band of endogenous Prp31 runs very close to the antibody heavy chain and is therefore difficult to visualize, we expressed C-terminally FLAG-tagged Prp31 in HEK293 cells and performed coimmunoprecipitations after formaldehyde crosslinking of cells to stabilize weak interactions.40,41 Immunoprecipitation and washes were performed using radioimmunoprecipitation assay (RIPA) buffer along with RNaseA treatment. Mtr4 coimmunoprecipitated with FLAG-tagged Prp31 even in the absence of formaldehyde (Fig. 2, lane 8), indicating that the inability of rabbit anti-Prp31 to coimmunoprecipitate Mtr4 is not simply due to a particularly weak interaction between Mtr4 and Prp31. Moreover, FLAG-tagged Prp31 coimmunoprecipitated the exosome subunit PM/Scl-100 (Rrp6), which is possibly associated through Mtr4,42 even in the absence of formaldehyde crosslinking (Fig. 2, lanes 7 and 8). In contrast, Prp6, a U5-associated protein that interacts directly with Prp31,35 was present at much higher levels in the anti-FLAG IP after formaldehyde treatment (compare lanes 7 and 8). Likewise, visualization of the PM/Scl-75 component of the exosome in the FLAG-tagged Prp31 IP was dependent on formaldehyde (lanes 7 and 8). Even though the interactions of Prp6 and PM/Scl-75 seem weak, their presence suggests involvement of mature and functionally relevant complexes in this association. As a control, we used a FLAG-tagged protein with known nuclear localization, METTL1, which was unable to pull down Mtr4, Prp6, PM/Scl-100, or -75 (lane 6).
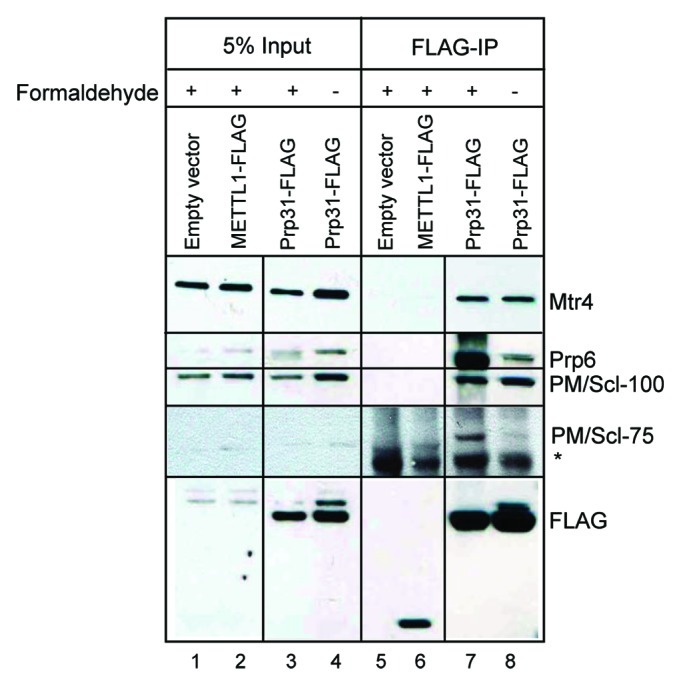
Figure 2. Prp31 forms a complex with both Mtr4 and PM/Scl-100. FLAG-tagged Prp31 was expressed in HEK293 cells and cell extracts were prepared with or without prior formaldehyde crosslinking using RIPA buffer followed by sonication. Anti-FLAG M2 conjugated agarose beads (Sigma) were used for immunoprecipitation followed by western blot analysis for the tri-snRNP-associated protein Prp6 and the exosome-associated proteins PM/Scl-100, PM/Scl-75 and Mtr4, as indicated on the right. * represents an unknown non-specifically reacting protein. Both lanes 2 and 3 and lanes 6 and 7 were initially separated by one lane, which was removed for presentation.
After confirming the interaction between Prp31 and Mtr4, we wanted to establish whether Mtr4 binding is specific for a posttranslationally modified form of Prp31. Since Prp31 has multiple potential phosphorylation sites, we introduced mutations into each cluster of sites in two Prp31 variants deleted at the C-terminus (aa1-483 and 1-438, Fig. 3A, top). Specifically, we first generated the truncated Prp31 (aa1-483) to ask whether the antibody-binding surface of Prp31 located in the C-terminus between aa484 and aa49735 is required for the association of Prp31 with Mtr4. This would explain the lack of Mtr4 coimmunoprecipitation with rabbit anti-Prp31 raised against this epitope (Fig. 1). Second, we created a further deletion (leaving aa1-438) of the C-terminus to remove several putative phosphorylation sites and also generated alanine mutations to disrupt other sites: mutant 1 (mut1) carries alanine mutations at sites 205, 273, and 275, while mutant 2 (mut2) has additional mutations to alanine in residues 439, 440, and 455. Mutants 1 and 2 were created in deletion mutant 1-483 (483mut1 and 483mut2), and mutant 1 was generated in the 1-438 (438mut1) truncated Prp31 (Fig. 3A).
Figure 3A shows that both Prp31 deletion mutants (483 and 438aa long) bind Mtr4 comparably to the wild-type protein (compare lanes 10 and 13 with lane 9), suggesting that neither the antibody binding site (aa484-497) nor the C-terminal phosphorylation sites (aa439-455) contribute to Mtr4 binding. A similar profile was obtained for PM/Scl-100 binding (Fig. 3A). However, mutations of the phosphorylation sites in the middle of the 483aa truncated protein (Y205A, Y273A, Y275A) resulted in diminished binding of Mtr4 (lane 11), suggesting that phosphorylation of these Prp31 residues strengthens the interaction with Mtr4. As expected from lane 13, additional mutations in the C-terminal phosphorylation sites (S439A, T440A, T445A) did not hinder the binding capacity further (compare lanes 11 and 12). Similar to the mutations in the 483aa protein, mutations of aa205, 273, and 275 in the 438aa Prp31 protein reduced binding of Mtr4, but even more efficiently (lane 14). We therefore conclude that the phosphorylation sites in the middle of the Prp31 molecule contribute significantly to its interaction with Mtr4 and PM/Scl-100. Quantification of the western blot data are shown in Figure 3B.
Mtr4 associates with other components of the tri-snRNP independent of Prp31
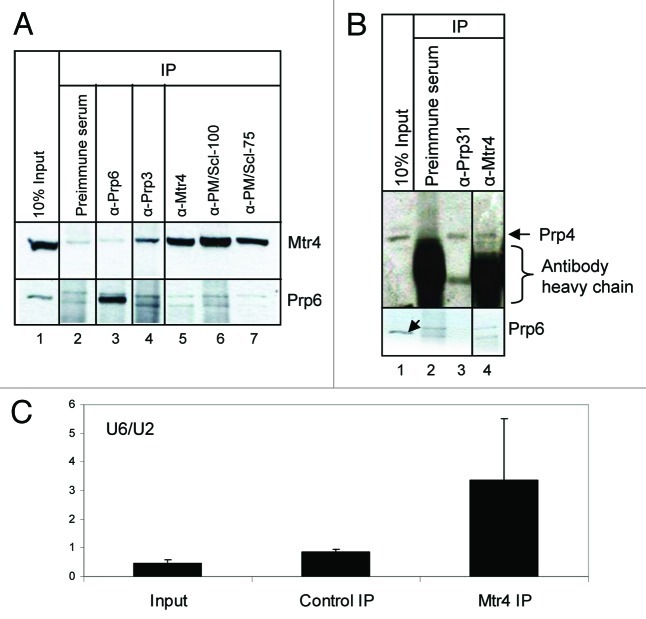
Since Prp31 is part of the U4/U6.U5 tri-snRNP, we asked whether Mtr4 interaction extends to other components of the tri-snRNP complex. We used available antibodies to probe the interactions of endogenous (rather than tagged) tri-snRNP proteins (Prp4 and Prp6) with Prp3, Prp31, Mtr4, PM/Scl-100, and PM/Scl-75 in nuclear extract treated with RNaseA (Fig. 4A). The anti-Prp3 IP contained significant amounts of Mtr4 (Fig. 4A, lane 4). Similarly, anti-Mtr4 appears to coimmunoprecipitate Prp4 (Fig. 4B, lane 4), a conclusion that is confirmed below (see Fig. 5). In contrast, none of the proteins detectably coimmunoprecipitated Prp6 (Fig. 4A), consistent with Prp6’s weak interaction with the Mtr4-Prp31 complex in the absence of formaldehyde (Fig. 2, lane 8). These results argue that Mtr4 associates with at least two other proteins of the U4/U6 complex (Prp3 and Prp4) and not just with a fraction of Prp31 that is not part of the tri-snRNP. We further assessed the association of snRNAs with these complexes (Fig. 4C). The anti-Mtr4 IP in the absence of RNaseA showed enrichment of U6 snRNA over U2, suggesting the presence of tri-snRNP rather than the whole spliceosome.
Figure 4. Mtr4 associates with tri-snRNP proteins Prp3 and Prp4. (A) Coimmunoprecipitation reveals interaction of Mtr4 with Prp3. Antibodies against several Prp proteins and decay factors were used for IP from RNase-treated HeLa nuclear extract followed by western blot analysis. (B) Prp4 associates with Mtr4 in a similar IP conducted using anti-Mtr4 and probing the precipitate with anti-Prp4. Lane 4 is from the same gel as lanes 1–3, with the same exposure time. (C) U6 and U2 snRNAs were analyzed using quantitative RT PCR of Mtr4 IPs performed without RNase treatment. Results are presented as an average of two experiments.
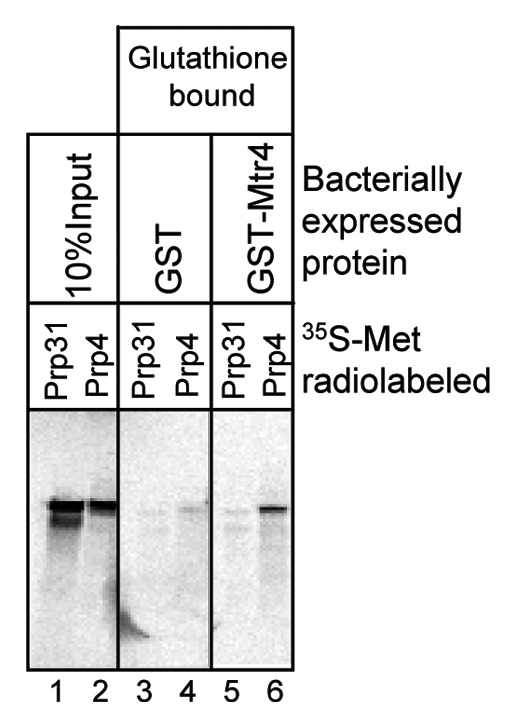
Figure 5. Prp4 interacts with Mtr4. Bacterially expressed GST-Mtr4 polypeptides were isolated on glutathione beads and incubated with 35S-Met radiolabeled Prp31 or Prp4 transcribed and translated (lanes 1 and 2) in reticulocyte lysate (Promega). Bound fractions were washed and fractionated by SDS-PAGE followed by autoradiography. Prp4 showed significant binding to GST-Mtr4 compared with GST. In this experiment, Prp31 consistently exhibited two bands (lane 1), which could be either a stalled or phosphorylated product.
To provide mechanistic insight into the recruitment of the nuclear exosome complex to the spliceosome, we sought to identify the direct interaction partner(s) of Mtr4 using bacterially-expressed Mtr4 and 35S-Met radiolabeled Prp31 and Prp4 proteins synthesized in reticulocyte lysates. Even though it was difficult to generate full-length GST-tagged Mtr4 protein (because of stalled translation of the protein even in Rosetta cells), we were able to verify binding of Prp4 to the GST-Mtr4 partial proteins (Fig. 5, lane 6). 35S-labeled Prp31 did not bind to the mixture of GST-Mtr4 products (Fig. 5, lane 5), which is compatible with the results of the depletion experiment shown in Figure 6. However, since the comparable experiment with Prp3 did not reveal binding to GST-Mtr4 above the background level (data not shown), we cannot rule out additional direct interactions with other di-snRNP proteins.
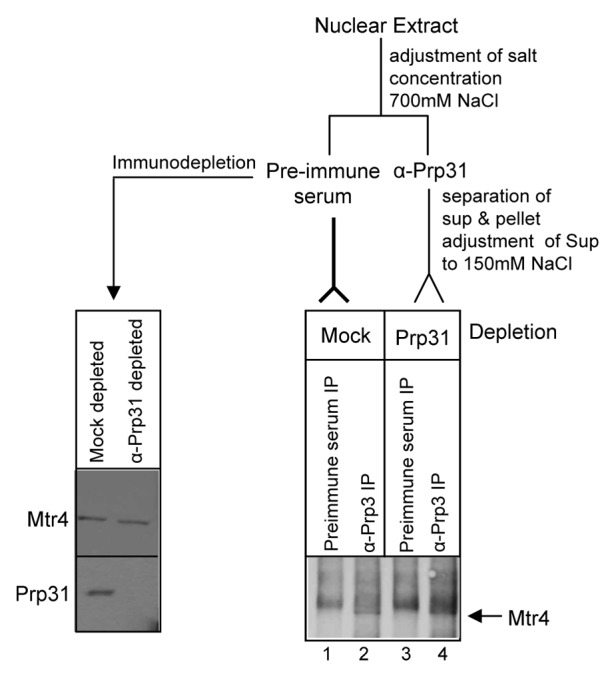
Figure 6. Mtr4 associates with tri-snRNP proteins independent of Prp31. Anti-Prp3 coimmunoprecipitates Mtr4 from extracts lacking Prp31. Prp31 was first immunodepleted from nuclear extract and the depleted extract was subjected to immunoprecipitation by anti-Prp3 antibody. Left panel: Immunodepletion was verified by checking with anti-Prp31 (bottom) and anti-Mtr4 (top). Right panel: Immunoprecipitated proteins were analyzed by western blotting with anti-Mtr4 antibody. The arrow indicates the position of Mtr4 in lanes 2 and 4, running just below the non-specific band appearing in all lanes. The smear above the Mtr4 band present in both control and Prp3 IP lanes (lanes 1–4) was consistently present when high salt conditions were used during the IP.
To determine whether Mtr4’s interaction with Prp3 and Prp4 proteins of the U4/U6 di-snRNP complex depends on Prp31, we performed immunoprecipitation of Prp3 from Prp31-depleted nuclear extract. Prp3 was chosen since it showed more efficient coimmunoprecipitation of Mtr4 than the other di-snRNP proteins (Fig. 4A and data not shown). At low salt concentration (150–200 mM KCl) the U4/U6.U5 tri-snRNP is stable and retains all its protein and RNA components. But at higher concentrations (300–400 mM) of KCl, the U5 snRNP dissociates from the U4/U6 complex; moreover, Prp31 dissociates from the U4/U6 di-snRNP.35,43 We performed immunodepletion of Prp31 at 700 mM NaCl where Prp31 can be immunoprecipitated alone.35,44 The salt concentration was then adjusted back to 150 mM NaCl, a condition in which Prp4 and Prp3 maintain their interaction.43 The resulting Prp31-depleted supernatant, as well as a mock-depleted supernatant, were subjected to immunoprecipitation by anti-Prp3 antibody. In both cases, Mtr4 appeared (Fig. 6, right panel, lanes 2 and 4, arrow), running just below the non-specific band present in the preimmune serum. This result confirms that Prp3 associates with Mtr4 even in the absence of Prp31. In combination with Figure 5, the data suggest that Mtr4 is possibly associated with U4/U6 snRNP components through Prp4, while its interaction with Prp3 and Prp31 could be through Prp4 or through other TRAMP complex components.
Discussion
We have demonstrated that components of the nuclear decay machinery, Mtr4 and PM/Scl-100, interact with several U4/U6-associated proteins (Prp31, Prp3, and Prp4) of the tri-snRNP complex within the spliceosome. These findings suggest a possible mechanism for recruitment of the decay machinery to the pre-mRNA processing apparatus to ensure generation of properly spliced RNA and decay of aberrant transcripts.
Communication between the splicing and decay machineries was anticipated from genetic interactions observed between the splicing factor Prp2 and subunits of the exosome in S. cerevisiae.8 Specifically, whereas a temperature-sensitive allele of Prp2 showed decreased levels of spliced RNA without an increase in the unspliced precursor at the non-permissive temperature, when combined with a deletion of Rrp6, unspliced precursor levels increased. This result suggested that unspliced precursors generated because of a defect in splicing were being degraded by the nuclear exosome complex.8 Likewise, defects in the THO/Sub2 complex, which is involved in mRNP biogenesis and export, have been reported to result in degradation of a specific pool of mRNAs by a pathway that includes both the TRAMP and exosome complexes.11 Finally, defects in mRNA cleavage and polyadenylation likewise result in decay of long read-through transcripts by an Rrp6-dependent mechanism.7,12
Our results offer a possible scenario for recruiting the exosome to the mammalian spliceosome through the U4/U6.U5 tri-snRNP complex, adding to our understanding of the mammalian decay process. Prp31 exhibited an RNaseA insensitive association with Mtr4 (Figs. 1–3). This interaction does not seem to be direct and is not itself principally responsible for the association between the tri-snRNP and TRAMP complexes since anti-Prp3 antibodies can immunoprecipitate Mtr4 from a Prp31-depleted extract (Fig. 6). Moreover, bacterially expressed GST-Mtr4 binds in vitro translated Prp4 (Fig. 5), suggesting that this is one of the principal interactions. Interestingly, mutations in three potential phosphorylation sites (Y205, Y273, and Y275) of Prp31 were able to disrupt association of Prp31 with Mtr4 significantly. The X-ray crystal structure of Prp31 shows that residue Y205 does not interact with the 15.5k protein of the U4 snRNP but is exposed and could bind some other component.38 Residues Y273 and Y275 are located in a loop within a cleft that is in close proximity to the U4 snRNA binding site; however, the side chains point away from the U4 RNA. Although the mechanism by which phosphorylation enhances Prp31 interaction with Mtr4 is not clear, it is possible that aa 205, 273, and 275 contribute to stability of the U4/U6 complex and that other proteins of this complex bridge the interaction of Prp31 with Mtr4. Recent work from the Lührmann laboratory suggests that phosphorylation of Prp31 by the Prp4 kinase induces assembly and stabilization of the spliceosomal B complex.37 Our work supports the idea that even though Mtr4 directly binds to the tri-snRNP through Prp4, phosphorylated Prp31 enhances the stability of the interaction between the spliceosome and the exosome.
Prp31 is evolutionarily conserved and the human protein shares 25% identity and 60% similarity with its homolog in budding yeast.35 Interestingly, the Prp31 homolog in S. cerevisiae is conserved in some residues including Y273 but lacks many other tyrosine, threonine, and serine residues present in hPrp31. Budding yeast also lacks the Prp4 kinase that is known to be responsible for phosphorylating both Prp31 and Prp6 in humans.37 Thus, even though our Y205A, Y273A, and Y275A triple mutant of human Prp31 is significantly deficient in binding Mtr4, in budding yeast there must exist an alternative way of recruiting the exosome complex. Perhaps with the lower complexity of splicing in yeast, fewer contacts are needed between the decay and the splicing machineries.
Mass spectrometric analysis of spliceosomal proteins at various stages has demonstrated recruitment of Mtr4 to the B complex and its continuous association with the C complex (supplementary data, ref. 43). The presence of exosome subunits in purified spliceosomes has also been reported [supplementary data, ref. 48]. Since activation of the spliceosomal B complex results in the dissociation of many U4/U6-associated proteins, including Prp4, Prp3, and Prp31, the presence of Mtr4 in the C complex suggests that its association with the active spliceosome is achieved through some other constituent(s), possibly hnRNPs.45 Whereas the tri-snRNP may act as the platform for initial recruitment of the nuclear exosome-mediated decay complex to spliceosomes, other proteins such as transcription factors can cotranscriptionally recruit the nuclear exosome to a pre-mRNA. For example, Andrulis et al. demonstrated interactions between the nuclear exosome subunits dRrp6 and dSpt6, elongation factor dSpt5, and RNA polymerase II in D. melanogaster.46 Using Chironomus and Drosophila, Hessle et al. showed that the core exosome subunit Rrp4 is associated with polytene chromosomes and that this association is dependent on hnRNP M.45
Observations that the exosome can interact with multiple machineries suggest that production of a fully functional mRNA may be ensured through several different checkpoints. The exosome makes several contacts with the transcription and RNA processing machineries, but whether these interactions are mediated by the TRAMP complex is unknown. Our results show that both the TRAMP component Mtr4 and the exosome subunit PM/Scl-100 (and -75) exhibit similar binding interactions with the U4/U6 complex. In addition, we cannot rule out the possibility that multiple members of the TRAMP complex make separate contacts with one or more tri-snRNP components. Our mass spectrometric analysis was unable to detect any Mtr4 in an anti-Trf4-1 IP. The absence of Mtr4 may be due to technical reasons, as evidenced by the inconsistent presence of exosome components in the spliceosome analyzed by mass spectrometry.47 Our understanding is limited by the absence of a fully-characterized human TRAMP complex. Further work will be necessary to understand the contribution of different Trf4 homologs and their isoforms to functional TRAMP complexes, and to the stability and integrity of these complexes in the absence of an RNA substrate.
It will be interesting to learn how and when the decay proteins dissociate after completion of a splicing event. Their association with the splicing machinery may simply reflect a coupled process whereby newly excised introns are rapidly degraded. Communication between cytoplasmic nonsense-mediated decay and alternative splicing is one avenue for achieving proper gene expression.26 Since the majority of mammalian transcripts are regulated by alternative splicing, the role of nuclear decay factors in determining the correct splice variant may be another important aspect of surveillance.
Materials and Methods
Plasmids and cloning
Primers used to generate cDNAs for Trf4-1, Mtr4, and Prp31 are listed in Table S2 along with the primers that were used for introducing mutations in Prp31. hTrf4-1 cDNA was generated by primers AN101 and AN102 and was introduced into the HindIII site of pcDNA3.1 carrying a FLAG tag upstream. METTL1-FLAG is described in Alexandrov et al.48 Primers AN189 and AN190 were then used to generate Prp31 cDNA with a C-terminal FLAG sequence, which was inserted in the BamHI-XhoI site. AN202, AN203, AN204, and AN205 were used to introduce the Y205A, Y273A, and Y275A mutations (mut1), whereas AN206, AN207, AN210, and AN211 were used to generate additional S439A, T440A, and S455A mutations (mut2) into the C-terminally FLAG-tagged Prp31 by site-directed mutagenesis. Primers AN189/AN225 and AN189/AN226 were used to generate truncated Prp31 proteins, aa1–483 and aa1–438. Mtr4 cDNA was generated using AN110 and AN232 primers and was inserted into the XhoI site of the pGEX-6P-3 construct (GE Life Healthcare Sciences). Plasmids used for in vitro transcription/translation of Prp proteins were obtained from Open Biosystems.
Extract preparation and immunoprecipitation
FLAG-tagged proteins were expressed in HEK293 cells by transfecting appropriate plasmids. 48 h later, cells were harvested and washed with ice-cold PBS. Cells were then incubated in 1 packed cell volume (PCV) of buffer A (10 mM Hepes, 1.5 mM MgCl2, 100 mM KCl, 0.1 mM DTT), followed by passage through a 25 5/8G needle attached to a 1 ml syringe 8 times. The cell lysate was centrifuged at 12000 g. The supernatant was discarded and the pellet that contains nuclei was collected. The pellet was incubated with 2/3 PCV of buffer C (20 mM Hepes, 25% glycerol, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.1 mM DTT, and 1× protease inhibitor cocktail from Calbiochem) for 15 min on ice. Finally, the nuclear fraction was centrifuged at 12000 g for 5 min and the supernatant collected for immunoprecipitation. Extract from four 10 cm plates (approximately 20 × 106 cells) was mixed with 40 μl FLAG-M2 beads (Sigma) for immunoprecipitation for 2–4 h at 4°C. Beads were washed with 20 volumes of wash buffer (20 mM Tris pH7.5, 0.5 mM MgCl2, 0.5 M NaCl, and 0.1% NP-40) 5 times. Proteins were eluted using 60 μg FLAG peptide. LC-MS/MS was analyzed by the protein core facility at Columbia University Medical Center.
For formaldehyde cross-linking experiments, approximately 5 × 10 6 cells (one 90% confluent 10 cm plate) were incubated with 0.1% formaldehyde for 10 min at 37°C. Cells were washed with ice-cold PBS to eliminate extra formaldehyde and the cross-linking reaction was quenched with 0.5 ml 0.25 M glycine for 5 min. The cells were washed twice with ice-cold PBS and resuspended in RIPA buffer containing protease inhibitor (Calbiochem) and 1 mM DTT. After incubation on ice for 10 min, the cell suspension was sonicated 3 times for 10 sec at 30% efficiency. The lysate was centrifuged at 10000 rpm for 10 min and the supernatant was used for immunoprecipitation. Proteins were precipitated overnight at 4°C using 20 μl FLAG-M2 beads in the presence of 50 μg BSA and 10 μg RNaseA. The IP was washed with 1 ml RIPA buffer 8 times and proteins were eluted with SDS buffer. Experiments without crosslinking were done exactly the same way but without formaldehyde and glycine. We were able to obtain significant binding between FLAG-tagged Prp31 and Mtr4 reproducibly only when cell lysates were prepared in RIPA buffer followed by sonication. An extract prepared by the milder Dignam protocol failed to show significant interaction.
Prp31 immunodepletion
An amount of 40 μl Dignam nuclear extract was adjusted to 700 mM NaCl, followed by immunodepletion using 6 mg protein A beads bound to anti-Prp31 antibody for 2 h. Supernatant from the depletion was incubated with protein A beads without antibody for 30 min. A mock depletion was performed in parallel using rabbit preimmune serum. Depleted extract was diluted with buffer D to bring the final salt concentration to 150 mM NaCl. Extract was divided into two sets for anti-Prp3 and rabbit preimmune serum-mediated immunoprecipitation using 2.5 mg protein A beads. After 2 h, the IP was washed four times with 500 μl NET-2 buffer. Proteins were eluted with SDS sample buffer and were run on an 8% gel followed by probing with anti-Mtr4 antibody.
GST-Mtr4 expression and immunoprecipitation
The GST-Mtr4 expression plasmid (see Plasmids and cloning) was transformed into Rosetta cells and protein was expressed at 37°C for 3 h following IPTG treatment. Proteins were isolated following the protocol for the GST expression system (GE Healthcare Life Sciences) and were purified on 100 μl glutathione beads. Twenty microliter glutathione bead-bound protein was subsequently incubated with 45 μl T7/SP6 reticulocyte lysate-generated (TNT T7/SP6 Quick Coupled Transcription/Translation System, Promega) 35S-Met radiolabeled protein from individual plasmids expressing Prp proteins overnight at 4°C. Beads were washed with 500 μl IPP150 buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris pH 8.0) three times, and proteins were eluted in SDS buffer and run on SDS-PAGE.
RNA analysis
IP was conducted with anti-Mtr4 antibody on protein A/G beads (Calbiochem) without RNase treatment followed by DNase treatment and recovery of the RNA using Trizol (Invitrogen) following the manufacturer’s protocol. This RNA was subjected to qPCR analysis using primers AN244/AN245 for U2 and AN246/247 for U6 (Table S2).
Antibodies
Anti-Mtr4 (cat no. 1574, 1575) was obtained from Novus Biologicals, mouse polyclonal anti-Prp31 (H00026121-A01) from Abnova, and anti-PM/Scl-75 from Santa Cruz Biotechnology. Antibodies for Prp3, Prp4, Prp6, and Prp31 (raised against a peptide corresponding to aa 484–497 of Prp31) were the generous gift of Dr. R. Lührmann (Max-Planck Institute for Biophysical Chemistry, Goettingen, Germany). PM/Scl-100 antibody was a gift from Dr. G. Pruijn (Nijmegen Center for Molecular Life Sciences, Netherlands). Gemin4 antibody was a gift from Dr. Gideon Dreyfuss (University of Pennsylvania).
Supplementary Material
Acknowledgments
We thank Drs. Kristina Herbert, Andrei Alexandrov, and Kazimierz Tycowski for their helpful comments on the manuscript; Mei-Di Shu and Yingchun Tong for technical help; and Drs. Lührmann and Pruijn for providing antibodies. A.N. was supported by an Anna Fuller Cancer Research fellowship, a Leukemia Lymphoma Postdoctoral fellowship, and by grant GM026154 to J.A.S. from the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Glossary
Abbreviations:
- TRAMP
Trf4/Air2/Mtr4 Polyadenylation
- PM/Scl
Polymyositis/scleroderma
- NE
Nuclear extract
- snRNP
Small nuclear ribonucleoprotein
- RIPA
Radioimmunoprecipitation assay
- IP
Immunoprecipitation
- RNase
Ribonuclease
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/19431
References
- 1.Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–72. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–11. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–17. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–40. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol. 2002;22:8254–66. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–75. doi: 10.1016/S0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 9.Luna R, Jimeno S, Mari´n M, Huertas P, Garci´a-Rubio M, Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell. 2005;18:711–22. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 11.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, et al. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–26. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol Cell Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/S0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 14.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–42. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 15.Vanácová S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–7. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:239–46. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–49. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–76. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van´cová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–24. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 23.Chlebowski A, Tomecki R, Lo´pez ME, Se´raphin B, Dziembowski A. Catalytic properties of the eukaryotic exosome. Adv Exp Med Biol. 2011;702:63–78. doi: 10.1007/978-1-4419-7841-7_6. [DOI] [PubMed] [Google Scholar]
- 24.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–7. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–58. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–15. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 29.Cristodero M, Clayton CE. Trypanosome MTR4 is involved in rRNA processing. Nucleic Acids Res. 2007;35:7023–30. doi: 10.1093/nar/gkm736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etheridge RD, Clemens DM, Gershon PD, Aphasizhev R. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol Biochem Parasitol. 2009;164:66–73. doi: 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura R, Takeuchi R, Takata K, Shimanouchi K, Abe Y, Kanai Y, et al. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Mol Cell Biol. 2008;28:6620–31. doi: 10.1128/MCB.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasudevan S, Peltz SW. Nuclear mRNA surveillance. Curr Opin Cell Biol. 2003;15:332–7. doi: 10.1016/S0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 33.West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell. 2006;21:437–43. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–11. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova OV, Makarov EM, Liu S, Vornlocher HP, Lührmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–57. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidenhammer EM, Ruiz-Noriega M, Woolford JL., Jr Prp31p promotes the association of the U4/U6 x U5 tri-snRNP with prespliceosomes to form spliceosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3580–8. doi: 10.1128/mcb.17.7.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider M, Hsiao HH, Will CL, Giet R, Urlaub H, Lührmann R. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat Struct Mol Biol. 2010;17:216–21. doi: 10.1038/nsmb.1718. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–20. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 39.Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol Cell. 2001;8:375–81. doi: 10.1016/S1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 40.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–4. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–18. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–23. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lauber J, Plessel G, Prehn S, Will CL, Fabrizio P, Gröning K, et al. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–41. [PMC free article] [PubMed] [Google Scholar]
- 44.Makarov EM, Makarova OV, Achsel T, Lührmann R. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J Mol Biol. 2000;298:567–75. doi: 10.1006/jmbi.2000.3685. [DOI] [PubMed] [Google Scholar]
- 45.Hessle V, Björk P, Sokolowski M, González de Valdivia E, Silverstein R, Artemenko K, et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol Biol Cell. 2009;20:3459–70. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–41. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 47.Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexandrov A, Colognori D, Steitz JA. Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex. Genes Dev. 2011;25:1078–90. doi: 10.1101/gad.2045411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.