Abstract
Satellite cells (SCs) are myogenic stem cells found in skeletal muscle that function to repair tissue damaged by injury or disease. SCs are quiescent at rest, although the signaling pathways required to maintain quiescence are unknown. Using a transgenic Notch reporter mouse and quantitative reverse-transcription polymerase chain reaction analysis of Notch target genes, we determined that Notch signaling is active in quiescent SCs. SC-specific deletion of recombining binding protein-Jκ (RBP-Jκ), a nuclear factor required for Notch signaling, resulted in the depletion of the SC pool and muscles that lacked any ability to regenerate in response to injury. SC depletion was not due to apoptosis. Rather, RBP-Jκ-deficient SCs spontaneously activate, fail to self-renew, and undergo terminal differentiation. Intriguingly, most of the cells differentiate without first dividing. They then fuse with adjacent myofibers, leading to the gradual disappearance of SCs from the muscle. These results demonstrate the requirement of Notch signaling for the maintenance of the quiescent state and for muscle stem cell homeostasis by the regulation of self-renewal and differentiation, processes that are all critical for normal postnatal myogenesis.
Keywords: Notch, RBP-J, Satellite Cell, Quiescence
INTRODUCTION
Myogenic stem cells (satellite cells [SCs]) are the primary cells in muscle tissue required for the regeneration that occurs in response to injury or disease [1]. In response to injury, SCs are activated and they proliferate and differentiate into myoblasts that undergo further differentiation and fusion to form muscle fibers. Under resting conditions, SCs are quiescent, a common feature of stem cells characterized by reversible mitotic arrest and reduced metabolic activity that protects stem cells against endogenous stress caused by DNA replication and cellular respiration [2]. Stem cell populations are maintained throughout the lifetime of an animal through self-renewal, which is characterized by a return to quiescence [2-6]. Although quiescence is an essential property of SCs, and stem cells in general, regulatory mechanisms necessary to maintain stem cells in the quiescent state are largely unknown.
In adults, Notch signaling is an important regulator of stem cells from a variety of tissues including skeletal muscle, brain, skin, intestine, and the vasculature [7-13]. In mammals, there are four Notch receptors and five ligands [14]. Binding of the Notch receptor to its ligand leads to a series of proteolytic cleavages of the Notch receptor and liberation of the notch intracellular domain (NICD) [14]. NICD is translocated to the nucleus where it binds recombining binding protein-Jκ (RBP-Jκ) [15]. RBP-Jκ (hereafter referred to simply as RBP-J) is a key mediator of canonical Notch signaling and acts downstream of all four Notch receptors [16]. In the absence of a Notch signal, RBP-J inhibits transcription of target genes by binding transcriptional corepressors [17, 18], while in the presence of a Notch signal, NICD binds to RBP-J and displaces corepressors leading to transcriptional activation [19-22]. Genetic deletion of RBP-J is embryonically lethal [23]. Therefore, a conditional knockout of RBP-J is required to evaluate its loss, and by association the loss of Notch signaling, in adult cells and tissues [9 24-26].
We have previously shown that Notch signaling features prominently in regulation of proliferation and differentiation of activated SCs [7]. Interestingly, Notch3 is expressed by quiescent SCs [10], and Notch3 disruption leads to misregulation of SC proliferation [27]. Furthermore, Hes1, Hey1 and HeyL, which are all downstream targets of Notch signaling, are highly expressed at the transcript level in quiescent SCs, and HeyL protein is expressed by quiescent SCs [10]. Therefore, in addition to regulating proliferative expansion of activated SCs, Notch may also be important in the regulation of SC quiescence.
In this study, we investigated the role for Notch signaling in adult SCs by eliminating RBP-J specifically in those cells using an inducible genetic system. We find that RBP-J is required to maintain the SC population in a state of quiescence by preventing SC activation, demonstrating that Notch signaling plays an additional and unexpected role in the regulation of SC quiescence in adults. Furthermore, RBP-J-deficient SCs, once activated, fail to self-renew suggesting that Notch signaling not only regulates the maintenance of the quiescent state but also the return to the quiescent state.
METHODS
Mouse Strains
Mice harboring an allele of RBP-J (RBP-Jf mice) in which exons 6 and 7 are flanked by loxP sites [25] were used to inactivate RBP-J in SCs. RBP-Jf/f mice were crossed with Pax7-CreERtm (herein referred to as Pax7CreER/+) mice [28] to generate Pax7CreER/+; RBP-Jf/+ mice. F1 mice were crossed with RBP-Jf/f mice to generate Pax7CreER/+;RBP-Jf/f (RBP-Jcko, experimental) and Pax7+/+; RBP-Jf/f (control) mice. For lineage tracing, tamoxifen-regulated enhanced yellow fluorescent protein (YFP) was introduced into the experimental and control genetic backgrounds using ROSA26tm1(EYFP)/Cas mice [29] (referred to as ROSAeYFP/+ in the text) to generate Pax7CreER/+; RBP-Jf/f;ROSAeYFP/+. All mice used in these studies were between 2 and 4 months of age. Animal strain maintenance, surgical procedures, drug treatments, and husbandry were carried out at the Veterinary Medical Unit of the Veterans Affairs Health Care System in Palo Alto, and all procedures were approved by the Institutional Animal Care and Use Committee.
Mouse Treatments
Tamoxifen (Sigma-Aldrich, St. Louis, MO, http://www.sigmaal drich.com) was dissolved in corn oil at a concentration of 20 mg/ mL, and experimental and control mice were injected intraperitoneally with 150 μL (3 mg) once a day for 5 days to induce Cremediated excision. For detection of SC proliferation, 200 μL of a 4.0 mg/mL solution of 5-ethynyl-2′-deoxyuridine (EdU) (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) dissolved in sterile phosphate buffered saline (PBS) was injected intraperitoneally daily. Resuspended EdU was stored at −20°C.
Myogenic Cell Preparation
To isolate SCs, dissected hind limb muscles from treated animals were digested in 0.2% type II collagenase in Dulbecco’s modified Eagle’s medium (DMEM) for 90 minutes at 37 °C, followed by a second digest in 1% Dispase/0.2% type II collagenase for 30 minutes at 37 °C (all enzymes from Invitrogen). SCs were mechanically dissociated from myofibers after the final digestion by passing the tissue suspension through a 20-gauge needle. SCs were further purified by fluorescence activated cell sorting (FACS) [10] using a FACSAria II (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). Primary antibodies used were rat anti-CD31 conjugated to allophycocyanin (APC; 1:100, BD Biosciences), rat anti-CD45 antibodies conjugated to allophycocyanin (APC;1:100, BD Biosciences), rat anti-Sca1 antibodies conjugated to Pacific Blue (1:100, BioLegend, San Diego, CA, http://www.biolegend.com), biotinylated rat anti-CD106 antibody (vascular cell adhesion molecule; 1:100, BD Biosciences), phycoerythrin-Cy7 Streptavidin (1:100, BD Biosciences).
Single Fiber Preparations
To isolate single myofibers [30], extensor digitorum longus (EDL) muscles were digested using 0.2% type II collagenase (Invitrogen) in DMEM for 70 minutes at 37 °C with shaking. Muscles were mechanically dissociated and then washed five times to eliminate debris and contaminating cells. EDL myofibers were immediately fixed using 2% paraformaldehyde (PFA, Sigma-Aldrich) in PBS.
Quantitative Polymerase Chain Reaction (PCR)
Quantitative reverse-transcription PCR (RT-PCR) was performed using the ABI Prism 7900 (ABI Biosystems, Foster City, CA), with SYBR Green qPCR Super Mix (Invitrogen). The following PCR conditions were used: one cycle of 95 °C for 10 minutes; 45 cycles of the two-step reaction 95 °C (30 seconds), 60 °C (30 seconds). Primer sets used were Hes1 (5′-GCCAATTTGCCTTTCTCATC-3′, 5′-G AGAGGTGGGCTAGGGACTT-3′), Hes5 (5′-CGTGGGGTTGTT TTGTGTTT-3′, 5′-ATGTGGACCTTGAGGTGAGG-3′), Hes6 (5′-CCCTAGAGCTCTGGATGGTG-3′, 5′-GCGCAACTGTGTTA CAAACG-3′), Hey1 (5′-TCAGCGTGGGGAATCTTAAC-3′, 5′-GATTCAGGGCACAGACACCT-3′), Hey2 (5′-TGGAAAAGGA AAACGCCATA-3′, 5′-ATCTGCAGCCTGACACATTG-3′), HeyL (5′-GCGATTGAAGTCCCCAGATA-3′, 5′-ACTGGGGTCACCAG ACTGAG-3′), RBP-J (5′-GGTCCCAGACATTTCTGCAT-3′, 5′-G GAGTTGGCTCTGAGAATCG-3′), Pax7 (5′-CCCTCCATGTCAC CTCAAGT-3′, 5′-CCAGCGGGTTTTTGTTTTTA-3′), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (5′-(TGCGACTT-CAACAGCAACTC-3′, 5′-ATGTAGGCCATGAG GTCCAC-3′). Expression levels were normalized to GAPDH.
Muscle Injury
To the tibialis anterior (TA) muscles of mice anesthetized using an isoflurane/O2 mixture, 30 μL of a 1.2% solution of BaCl2 (w/v in ddH2O, Sigma-Aldrich) was injected.
Immunofluorescence and Histology
TA muscles were harvested, fixed by rocking in 0.5% electron microscopy-grade PFA (Electron Microscopy Sciences, Hatfield, PA, http://www.emsdiasum.com), frozen in Optimal Cutting Temperature mounting media (Sakura Finetek, Torrance, CA, http://www.sakuraus. com/), and cryosections were collected. Histological analysis was performed using hematoxylin and eosin (Sigma-Aldrich) staining. For immunofluorescence, cryosections were postfixed using 0.2% PFA, permeabilized using 0.2% TritonX-100/PBS/5% donkey serum for 10 minutes at room temperature, and further processed using the MOM kit (Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com) or EdU visualization kit (Click-IT, Invitrogen) according to the manufacturer’s specifications. For processing cells for immunofluorescence, FACS-purified myoblasts were plated on BioCoat laminin-coated chamber slides (BD Biosciences) coated with extracellular matrix gel (1:500, Sigma-Aldrich) in F10 media with 10% horse serum for 30 minutes at 37 °C. Plated myoblasts were fixed in 2% PFA for 10 minutes at room temperature and treated with 0.2% TritonX-100/ PBS/5% donkey serum for 10 minutes at room temperature. Both tissue sections and purified myoblasts were incubated with primary antibodies overnight at 4 °C. Primary antibodies used include rabbit anti-GFP (1:500, Invitrogen), mouse anti-MyoD (1:1000, DAKO, Carpintaria, CA, http://www.dako.com), rat anti-RBP-J (1:500, Cosmo Bio Ltd., Kyoto, Japan, http://www.cosmobiousa.com), mouse anti-Myogenin (1:200, Becton/Dickson), mouse anti-Pax7 (1:5–1:500, DSHB, http:// dshb.biology.uiowa.edu/), rat anti-Laminin (1:10000, Sigma-Aldrich), and rabbit antiactivated Caspase-3 (1:50, Cell Signaling Technologies, http://www.cellsignal.com/). Species-specific secondary antibodies (donkey) were conjugated to Alexa 488, 594, and 647 and used at a concentration of 1:1,000 (Invitrogen), and 4′,6-diamidino-2-phenylindole (1:2,000) was used to visualize nuclei. Fluorescence was visualized and photographed using a Zeiss Axiovert microscope (Carl Zeiss, Thornwood, NY), and images were processed using Volocity software (PerkinElmer, Waltham, MA).
Statistical Analysis
For all quantitative analyses presented, a minimum of three replicates was performed in terms of individual animals. Data are presented as means ± standard errors. Two-tailed Student’s t tests were used to test for statistical significance between groups. Differences were considered statistically significant at p < .05.
RESULTS
Notch Signaling Is Active in Quiescent SCs
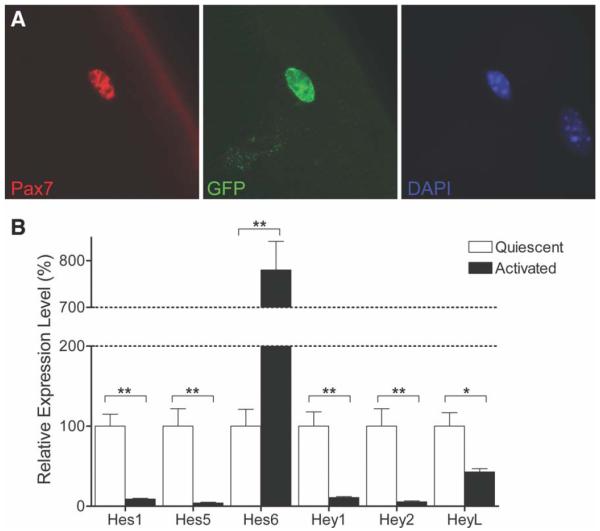
To determine whether Notch signaling is active in quiescent SCs, we used a transgenic Notch reporter (TNR) mouse in which enhanced green fluorescent protein expression is regulated by four tandem RBP-J binding sites [31]. In immunostained single EDL myofiber preparations, we observed GFP expression in SCs (Fig. 1A). Although the expression of GFP was not uniformly high among all SCs, these observations suggest that Notch signaling may be active even in the quiescent state (Fig. 1A).
Figure 1.
Notch signaling is active in quiescent satellite cells (SCs). (A): Immunostaining for Pax7 and GFP in SCs associated with freshly isolated myofibers from a transgenic Notch reporter mouse (×63 magnification). GFP expression in Pax7+ve cells indicates active Notch signaling in quiescent SCs. (B): Quantitative RT-PCR analysis of Notch target gene expression in quiescent and activated SCs. Hes1, Hes5, Hey1, Hey2, and HeyL levels were higher in quiescent SCs. Hes6 was more highly expressed after activation. RNA expression levels are normalized to those at quiescence. (*, p < .05, **, p < .01). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; Pax7, paired box protein 7; RT-PCR, reverse-transcription polymerase chain reaction.
As a further test for activation of Notch signaling in the quiescent state, we compared the expression of Notch target genes in quiescent and activated SCs. SCs were FACS purified from uninjured hind limb muscles (quiescent SCs) or from muscles 3.5 days after BaCl2-induced injury (activated SCs). Quantitative RT-PCR analysis was performed and expression levels for Notch target genes were normalized to levels found at quiescence (Fig. 1B). Consistent with the reporter gene expression, a subset of Notch target genes (Hes1, Hes5, Hey1, Hey2, and HeyL) are highly expressed in the quiescent state and then downregulated during activation (Fig. 1B). Intriguingly, the expression of Hes6 exhibits the opposite pattern, increasing with SC activation and consistent with our previous results showing that Notch signaling promotes proliferative amplification of activated SCs [7]. Therefore, it appears that Notch target genes are not necessarily coordinately regulated and that there may be parallel pathways mediating quiescence and activation and regulated by different Notch targets.
In Vivo Deletion of Notch Signaling in Quiescent SCs
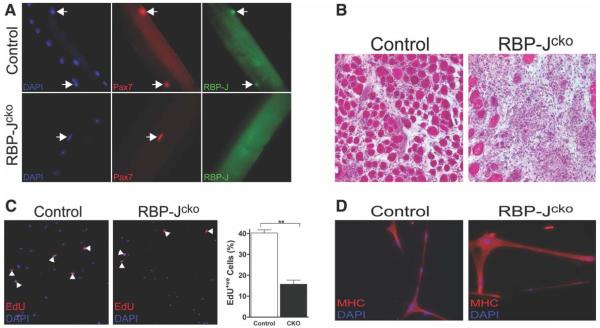
A conditional knockout of RBP-J in muscle progenitor cells during development causes premature myogenic differentiation and results in fewer SCs postnatally [32]. Therefore, to circumvent the dependence of embryonic myogenic development on Notch signaling, an inducible and conditional Cre driver was used. Previous work has demonstrated that the tamoxifen-inducible CreER protein expressed from a modified Pax7 locus (inserted into the 3′-untranslated region; Pax7CreER/+) is spatially restricted to SCs and is effective in lineage tracing and gene disruption studies [28, 33, 34]. Therefore, to eliminate Notch signaling in SCs, a mouse with the Pax7CreER/+ allele was crossed to a mouse carrying a conditionally mutant “floxed” RBP-J allele [25], to generate Pax7CreER/+;RBP-Jf/f mice (RBP-Jcko). After 10 days of tamoxifen treatment of RBP-Jcko mice, the expression of RBP-J protein was eliminated from SCs (Fig. 2A). Quantitative RT-PCR analysis confirmed the transcript levels for RBP-J, and the Notch target genes Hes1, Hey1 and HeyL, which are highly expressed during SC quiescence (Fig. 1B), were significantly reduced in quiescent SCs from tamoxifen-treated RBP-Jcko mice (Supporting Information Fig. S1).
Figure 2.
Deletion of RBP-J in SCs leads to a failure of regeneration. (A): Satellite cells (SCs) assayed by immunostaining for the presence of RBP-J protein in control and RBP-Jcko mice after tamoxifen treatment. RBP-J protein is eliminated from Pax7+ve SCs (arrows) in RBP-Jcko mice (−63 magnification). (B): After 4 weeks of the initiation of tamoxifen treatment, RBP-Jcko and control mice were injured using BaCl2 injection and muscles were harvested 7 days later. Cryosections were stained with hematoxylin and eosin and reveal a complete lack of regeneration in the RBP-Jcko muscle. (C): Fluorescence activated cell sorting-purified control and RBP-Jcko SCs from tamoxifen-treated animals 9 days after the completion of tamoxifen injections were tested in vitro for proliferation by the incorporation of EdU. The panels on the left show that both populations underwent proliferative amplification. The graph on the right shows that the population of control SCs expanded at a greater rate than that of the RBP-J-deficient SC population. (**, p < .01). (D): Equivalent numbers of control and RBP-J-deficient SCs were plated and induced to differentiate by culture in low serum medium. Both populations differentiated to produce multinucleate myotubes expressing MHC with no obvious difference between the two populations. Abbreviations: CKO, conditional knockout; DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′-deoxyuridine; MHC, myosin heavy chain; Pax7, paired box protein 7; RBP-J, recombining binding protein-J.
Functional Significance of RBP-J Deletion in Quiescent SCs
We analyzed muscle histology for any functional consequences of RBP-J deletion in the SC compartment. Muscles from RBP-Jcko mice appeared grossly normal 9 weeks after RBP-J deletion (Supporting Information Fig. S2). To test for SC functionality, muscles of RBP-Jcko mice were injured 4 weeks after tamoxifen injection. After 7 days of injury, muscles were analyzed histologically and exhibited a complete absence of any regenerative myogenesis (Fig. 2B). Given the role of Notch signaling in regulating both the proliferation and differentiation of myogenic progenitors [7, 35], we isolated RBP-J-deficient SCs by FACS and assessed their ability to proliferate or differentiate to test for any defect in these critical functions that could potentially explain the failure of these cells to support regenerative myogenesis in vivo. SCs were isolated 9 days after tamoxifen treatment and analyzed in assays of proliferation and differentiation in vitro. RBP-J-deficient SCs were capable of proliferative amplification, albeit at a reduced rate compared to control SCs (Fig. 2C). Furthermore, myogenic progenitors lacking RBP-J were fully capable of myogenic differentiation, forming multinucleated myotubes (Fig. 2D). These data therefore exclude the possibility that the failure of regeneration of muscle in which RBP-J has been depleted from the SC population (Fig. 2B) is attributable to a failure of those cells either to proliferate or differentiate, although the reduced proliferative response of RBP-J-deficient SCs could have contributed to the impaired regenerative response.
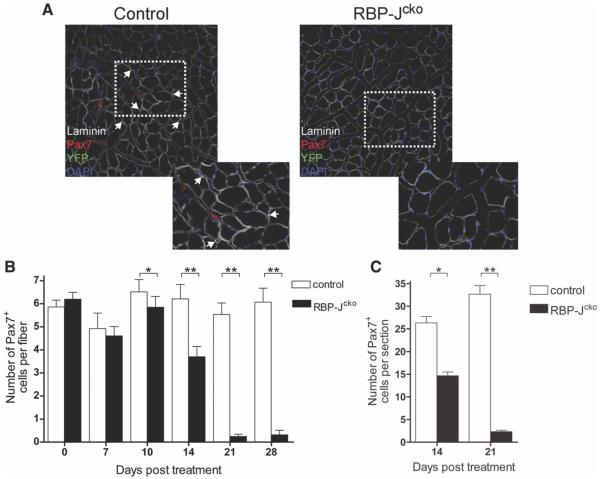
We then analyzed muscles of RBP-Jcko mice for any changes in SC number that could account for the defect in regeneration observed. Although muscle regenerates perfectly well even when SC numbers have been depleted to 10% or 20% of normal levels [36], the muscle regeneration defect in RBP-Jcko mice could possibly be due to a less severe depletion in combination with functional changes of proliferation noted above. To identify and quantify RBP-J-deficient SCs, we genetically tagged recombined SCs from RBP-Jcko mice with eYFP using the ROSAeYFP/+ allele. Mice were treated with tamoxifen, and TA muscles were harvested 3 weeks later. In control mice and in RBP-Jcko mice prior to tamoxifen treatment, YFP expression revealed normal numbers of SCs, confirmed by costaining for Pax7 (Fig. 3A). By contrast, no YFP-labeled cells were observed in RBP-Jcko mice 3 weeks after tamoxifen treatment (Fig. 3A). Therefore, the deletion of RBP-J from the SC compartment resulted in a complete depletion of the SC pool by 3 weeks after tamoxifen treatment.
Figure 3.
Deletion of RPB-J in satellite cells (SCs) leads to their depletion. (A): Laminin, Pax7, and YFP expression in tibialis anterior muscles from control and RBP-Jcko mice 21 days following tamoxifen treatment. Images to the lower right are higher power views of the areas indicated by the dotted rectangles. YFP+ve SCs are absent in RBP-Jcko animals. (B): Average number of Pax7+ve cells per myofiber as assessed in single fiber cultures from control and tamoxifen-treated RBP-Jcko mice as a function of time after tamoxifen administration. (C): Average number of Pax7+ve cells per cryosection in control and tamoxifen-treated RBP-Jcko mice 14 and 21 days following tamoxifen administration. The decline in SC number paralleled exactly that seen in single myofibers preparations (panel B). (*, p < .05, **, p < .01). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; Pax7, paired box protein 7; RBP-J, recombining binding protein-J; YFP, yellow fluorescent protein.
To assess the temporal pattern of SC loss following deletion of RBP-J, the numbers of SCs were quantified in EDL myofiber explant cultures from tamoxifen-treated RBP-Jcko and control animals. A week following tamoxifen treatment, there was no difference in the number of SCs present on single myofibers harvested from the two groups (Fig. 3B). However, 10 days following tamoxifen treatment there was a small, but statistically significant, decrease in the number of SCs found in RBP-Jcko animals (Fig. 3B). The number of SCs per fiber continued to decrease in tamoxifen-treated RBP-Jcko mice thereafter, reaching the point of near total (>95%) elimination of all SCs by 3 weeks after the initiation of treatment (Fig. 3B). SC depletion was also observed in TA cryosections 14 and 21 days following tamoxifen treatment of RBP-Jcko animals with almost identical kinetics (Fig. 3C). Collectively, our data show that Notch signaling has a significant role in maintaining the SC population as a loss of RBP-J causes their progressive depletion.
SC Loss from RBP-J Deletion Is Not Due to Apoptosis
To test whether the disappearance of RBP-J-deficient SCs was due to apoptosis, muscle sections from tamoxifen-treated RBP-Jcko mice were immunostained for activated Caspase-3 either 10 or 14 days after tamoxifen treatment. Our time course studies established that, at these time points, SCs were in the process of disappearing (Fig. 2B, 2C). We did not detect any evidence of immunostaining for activated Caspase-3 in SCs from RBP-Jcko animals at either time point. Of the cells that stained positive for activated Caspase-3, none were located beneath the basal lamina (Supporting Information Fig. S3A, S3B), the anatomical position of SCs [37]. Therefore, it does not appear that RBP-J deletion leads to SC loss because of apoptotic cell death.
Loss of RBP-J in SCs Causes Proliferation
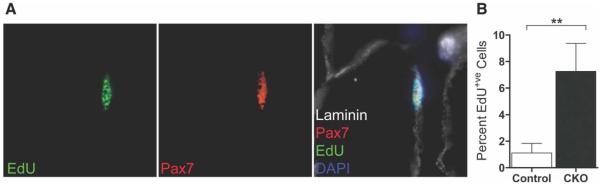
Quiescent cells are characterized by reversible mitotic arrest [4]. Given the absence of evidence of apoptotic cell death, we hypothesized that the loss of SCs following RBP-J deletion might be due to their exit from the quiescent state, entry into the cell cycle, and failure of self-renewal. Therefore, we tested for proliferation of RBP-J-deficient SCs. In adult uninjured mouse muscle, proliferating SCs are rare [38]. We therefore used a long-term EdU labeling approach to identify rarely dividing SCs. Tamoxifen-treated RBP-Jcko and control animals received daily injections of EdU for 14 days, beginning at the onset tamoxifen treatment. Hind limb muscles were then harvested and SCs were purified by FACS, plated on coated chamber slides, and processed for immunocytochemistry. At this time point, we could identify EdU+ve cells that were both Pax7+ve and in the SC position (Fig. 4A). Quantitative analysis revealed that there was more than a sevenfold increase in the percentage of EdU+ve SCs in the RBP-Jcko muscles compared to wild-type muscles (Fig. 4B). These results indicate that the loss of RBP-J induces at least some SCs to enter the cell cycle, and that the frequency is much higher than in wild-type muscle.
Figure 4.
Loss of RBP-J in quiescent satellite cells (SCs) induces cell cycle entry. (A): Control and RBP-Jcko animals were treated with tamoxifen for 5 days and EdU for 14 days, both initiated at the same time. Cryosections of tibialis anterior muscles were assessed for evidence of SC proliferation by EdU incorporation. An EdU+ve cell is shown that is both Pax7+ve and beneath the basal lamina as outlined by laminin staining. (B): Control and RBP-Jcko animals were treated with tamoxifen for 5 days and EdU for 14 days as in panel (A). SCs were purified by fluorescence activated cell sorting, plated, and stained to assess EdU incorporation. The percentages of SCs that were EdU+ve are shown. (**, p < .01). Abbreviations: CKO, conditional knockout; DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′-deoxyuridine; Pax7, paired box protein 7; RBP-J, recombining binding protein-J.
RBP-J-Deficient SCs Progress Along the Myogenic Lineage
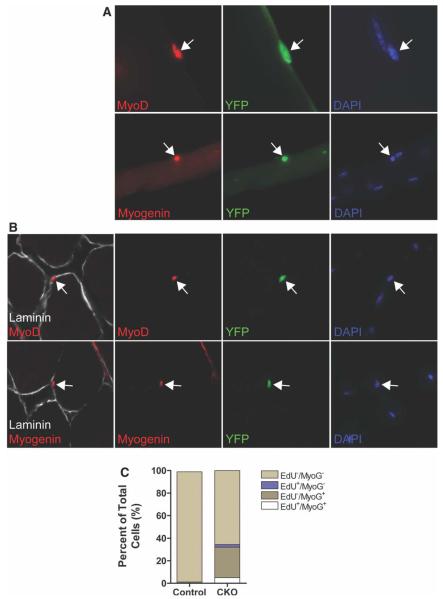
As deletion of RBP-J appeared to induce SCs to break quiescence, we sought to determine the fate of the SCs following activation. We analyzed muscle from tamoxifen-treated RBP-Jcko mice 10 days after the administration of tamoxifen for evidence of myogenic lineage progression and observed MyoD+ve cells in the SC position both on myofibers ex vivo and in muscle cryosections (Fig. 5A, 5B), consistent with the expression of MyoD in SCs that have activated and begun to proliferate but rarely observed in uninjured muscle [39]. Even more surprisingly, we found Myogenin expression, which is a marker of myogenic differentiation [40], in YFP+ve cells adjacent to myofibers ex vivo and in vivo (Fig. 5A, 5B). Although Myogenin+ve cells were never detected in control animals, more than 20% (22.4 ± 2.5%, n1/4 3) of YFP+ve cells expressed Myogenin in muscle from RBP-Jcko mice. We FACS purified SCs from control and tamoxifen-treated RBP-Jcko animals and performed quantitative RT-PCR analysis. We found that the expression of Pax7 was markedly reduced in RBP-J-deficient SCs (16.3 ± 1.4% of control levels; n = 3), consistent with the decline of Pax7 expression as SCs activate 1/4 and their progeny differentiate [39, 41]. When we isolated YFP+ve cells from mice that had received continuous EdU injections (as in Fig. 4), we found that approximately 25% of those cells were Myogenin+ve (Fig. 5C). Interestingly, the vast majority of those were EdU −ve; of the Myogenin+ve cells, less than 10% were also EdU+ve. This suggests the surprising possibility that most SCs undergo differentiation without dividing.
Figure 5.
Loss of RBP-J induces satellite cell (SC) activation and differentiation. (A): Muscles were harvested from RBP-Jcko and control mice 10 days after tamoxifen treatment, and myofiber cultures were analyzed for the expression of myogenic activation (by the expression of MyoD) and differentiation (by the expression of Myogenin) in SCs or their progeny identified by YFP expression. Arrows indicate YFP+ve cells that are positive for either MyoD (above) or Myogenin (below). (B): Cryosections from muscles of mice treated as in panel (A), also analyzed for the same markers of activation and differentiation. Arrows indicate YFP+ve cells that express either of the myogenic lineage markers. (C): Fluorescence activated cell sorting-purified SCs from tamoxifen-treated control and RBP-Jcko mice administered EdU for 14 days were plated and immediately assayed for the incorporation of EdU and the expression of Myogenin. In the RBP-J-deficient SC population, more than 25% of the cells were Myogenin+ve, and of those, the vast majority did not incorporate EdU. Abbreviations: CKO, conditional knockout; DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′-deoxyuridine; RBP-J, recombining binding protein-J; YFP, yellow fluorescent protein.
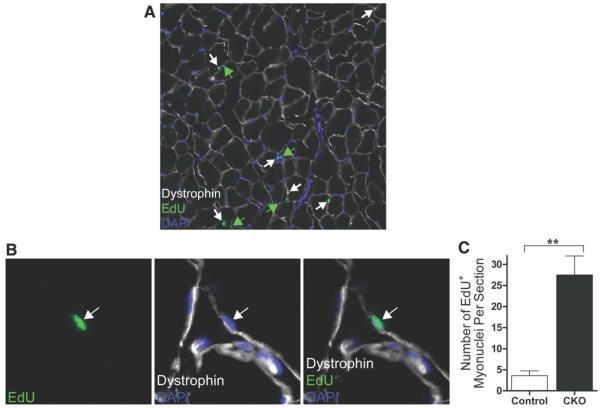
Given that we observed no evidence of apoptosis of SCs or their progeny and that, by several criteria, the SCs or their progeny appeared to be undergoing myogenic differentiation, we postulated that the gradual depletion of the SC pool that we observed could be due to the fusion of the differentiating cells with associated myofibers. To test this directly, we administered EdU to tamoxifen-treated RBP-Jcko mice for more than 2 weeks and analyzed muscles for the fate of any SC progeny that might have entered the cell cycle and incorporated EdU. In particular, we examined cryosections for any EdU+ve myonuclei, indicative of fusion of cells that had incorporated EdU. In contrast to control muscles where evidence of incorporation of nuclei from adjacent SCs that have recently divided is exceedingly rare, we observed many EdU+ve myonuclei in muscles of tamoxifen-treated RBP-Jcko mice (Fig. 6A, 6B). Quantitative analysis revealed that the number of EdU+ve myonuclei was more than sevenfold higher in RBP-Jcko mice compared to controls (Fig. 6C), demonstrating that at least some of the SCs that are RBP-J deficient undergo at least one round of cell division before differentiating. Therefore, the deletion of RBP-J from the SC population appears to be the result of the spontaneous activation and differentiation of the cells and their progeny, with at least some and perhaps all of those differentiated cells then fusing with adjacent myofibers. This process results in a gradual depletion of the SC pool as the activation and myogenic lineage progression occurs spontaneously and without self-renewal to maintain the SC pool.
Figure 6.
Incorporation of RBP-J-deficient satellite cells (SCs) into adjacent myofibers. (A): Tibialis anterior (TA) muscle cryosection from a RBP-Jcko animal treated daily with EdU for 14 days, beginning at the onset of tamoxifen administration. EdU-labeled myonuclei located beneath the myofiber membrane as delineated by dystrophin staining are highlighted by white arrows. EdU+ve nuclei that are outside the muscle fiber membrane are highlighted by green arrows. (B): Higher power magnification showing a single EdU+ve myonucleus in a cryosection as in panel (A). (C): Quantification of EdU+ve myonuclei, expressed as the absolute number per TA cryosections, from control and RBP-Jcko mice treated as described for panel (A). (**, p < .01). Abbreviations: CKO, conditional knockout; DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′-deoxyuridine; RBP-J, recombining binding protein-J.
DISCUSSION
Our results demonstrate that RBP-J, an essential component of Notch signaling, is required to maintain SC quiescence. Notch signaling appears to be active in the quiescent state based both on reporter gene expression and on the expression of Notch target genes that decline on activation. We show that a loss of RBP-J leads to SC activation and differentiation, leading to a gradual depletion of the SC pool, indicative of a failure of SCs to self-renew. Not only does the loss of RBP-J lead to an exit of SCs from the quiescent state but also the progression to differentiation of the SC progeny occurs while the cell is still in the SC niche. The depletion of cells from the niche occurs when the differentiated cells fuse with the adjacent fibers.
Our results suggest at least three distinct effects of inhibition of Notch signaling on SC biology—(1) a loss of maintenance of quiescence; (2) a failure of self-renewal among activated SCs that enter the cell cycle; and (3) spontaneous differentiation of the activated SCs with minimal or no proliferative expansion. Clearly, the consequences of Notch activation depend on the specific cellular state, whether it be quiescent, activated, or proliferative. Such a state dependence may be due to the levels of activation of other pathways that may interact with and influence Notch signaling, resulting in different transcriptional outputs and different cellular consequences. Combinatorial regulation of Notch targets has been described in multiple systems, such as endodermal expression of ref-1 by GATA and Notch signaling in C. elegans [42], crosstalk between nuclear factor κ B family members and Notch signaling in the development of B-cells in mice [43], and the requirement of the myogenic gene, Twist, in combination with Notch signaling in muscle precursor cells in Drosophila [44]. Context-specific output for Notch signaling has been described in Xenopus neurogenesis where the presence of an E-box, which binds an unidentified transcription factor, in the Notch target gene, Esl10, is necessary for expression [45]. Crosstalk between Notch and other signaling pathways such as ras [46], Wnt [47, 48], and hedgehog [49] also provide examples of the modulation of Notch signaling in the context of the activation of other pathways. Clearly, understanding the multiple levels of regulation of the Notch pathway by other signaling cascades will be essential for understanding how activation of Notch signaling may have divergent effects depending on the state of muscle stem cells or their progeny.
The spontaneous activation of quiescent SCs following deletion of RBP-J and inhibition of further Notch signaling adds to the growing list of signaling pathways that have been described to promote SC activation. Hepatocyte growth factor (HGF) [50], members of the fibroblast growth factor family [51, 52], and tumor necrosis factor-α (TNF-α) [53] are potent activators of SCs that stimulate cell cycle entry. Because HGF and TNF-α signal transduction activate p38/mitogen-activated protein kinase (MAPK) [54, 55], a requirement for p38/MAPK pathway inhibition has been proposed to maintain SC quiescence [56]. Notch signaling inhibits the p38/MAPK pathway through induction of MAPK phosphatase-1 [57]; therefore, regulation of an antagonistic relationship between Notch signaling and the p38/MAPK pathways may be a critical component of SC activation.
Our data demonstrating Notch reporter gene expression in quiescent SCs (Fig. 1A), as well as the downregulation of certain Notch target genes on activation (Fig. 1B), led us to hypothesize that active Notch signaling is essential for SCs to maintain the quiescent state. This was supported by the surprising result that RBP-J-deficient SCs exit spontaneously from the quiescent state. The report of Notch3 and Notch target gene expression in quiescent SCs further supports the role of Notch signaling in maintaining quiescence [10]. Notch signaling may be an important regulator of ependymal cell quiescence in the central nervous system as demonstrated by the entry of ependymal cells into the cell cycle following the deletion of RBP-J in cells around the lateral ventricle [11]. However, this effect may be indirect as the deletion of RBP-J using the neural-specific driver NestinCreER resulted in no detectable increase in ependymal cell entry into the cell cycle [58]. Additionally, when Notch signaling is inhibited in mammary stem cells, population expansion occurs as assayed by FACS analysis and ductal outgrowth studies demonstrating that Notch signaling is required to suppress proliferation and maintain quiescence [59]. Furthermore, neural stem cells in zebrafish do not proliferate and are maintained in quiescence through the activation of Notch signaling by Delta, which is expressed by mitotically active and adjacent neuroblasts [60]. Notch target genes that are expressed at highest levels in quiescent SCs are likely to be important components of the quiescence signaling network, but it remains to be determined whether there are common signaling networks that maintain quiescence in different cell populations.
The studies of spontaneous activation and proliferation of SCs following RBP-J deletion also provided the opportunity to examine the spontaneous rate of turnover of SCs in the controls. SC turnover has been estimated based on many different techniques, including the frequency of indicators of ongoing mitoses (such as Ki67 or proliferating cell nuclear antigen expression) and on the analysis of probes (such as 3H or BrdU) that are incorporated into dividing cells. Estimates vary widely depending on the species, the muscle studied, the age of the animal, and the technique for assessing turnover rate [38, 61-63]. In adult rodent muscle, the turnover of SCs has been estimated at approximately 0.5% per week [38]. This is consistent with our results showing that approximately 1% of SCs in the TA muscle of the adult mouse were EdU+ve after 2 weeks of daily EdU injections. Based on general assumptions such as population homogeneity and typical cell cycle times, these data indicate that less than 1 in a 1,000 SCs would be dividing at any time, that the average duration of quiescence is greater than 1 year, and that the entire population would not be expected to turn over during the full lifespan of the mouse.
Factors known to regulate SC self-renewal include Sprouty1 (an inhibitor of receptor tyrosine kinase activators of MAPK) [34], angiopoietin1/Tie2 [64], and Myostatin [65]. Our data demonstrate that at least some subset of quiescent SCs begin to proliferate when Notch signaling is inhibited, but they fail to self-renew in the process. Rather, it appears that all the proliferating SC progeny differentiate resulting in a depletion of the stem cell pool. This is similar to the recent report showing that deletion of RBP-J results in failure of selfrenewal of neural stem cells which are, instead, induced to differentiate into neurons [58]. Furthermore, our data are consistent with a recent study showing that pharmacological inhibition of Notch signaling using cultured myofiber explants also inhibits SC self-renewal [66]. The pathways activated by Notch signaling that mediate self-renewal, and thus the return to quiescence, may overlap with those that maintain the quiescent state.
We observed the incorporation of SCs that had undergone at least one round of mitosis into adjacent myofibers by testing for EdU+ve myonuclei (Fig. 6A-6C). However, several pieces of evidence suggest that this is not the fate of all SCs, and that a significant proportion of the cells may undergo differentiation and fusion without first entering the cell cycle. If so, this would represent the first report to our knowledge of a stem cell undergoing differentiation without dividing. The evidence in support of this includes the fact that we observed a surprisingly small percentage of SCs incorporating EdU during the time when the cells are beginning to be depleted (Fig. 4). Although there is no reason to expect there to be synchronous activation of the SC population, we would still have expected to see a much higher percentage of SCs undergoing mitosis at different times after tamoxifen administration. Second, and even more compelling, is the observation that the vast majority of SCs (or their progeny) that were Myogenin+ve were EdU −ve, despite continuous EdU administration (Fig. 5C). Unless the quantitation of EdU+ve cells is a vast underestimate of the number of cycling SCs, these data suggest directly that SCs may undergo terminal differentiation without dividing. Supporting this conclusion are the data quantifying the number of EdU+ve myonuclei in cross-sections of RBP-Jcko muscles (Fig. 6C). Although these studies were designed primarily to test for the fate of activated SCs, the quantitative analysis revealed the surprising result that the number of EdU+ve myonuclei was in the very same range (i.e., ~25–30/cryosection of a TA muscle) as the number of SCs before RBP-J deletion (Fig. 3C), not two or four times the number that would be expected if the fate of the SCs was to divide once or twice before differentiation and fusion. Given the absence of evidence of apoptosis of SCs or their progeny at any point during the time course of SC depletion, the most consistent interpretation of all of these data is that, following RBP-J deletion, most (at least 75% or more) SCs undergo terminal differentiation without dividing and then subsequently fuse with adjacent myofibers. A minority of SCs does divide at least once and then also undergoes terminal differentiation and fusion. Our studies shed no light on the differential characteristics of these two populations that might predispose them to different fates, but increasingly there are studies suggesting heterogeneity among the SC pool [66, 67].
One of the most intriguing aspects of our findings is the observation that the deletion of RBP-J, thereby inhibiting Notch signaling, does not coordinately regulate downstream target genes (Fig. 1B). Our data demonstrate the downregulation of some targets and upregulation of other targets in response to the same manipulation of Notch signaling (in this case, inhibiting) in a single population. However, it should be noted that in the central nervous system there is an upregulation of Notch target genes following the deletion of RBP-J, suggesting a repressive role of RBP-J in regulating transcription [58]. Nevertheless, our data suggest that the activation (or inhibition) of Notch signaling does not necessarily lead to a concordant regulation of poised Notch target genes and that there may be parallel Notch pathways, for example, mediated by distinct ligand/receptor pairs in the same cell, one of which could suppress and the other of which could activate target genes. However, as noted [68], there have been few studies that have demonstrated clear differences in transcriptional output based on the specific Notch ligand/receptor pairs. Of course, we cannot exclude the possibility that this result is explained by heterogeneity among the SC population, with the different patterns of Notch target gene expression reflecting different responses in subsets of SCs. Analysis of gene expression at the single cell level would be necessary to distinguish between these two possibilities.
CONCLUSION
Quiescence is a property of most, but not all, somatic stem cells [69-71]. A failure of the maintenance of quiescence poses the threat of stem cell depletion if continuous stem cell activation results in proliferative exhaustion or replicative senescence [72]. The molecular regulation of quiescence is much less well understood than the regulation of the cell cycle itself, and even the characterization of the state of quiescence remains quite limited. For example, activation of SCs from the quiescent state occurs with different kinetics depending on the age of the organism from which the SCs were derived [69], but it is not known if that is due to the existence of more than one quiescent G0 state that vary in terms of the progression to G1 of the cell cycle. Understanding the molecular signaling of quiescent cells is important to be able to regulate their transitions into the cell cycle and the transition of cycling cells back to a state of quiescence.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Rando laboratory for helpful comments and discussions and Linda Ste. Marie for critical comments on earlier versions of the manuscript. Pax7CreER mice were kindly provided by Dr. Charles Keller (OHSU, Portland, OR); RBP-Jκf/f mice were kindly provided by Drs. Tasuku Honjo (Kyoto University, Japan) and Michael Rosenfeld (UCSD, San Diego, CA); TNR mice were kindly provided by Dr. Nicholas Gaiano (Johns Hopkins University, Baltimore, MD). This work was supported by the Glenn Foundation for Medical Research and by grants from the NIH (P01 AG036695, R37 AG23806, R01 AR056849, and an NIH Director’s Pioneer Award) and the Department of Veterans Affairs (Merit Review) to T.A.R.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Author contributions: C.R.R.B.: project conception and design, data collection, data analysis and interpretation, and manuscript writing; T.H.C. and L.L.: experiment design and data collection, analysis, and interpretation; P.V.T. and K.M.S.: technical assistance and data collection and assembly; T.A.R.: project conception and design, data analysis and interpretation, manuscript writing, and financial support.
REFERENCES
- 1.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 2.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 3.Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends Immunol. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 4.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Liu Y, Liu R, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohrin M, Bourke E, Alexander D, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 8.Fre S, Huyghe M, Mourikis P, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 9.Blanpain C, Lowry WE, Pasolli HA, et al. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukada S, Uezumi A, Ikemoto M, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 11.Carlen M, Meletis K, Goritz C, et al. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- 12.Lugert S, Basak O, Knuckles P, et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Fukuhara S, Sako K, et al. Angiopoietin-1/Tie2 signal augments basal Notch signal controlling vascular quiescence by inducing delta-like 4 expression through AKT-mediated activation of beta-catenin. J Biol Chem. 2011;286:8055–8066. doi: 10.1074/jbc.M110.192641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarriault S, Brou C, Logeat F, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Sakai T, Tamura K, et al. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 17.Oswald F, Kostezka U, Astrahantseff K, et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda K, Han H, Tani S, et al. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- 19.Kao HY, Ordentlich P, Koyano-Nakagawa N, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 21.Fryer CJ, Lamar E, Turbachova I, et al. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubman OY, Ilagan MX, Kopan R, et al. Quantitative dissection of the Notch:CSL interaction: Insights into the Notch-mediated transcriptional switch. J Mol Biol. 2007;365:577–589. doi: 10.1016/j.jmb.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oka C, Nakano T, Wakeham A, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 24.Hitoshi S, Alexson T, Tropepe V, et al. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanigaki K, Han H, Yamamoto N, et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 26.Han H, Tanigaki K, Yamamoto N, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 27.Kitamoto T, Hanaoka K. Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem Cells. 2010;28:2205–2216. doi: 10.1002/stem.547. [DOI] [PubMed] [Google Scholar]
- 28.Nishijo K, Hosoyama T, Bjornson CR, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenblatt JD, Lunt AI, Parry DJ, et al. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- 31.Mizutani K, Yoon K, Dang L, et al. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 32.Vasyutina E, Lenhard DC, Wende H, et al. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci USA. 2007;104:4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brack AS, Conboy MJ, Roy S, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 34.Shea KL, Xiang W, LaPorta VS, et al. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shawber C, Nofziger D, Hsieh JJ, et al. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 36.Gayraud-Morel B, Chretien F, Flamant P, et al. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 37.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmalbruch H, Lewis DM. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle Nerve. 2000;23:617–626. doi: 10.1002/(sici)1097-4598(200004)23:4<617::aid-mus22>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 39.Zammit PS, Golding JP, Nagata Y, et al. Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasty P, Bradley A, Morris JH, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 41.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: A potential mechanism for self-renewal. Dev Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neves A, English K, Priess JR. Notch-GATA synergy promotes endoderm-specific expression of ref-1 in C. elegans. Development. 2007;134:4459–4468. doi: 10.1242/dev.008680. [DOI] [PubMed] [Google Scholar]
- 43.Moran ST, Cariappa A, Liu H, et al. Synergism between NF-kappa B1/p50 and Notch2 during the development of marginal zone B lymphocytes. J Immunol. 2007;179:195–200. doi: 10.4049/jimmunol.179.1.195. [DOI] [PubMed] [Google Scholar]
- 44.Bernard F, Krejci A, Housden B, et al. Specificity of Notch pathway activation: Twist controls the transcriptional output in adult muscle progenitors. Development. 2010;137:2633–2642. doi: 10.1242/dev.053181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamar E, Kintner C. The Notch targets Esr1 and Esr10 are differentially regulated in Xenopus neural precursors. Development. 2005;132:3619–3630. doi: 10.1242/dev.01937. [DOI] [PubMed] [Google Scholar]
- 46.Hurlbut GD, Kankel MW, Artavanis-Tsakonas S. Nodal points and complexity of Notch-Ras signal integration. Proc Natl Acad Sci USA. 2009;106:2218–2223. doi: 10.1073/pnas.0812024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 48.Brack AS, Conboy IM, Conboy MJ, et al. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Dave RK, Ellis T, Toumpas MC, et al. Sonic hedgehog and notch signaling can cooperate to regulate neurogenic divisions of neocortical progenitors. PLoS ONE. 2011;6:e14680. doi: 10.1371/journal.pone.0014680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatsumi R, Anderson JE, Nevoret CJ, et al. HGF/SF is present in normal adult skeletal muscle and is capable of activating satellite cells. Dev Biol. 1998;194:114–128. doi: 10.1006/dbio.1997.8803. [DOI] [PubMed] [Google Scholar]
- 51.Johnson SE, Allen RE. Activation of skeletal muscle satellite cells and the role of fibroblast growth factor receptors. Exp Cell Res. 1995;219:449–453. doi: 10.1006/excr.1995.1251. [DOI] [PubMed] [Google Scholar]
- 52.Yablonka-Reuveni Z, Seger R, Rivera AJ. Fibroblast growth factor promotes recruitment of skeletal muscle satellite cells in young and old rats. J Histochem Cytochem. 1999;47:23–42. doi: 10.1177/002215549904700104. [DOI] [PubMed] [Google Scholar]
- 53.Li YP. TNF-alpha is a mitogen in skeletal muscle. Am J Physiol Cell Physiol. 2003;285:C370–C376. doi: 10.1152/ajpcell.00453.2002. [DOI] [PubMed] [Google Scholar]
- 54.Geng Y, Valbracht J, Lotz M. Selective activation of the mitogen-activated protein kinase subgroups c-Jun NH2 terminal kinase and p38 by IL-1 and TNF in human articular chondrocytes. J Clin Invest. 1996;98:2425–2430. doi: 10.1172/JCI119056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Recio JA, Merlino G. Hepatocyte growth factor/scatter factor activates proliferation in melanoma cells through p38 MAPK, ATF-2 and Cyclin D1. Oncogene. 2002;21:1000–1008. doi: 10.1038/sj.onc.1205150. [DOI] [PubMed] [Google Scholar]
- 56.Jones NC, Tyner KJ, Nibarger L, et al. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. J Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kondoh K, Sunadome K, Nishida E. Notch signaling suppresses p38 MAPK activity via induction of MKP-1 in myogenesis. J Biol Chem. 2007;282:3058–3065. doi: 10.1074/jbc.M607630200. [DOI] [PubMed] [Google Scholar]
- 58.Imayoshi I, Sakamoto M, Yamaguchi M, et al. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Chapouton P, Skupien P, Hesl B, et al. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spalding KL, Bhardwaj RD, Buchholz BA, et al. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 62.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackey AL, Kjaer M, Charifi N, et al. Assessment of satellite cell number and activity status in human skeletal muscle biopsies. Muscle Nerve. 2009;40:455–465. doi: 10.1002/mus.21369. [DOI] [PubMed] [Google Scholar]
- 64.Abou-Khalil R, Mounier R, Chazaud B. Regulation of myogenic stem cell behavior by vessel cells: The “menage a trois” of satellite cells, periendothelial cells and endothelial cells. Cell Cycle. 2010;9:892–896. doi: 10.4161/cc.9.5.10851. [DOI] [PubMed] [Google Scholar]
- 65.McCroskery S, Thomas M, Maxwell L, et al. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuang S, Kuroda K, Le GF, et al. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biressi S, Rando TA. Heterogeneity in the muscle satellite cell population. Semin Cell Dev Biol. 2010;21:845–854. doi: 10.1016/j.semcdb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersson ER, Sandberg R, Lendahl U. Notch signaling: Simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 69.Dhawan J, Rando TA. Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 70.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 71.Li J. Quiescence regulators for hematopoietic stem cell. Exp Hematol. 2011;39:511–520. doi: 10.1016/j.exphem.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 72.Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.