Synapse formation is a complex process requiring a high degree of coordination between presynaptic and postsynaptic cells. Speed and reliability of synaptic transmission demand that the appropriate neurotransmitter receptors be highly concentrated in the postsynaptic membrane precisely beneath transmitter release sites in the presynaptic cell. In the central nervous system, a single neuron may receive thousands of synaptic contacts from other neurons releasing a variety of neurotransmitters. Thus, neurons must possess the ability not only to cluster, but also to sort, the proper transmitter receptors to the correct synaptic site. Understanding, on a molecular level, how such specializations develop is a major goal of neurobiological research. This commentary will focus on the synaptic clustering and anchoring of neurotransmitter receptors through their association with a variety of cytoplasmic proteins.
Many details of the mechanisms of synaptogenesis have been gleaned from studies of the neuromuscular junction. Muscle nicotinic acetylcholine receptors (AChRs) are tightly clustered in the postsynaptic region of the muscle, reaching a density that approaches 10,000 receptors per square micrometer (1). The remarkable nature of this specialization is underscored by the fact that greater than 90% of AChRs in muscle are localized at the postsynaptic membrane, a region that accounts for less than 0.01% of the muscle surface. The cytoplasmic peripheral membrane protein rapsyn (43-kDa protein) plays an essential role in the AChR clustering process. Rapsyn originally was discovered as a major component of AChR-rich postsynaptic membrane preparations from Torpedo electric organ (2). In vivo, rapsyn and the AChR are coextensively distributed at the crests of the postsynaptic folds (3). When expressed in heterologous cells, recombinant rapsyn forms membrane clusters and is capable of recruiting AChRs to these clusters (4, 5). Definitive evidence for a critical role for rapsyn in AChR clustering came from the disruption of the rapsyn gene in mice (6). The lack of rapsyn abolishes AChR clustering at the synapse. Other postsynaptic proteins, including syntrophin, utrophin, and the dystroglycans, also fail to become localized postsynaptically in rapsyn-deficient mice, indicating that rapsyn is essential for organizing many components of the postsynaptic apparatus. Subtle, but important, presynaptic abnormalities in these mice suggest that retrograde signaling mechanisms also depend on rapsyn.
Twenty years after rapsyn’s discovery (2), however, the molecular mechanisms by which it forms self-clusters, associates with the AChR, and recruits other proteins to the postsynaptic cytoskeletal specialization, still remain a mystery. Although it generally is believed that rapsyn directly associates with cytoplasmic domains of the AChR subunits, biochemical proof is lacking. Rapsyn has proven to be notoriously difficult to deal with; conventional biochemical techniques, such as immunoaffinity purifications and protein blot overlays, have failed to reveal any binding partners for rapsyn. Even yeast two-hybrid screens have been largely unsuccessful. In this issue of the Proceedings, Ramarao and Cohen (7) have instead used a heterologous expression system to address the mechanisms of AChR clustering by rapsyn.
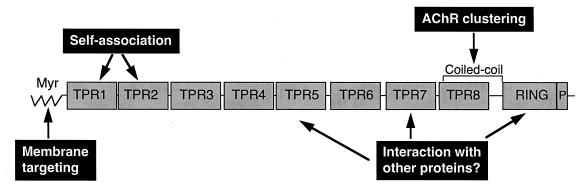
Significant insight into rapsyn’s domain organization was gained recently by the recognition that the first 75% of the protein is composed of eight tandem tetratricopeptide repeats (TPRs) (8). TPRs are 34-aa repeats predicted to form amphipathic α-helices, which mediate protein–protein interactions (9). The TPR domains in rapsyn are flanked by an N-terminal myristoylation site (10) and a C-terminal cysteine-rich region that conforms to a zinc ring finger motif (11, 12). Immediately following the zinc finger is a consensus site for serine phosphorylation. With knowledge of rapsyn’s domain organization, Ramarao and Cohen (7) created chimeric proteins containing various domains of rapsyn fused to green fluorescent protein (GFP) and coexpressed these along with the AChR in nonmuscle cells. The myristoylated N terminus was sufficient to target GFP to the plasma membrane, consistent with previous reports that mutation of an N-terminal glycine disrupts targeting of rapsyn to the membrane (13). Interestingly, whereas two TPR domains were sufficient to mediate rapsyn self-clustering, the presence of even seven TPRs was insufficient to recruit AChRs to these clusters. AChR clustering appears to depend, instead, on a previously unrecognized putative coiled-coil motif found within, and extending beyond, TPR8. These data indicate that membrane targeting, self-association, and AChR clustering are separable events mediated by distinct domains of rapsyn (summarized in Fig. 1).
Figure 1.
Structural domains of rapsyn. The N-terminal myristoylation site (Myr) targets rapsyn to the plasma membrane. The first two TPR domains are sufficient to mediate rapsyn self-clustering. Whether other TPR domains also can serve this function remains to be determined. The ability of rapsyn to promote AChR clustering appears to depend on a putative coiled-coil motif. TPR domains not involved in rapsyn self-association as well as the zinc ring finger (RING) domain are likely to be sites of interaction with other postsynaptic proteins. The function of the phosphorylation site (P) is unknown.
So, how do TPR domains mediate rapsyn self-clustering? TPRs are predicted to interact with other TPRs via a “knob and hole” mechanism, in which a bulky side-chain amino acid from one TPR (the knob) fits into a pocket created by the arrangement of a small uncharged amino acid in between two bulky amino acids (the hole) (9). In this manner, a TPR domain in one rapsyn molecule could contribute the hole into which a knob of the TPR from another rapsyn molecule could fit. Given the presence of eight tandem TPRs in rapsyn, a cluster of rapsyn molecules could readily be formed. It will be important to determine whether specific TPRs in rapsyn are necessary for self-association or whether the presence of any two (or perhaps even one) is sufficient to mediate this function.
Mutations in rapsyn that disrupt the propensity to form a coiled-coil structure abolish AChR clustering without affecting rapsyn self-association, suggesting that this structural motif is functionally important in mediating receptor clustering. Interestingly, Ramarao and Cohen (7) point out that there are stretches of 14–20 amino acids within the cytoplasmic loops of the AChR subunits predicted to form α-helices and possibly coiled-coil structures. In the simplest model, then, rapsyn may promote clustering via a direct coiled-coil-mediated interaction with the AChR.
Rapsyn is likely to interact with other postsynaptic proteins. When expressed in nonmuscle cells, rapsyn promotes the clustering of the muscle-specific receptor tyrosine kinase MuSK, a critical component of the agrin-induced AChR clustering pathway (14, 15). Curiously, rapsyn-induced MuSK clustering requires the ectodomain, but not the cytoplasmic domain of MuSK, implicating the existence of an unidentified transmembrane protein (termed RATL for rapsyn-associated transmembrane linker), which links MuSK extracellularly to rapsyn cytoplasmically (15, 16). Rapsyn also is able to cluster the transmembrane protein β-dystroglycan in heterologous expression systems, suggesting a potential connection between rapsyn and the dystrophin/utrophin protein complex at the postsynaptic membrane (17). One possibility is that RATL and/or β-dystroglycan may interact with rapsyn via TPR domains uninvolved in rapsyn self-association, because TPR domains also appear to be involved in protein associations with non-TPR-containing proteins. The zinc ring finger domain is also likely to mediate protein interactions with rapsyn. The observation that mutations in this domain alter cluster size without disrupting rapsyn’s ability to cluster AChRs is consistent with a role for this domain in linking rapsyn to other postsynaptic proteins (11).
The nicotinic AChR is the prototypic member of the family of ligand-gated ion channels, which also includes glycine receptors (GlyR), γ-aminobutyric acid receptors (GABAAR), and both NMDA (N-methyl-d-aspartate)- and AMPA (α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionic acid)-type glutamate receptors (GluR). Like AChRs at the neuromuscular junction, ionotropic glutamate receptors mediate excitatory electrical signaling at central synapses, whereas glycine and GABAA receptors underlie signaling at their respective inhibitory synapses. Given the homology between members of this receptor family, an expectation has been that they also may be clustered by rapsyn or proteins homologous to rapsyn. In fact, it recently has been reported that rapsyn is capable of clustering GABAA receptors when coexpressed in heterologous cells (18). It will be interesting to determine whether this clustering depends on rapsyn’s coiled-coil motif as it does for AChR clustering. That rapsyn induces GABAA receptor clustering, coupled with recent reports that rapsyn is expressed in neurons, suggests that it may play a broader role in clustering ionotropic receptors at central synapses (18, 19). On the other hand, channel-associated clustering proteins now have been identified for glycine and glutamate receptors, and these appear to share no similarity with rapsyn in terms of sequence, structure, or mode of clustering.
Gephyrin, a 93-kDa protein, originally was identified as a peripheral membrane protein that copurifies with the glycine receptor by affinity chromatography (20, 21). When neurons are depleted of gephyrin by treatment with antisense oligonucleotides, glycine receptor clusters fail to form, implicating an essential role for gephyrin in clustering glycine receptors at synapses (22). Recently gephyrin was shown to bind directly to a sequence in the cytoplasmic loop of the β subunit of the glycine receptor (23). Gephyrin also associates with polymerized tubulin (24) and may interact with microfilaments (25). So gephyrin may provide a link between glycine receptors and the cytoskeleton, thereby anchoring glycine receptors at synaptic sites (25). It is interesting to note that clusters of gephyrin have been identified at GABAergic synapses in hippocampal neurons, raising the possibility that gephyrin also may play a role in the synaptic anchoring of GABA receptors (26). In fact, the cloning of gephyrin revealed that several forms of gephyrin are produced by alternative splicing at sites near the N terminus of the protein. Thus, a reasonable model is that this part of gephyrin interacts with either glycine receptors or other proteins, such as GABA receptors, depending on the spliced form expressed, and that linkage to the microtubule-based cytoskeleton occurs through other parts of the molecule (27).
In contrast to rapsyn and gephyrin, which both were identified during biochemical preparations of their respective receptors, glutamate receptor-associated proteins recently have been isolated by using the yeast two-hybrid method. This approach has identified an important role for PDZ domain-containing proteins in the anchoring and clustering of glutamate receptors at synaptic sites in neurons. Like SH2 and SH3 domains, PDZ domains (named for the three proteins in which they were first found: PSD-95, discs large and ZO-1) are protein interaction modules. The role of PDZ proteins in clustering ion channels has been extensively reviewed elsewhere (28–32).
By using the cytoplasmic tails of NMDA receptor subunits as baits to screen brain cDNA libraries, an interaction was identified between an S/TXV motif at the extreme C terminus of NR2 subunits and members of the PSD-95 family of proteins (33–35). When coexpressed in heterologous cells, PSD-95 and its relative chapsyn-110 promote the clustering of NMDA receptors, in a manner analogous to rapsyn’s clustering of AChRs (36). Unlike rapsyn, however, PSD-95 and chapsyn-110 do not form membrane clusters when expressed alone. Whereas anchoring of the AChR at the synapse presumably occurs via interactions between rapsyn and the cytoskeleton, the NR1 subunits of NMDA receptors themselves appear to mediate direct associations with cytoskeletal elements (37–39). Interestingly, interaction between NR1 subunits and the cytoskeleton may be regulated by alternative splicing, protein phosphorylation, and/or calcium/calmodulin binding (37, 40).
By using a similar yeast two-hybrid approach as that taken with NMDA-type receptors, an AMPA receptor-associated protein recently was isolated (41). Although unrelated to the PSD-95 family, this protein, named GRIP (for glutamate receptor interacting protein), binds to C-terminal sequences of AMPA receptor subunits (SVKI) via its PDZ domains. GRIP is composed of seven PDZ domains; the fourth and fifth are involved in binding AMPA receptor subunits. The remaining PDZ domains presumably mediate interactions with other cytoskeletal and/or signaling proteins. GRIP colocalizes with AMPA receptors in cultured neurons and overexpression of the C terminus of the GluR2 subunit dramatically decreases the number of synaptic clusters of AMPA receptors. This experiment suggests that an interaction between the tail of AMPA receptor subunits and a synaptic protein, such as GRIP, is critical in clustering AMPA receptors.
These examples (summarized in Table 1) reveal a commonality in the mechanisms that cluster transmitter receptors at synapses—the involvement of a protein that interacts with cytoplasmic domains of the receptors. In fact, accumulating evidence suggests that these proteins may do much more than just cluster their respective receptors. The presence of multiple sites for protein interactions (PDZ domains, TPRs, etc.) implies that proteins such as rapsyn or PSD-95 may direct the assembly of multifunctional complexes at sites where high densities of ligand-gated channels accumulate. In some cases, the cytoplasmic protein machinery may respond directly to ions entering through the channel. For example, the linkage of nitric oxide synthase to NMDA receptors by PSD-95 (42) suggests a mechanism by which calcium entering a neuron at the synapse could produce nitric oxide for retrograde signaling. Thus, as more proteins that interact with rapsyn, gephyrin, PSD-95, and GRIP are identified, our understanding of the complexity of synaptically mediated biochemical events is certain to grow.
Table 1.
Summary of mechanisms involved in clustering ligand-gated ion channels
| Neurotransmitter receptor | Subunit composition | Clustering protein | Mode of interaction | Cytoskeletal attachment |
|---|---|---|---|---|
| AChR | α, β, γ/ɛ, and δ | Rapsyn | Coiled-coil region of rapsyn required for heterologous clustering of AChR | Actin via β-dystroglycan/utrophin? |
| GlyR | α and β | Gephyrin | Binding to gephyrin requires intracellular loop in β subunit of GlyR | Tubulin Microfilament? |
| GABAAR | α, β, and γ | Rapsyn? | ? | ? |
| Gephyrin? | ||||
| GluR-NMDA | NR1 and NR2 A–D | PSD-95 family members | PDZ domains of PSD-95 family members bind to C-terminal SXV sequences in NR2 subunits and induce clustering in heterologous cells | C0 cassette of NR1 binds α-actinin (37) C1 cassette binds neurofilament (38) and yotiao (39) |
| GluR-AMPA | GluR 1–4 | GRIP | PDZ domains in GRIP bind to C-terminal SKVI sequence in GluR subunits | ? |
References
- 1.Fertuck H C, Salpeter M M. Proc Natl Acad Sci USA. 1974;71:1376–1378. doi: 10.1073/pnas.71.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobel A, Weber M, Changeux J-P. Eur J Biochem. 1977;80:215–244. doi: 10.1111/j.1432-1033.1977.tb11874.x. [DOI] [PubMed] [Google Scholar]
- 3.Sealock R, Wray B E, Froehner S C. J Cell Biol. 1984;98:2239–2244. doi: 10.1083/jcb.98.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Froehner S C, Luetje C W, Scotland P B, Patrick J. Neuron. 1990;5:403–410. doi: 10.1016/0896-6273(90)90079-u. [DOI] [PubMed] [Google Scholar]
- 5.Phillips W D, Kopta C, Blount P, Gardner P D, Stenibach J H, Merlie J P. Science. 1991;251:568–570. doi: 10.1126/science.1703661. [DOI] [PubMed] [Google Scholar]
- 6.Gautam M, Noakes P G, Mudd J, Nichol M, Chu G C, Sanes J R, Merlie J P. Nature (London) 1995;377:232–236. doi: 10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- 7.Ramarao M K, Cohen J B. Proc Natl Acad Sci USA. 1998;95:4007–4012. doi: 10.1073/pnas.95.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting C C P, Phillips C. Biochem J. 1996;314:1053–1056. doi: 10.1042/bj3141053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goebi M, Yanagida M. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 10.Frail D E, Mudd J, Shah V, Carr C, Cohen J B, Merlie J P. J Biol Chem. 1988;263:15602–15607. [PubMed] [Google Scholar]
- 11.Freemont P S. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 12.Scotland P B, Colledge M, Melnikova I, Dai Z, Froehner S C. J Cell Biol. 1993;123:719–727. doi: 10.1083/jcb.123.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips W D, Maimone M M, Merlie J P. J Cell Biol. 1991;115:1713–1723. doi: 10.1083/jcb.115.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie S K H, Balasubramanian S, Fung E T, Huganir R L. Neuron. 1996;16:953–962. doi: 10.1016/s0896-6273(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 15.Apel E D, Glass D J, Moscoso L M, Yancopoulos G D, Sanes J R. Neuron. 1997;18:623–635. doi: 10.1016/s0896-6273(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 16.Glass D J, Apel E D, Shah S, Bowen D C, DeChiara T M, Stitt T N, Sanes J R, Yancopoulos G D. Proc Natl Acad Sci USA. 1997;94:8848–8853. doi: 10.1073/pnas.94.16.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apel E D, Roberds S L, Campbell K P, Merlie J P. Neuron. 1995;15:115–126. doi: 10.1016/0896-6273(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 18.Yang S-H, Armson P F, Cha J, Phillips W D. Mol Cell Neurosci. 1997;8:430–438. doi: 10.1006/mcne.1997.0597. [DOI] [PubMed] [Google Scholar]
- 19.Burns A L, Benson D, Howard M J, Margiotta J F. J Neurosci. 1997;17:5016–5026. doi: 10.1523/JNEUROSCI.17-13-05016.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt B, Knaus P, Becker C-M, Betz H. Biochemistry. 1987;26:805–811. doi: 10.1021/bi00377a022. [DOI] [PubMed] [Google Scholar]
- 21.Pfeiffer F, Graham D, Betz H. J Biol Chem. 1982;257:9389–9393. [PubMed] [Google Scholar]
- 22.Kirsch J, Wolters I, Triller A, Betz H. Nature (London) 1993;366:745–748. doi: 10.1038/366745a0. [DOI] [PubMed] [Google Scholar]
- 23.Meyer G, Kirsch J, Betz H, Langosch D. Neuron. 1995;15:563–572. doi: 10.1016/0896-6273(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 24.Kirsch J, Langosch D, Prior P, Littauer U Z, Schmitt B, Betz H. J Biol Chem. 1991;266:22242–22245. [PubMed] [Google Scholar]
- 25.Kirsch J, Betz H. J Neurosci. 1995;15:4148–4156. doi: 10.1523/JNEUROSCI.15-06-04148.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craig A M, Banker G, Chang W, McGrath M E, Serpinskaya A S. J Neurosci. 1996;16:3166–3177. doi: 10.1523/JNEUROSCI.16-10-03166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prior P, Schmitt B, Grenningloh G, Pribilla I, Multhaup G, Beyreuther K, Maulet Y, Werner P, Langosch D, Kirsch J, Betz H. Neuron. 1992;8:1161–1170. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 28.Sheng M, Wyszynski M. BioEssays. 1997;19:847–853. doi: 10.1002/bies.950191004. [DOI] [PubMed] [Google Scholar]
- 29.Sheng M. Nature (London) 1997;386:221–222. [Google Scholar]
- 30.Ehlers M D, Mammen A L, Lau L-F, Huganir R L. Curr Opin Cell Biol. 1996;8:484–489. doi: 10.1016/s0955-0674(96)80024-x. [DOI] [PubMed] [Google Scholar]
- 31.Ponting C P, Phillips C, Davies K E, Blake D J. BioEssays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 32.Kornau H-C, Seeburg P H, Kennedy M B. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 33.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 34.Muller B M, Kistner U, Kindler S, Chung W J, Kuhlendahl S, Fenster S D, Lau L-F, Veh R W, Huganir R L, Gundelfinger E D, Garner C C. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- 35.Niethammer M, Kim E, Sheng M. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E, Cho K-O, Rothschild A, Sheng M. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 37.Wyszynski M, Lin J, Rao A, Nigh E, Beggs A H, Craig A M, Sheng M. Nature (London) 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 38.Ehlers M D, Fung E T, O’Brien R J, Huganir R L. J Neurosci. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, J. W., Wyszynski, M., Madhavan, R., Sealock, R., Kim, J. U. & Sheng, M. (1998) J. Neurosci., in press. [DOI] [PMC free article] [PubMed]
- 40.Ehlers M D, Tingley W G, Huganir R L. Science. 1995;269:1734–1737. doi: 10.1126/science.7569904. [DOI] [PubMed] [Google Scholar]
- 41.Dong H, O’Brien R J, Fung T, Lanahan A A, Worley P F, Huganir R L. Nature (London) 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 42.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, et al. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]