Abstract
Fluorescent proteins (FPs) are useful tools for visualizing live cells and their behaviors. Protein domains that mediate subcellular localization have been fused to FPs to highlight cellular structures. FPs fused with histone H2B incorporate into chromatin allowing visualization of nuclear events. FPs fused to a glycosylphosphatidylinositol anchor signal sequence (GPI) label the plasma membrane, highlighting cellular shape. Thus, a reporter gene containing both types of FP fusions would allow for effective monitoring of cell shape, movement, mitotic stage, apoptosis and other cellular activities. Here we report a binary color-coding system using four different colored fluorescent protein reporters that generates 16 distinct color codes to label the nuclei and plasma membranes of live cells in culture and in transgenic mice. As an initial test of this system in vivo, the promoter of the human Ubiquitin C (UBC) gene was used to widely express one of the color code reporters. Widespread expression of the reporter was attained in embryos; however, both male and female transgenic mice were infertile. In contrast, the promoter of the mouse Oct4/Pou5f1 gene linked to two different color code reporters specifically labeled blastocysts, primordial germ cells, and postnatal germ cells and these mice were fertile. Time-lapse movies of fluorescently-labeled primordial germs cells demonstrate the utility of the color code system to visualize cell behaviors. This set of new fluorescent protein reporters should be a useful tool for labeling distinct cell populations and studying their behaviors in complex tissues in vivo.
Keywords: fluorescent protein, 2A, live imaging, transgenic mice, Oct4, embryo, primordial germ cells
Organ formation is a complex process involving cell proliferation, differentiation and morphogenesis. We are interested in understanding this process and therefore sought to generate new genetic tools to monitor cell behaviors in vivo. To monitor these behaviors, we aimed to generate transgenic mice in which specific cell types express both a fluorescent nuclear label and a fluorescent plasma membrane label. In this manner, mitosis, apoptosis, cell shape, and movements can be simultaneously monitored in live cells. Fluorescent proteins (FP) fused to either histone H2B or a glycosylphosphatidylinositol (GPI) signal sequence have been shown to label the nucleus and plasma membrane, respectively (Hadjantonakis and Papaioannou, 2004; Rhee et al., 2006). Thus, we created gene constructs for transgenesis in which a tissue-specific promoter of choice directs expression of both fusion proteins. To combine the advantages of both types of fluorescent markers, we designed co-expression cassettes in which the coding regions for the two FP-fusions are separated by the self-cleavage 2A peptide sequence. 2A or 2A-like peptides are conserved amino acid sequences mainly found in the genomes of several groups of animal viruses and mediate co-translational cleavage of multi-polypeptides from a common open reading frame (Szymczak and Vignali, 2005). 2A peptide mediated co-expression has a few advantages over that mediated by the internal ribosome entry site (IRES). First, 2A peptides are small, consisting of 18 to 25 amino acids. Second, 2A-linked coding regions are expressed at stoichiometric levels.
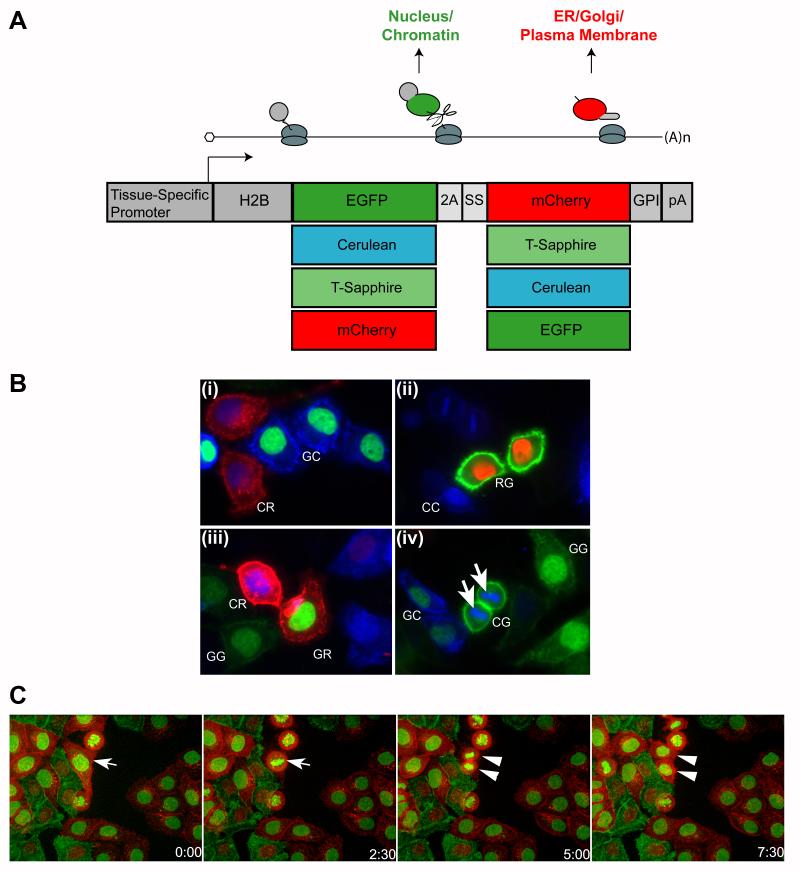
We sought to create a genetic “tool kit” of different FP color combinations for the purpose of distinguishing individual cell types within a given tissue. FPs in distinguishable excitation/emission wavelength categories were selected (one in each category based on brightness and photostability). Cerulean was chosen in the cyan category (Rizzo et al., 2004); enhanced green fluorescent protein (EGFP) in the green; monomeric Cherry fluorescent protein (mCherry) in the red (Shaner et al., 2004); and also T-Sapphire, a long-UV activated mutant EGFP (Zapata-Hommer and Griesbeck, 2003). In total, there are 16 possible color combinations using this system and we generated an individual construct for each. We use the nomenclature HS-XY to describe the cassettes where X and Y are either C for Cerulean (a CFP; cyan FP), G for EGFP, R for mCherry (an RFP; red FP), or T for T-Sapphire. X is the FP that is fused with histone H2B and Y is the FP tagged with the GPI signal sequence.
The basic design of the dual labeling constructs is illustrated in Fig. 1A. A promoter of choice directs expression of a single mRNA encoding two FP fusion proteins separated by a Thosea asigna viral 2A sequence (Szymczak and Vignali, 2005). The first FP is fused to histone H2B and therefore labels chromatin. The second FP is fused at the N-terminus with the acrosin secretion signal peptide for insertion into the endoplasmic reticulum and at the C-terminus with the GPI anchor signal sequence from the Thy1 gene for insertion into the plasma membrane. All combinations of the four chosen FPs were generated yielding a total of 16 gene constructs.
Fig. 1.
Generation and analysis of gene constructs for dual labeling. A, Schematic of gene constructs. A promoter of choice directs expression of a single transcript encoding both a histone H2B-fluorescent protein and a fluorescent protein with a C-terminal GPI anchor signal sequence. A 2A self-cleavage peptide placed between the two fluorescent proteins physically separates the two polypeptides during translation. SS = endoplasmic reticulum signal peptide; pA = bovine growth hormone polyadenylation signal. B, HeLa cells expressing different bicolor constructs. HeLa cells were transfected with different CAG-HS constructs separately, and then pooled to simulate labeling different cells with different color codes in the same tissue. (i) HS-GC and -CR-expressing cells. (ii) HS-RG and -CC-expressing cells. (iii) HS-CR, -GR, -GG and -CC-expressing cells. (iv) CG, -GC and -GG-expressing cells. Chromosomes in dividing cells can be observed (arrows). C, Frames at different hours from a time-lapse video of GR- and RG-expressing HeLa cells. Arrow indicates a cell that undergoes mitosis and daughter cells (arrowheads) during the video.
We tested the labeling quality and efficiency of the gene cassettes by transfecting HeLa cells with individual cassettes under the control of the CAG promoter (Niwa et al., 1991). Each well of HeLa cells was transfected with a different construct. The cells were then pooled for live imaging. The dual label cassettes behaved as designed. H2B-FPs localized to nuclei and chromosomes and GPI-FPs labeled the plasma membrane (Fig. 1B). The 2A peptide was very efficient at inducing cleavage because co-localization of H2B and GPI fusion FPs was never observed. Both FPs in individual cassettes appeared to be expressed at similar levels. Importantly, the expression of these distinct color codes could unambiguously identify different cells even if they were adjacent to each other.
To demonstrate the feasibility of imaging live dually-labeled cells over time, we stably transfected HeLa cells with CAG-HS-RG and CAG-HS-GR constructs. After two weeks of selection and four passages, virtually all of the surviving cells express the HS cassette, though with some variation in levels. We mixed the CAG-HS-RG and CAG-HS-GR cells together and imaged them with a spinning disc confocal microscope for 48 hours in a thermostatic chamber with a humidified CO2/air supply. We captured image stacks at different focal planes and time points. By assembling the images into a video, we were able to observe cell divisions and shape changes (Fig. 1C, Video S1). These in vitro experiments demonstrate the feasibility of using this system to differentially label distinct cell populations and observe their interactions in real time. Maintained in the G418 selection medium, the stably transfected HeLa cells were passaged more than ten times without observable loss of the FP signals or change of labeling characteristics.
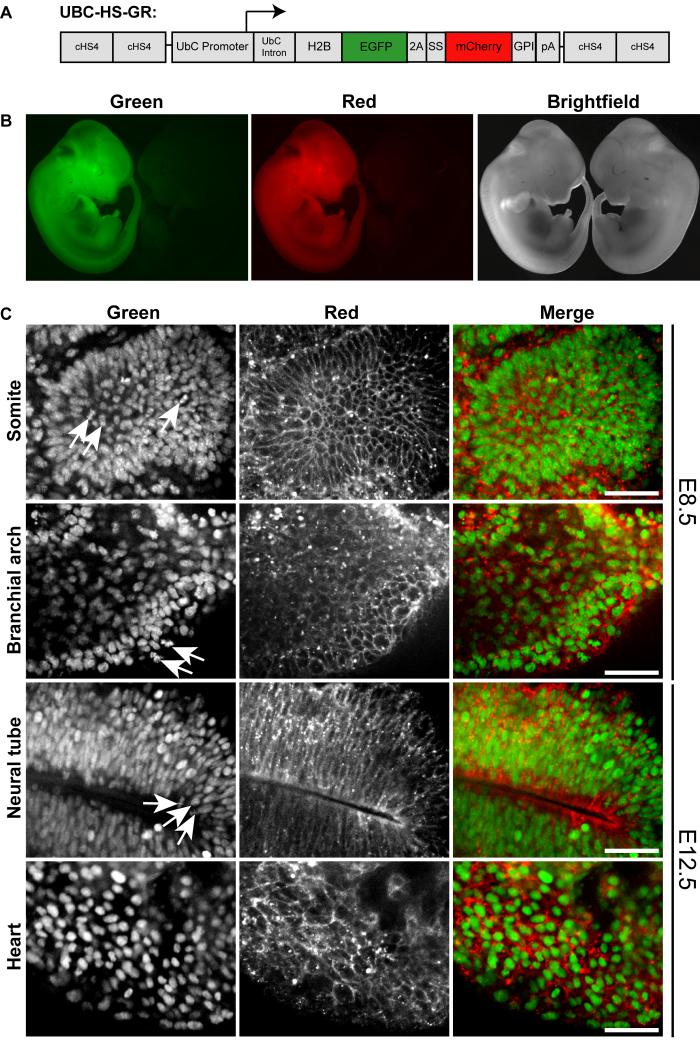
As an initial in vivo test of this dual fluorescent labeling system, we generated transgenic mice widely expressing H2B-EGFP and mCherry-GPI. The human Ubiquitin C (UBC) promoter fragment was used to direct expression of the HS-GR cassette (UBC-HS-GR) in a wide variety of embryonic cell types (Schorpp et al., 1996). To reduce position effects at the site of transgene integration, the transgene was flanked by two copies of cHS4 (chicken hypersensitive site 4) transcriptional insulators (Potts et al., 2000). The gene construct is illustrated in Fig. 2A.
Fig. 2.
Generation and characterization of UBC-HS-GR transgenic mouse embryos. A, Schematic of the UBC-HS-GR gene construct. cHS4 = chicken hypersensitive site 4; SS = endoplasmic reticulum signal peptide; pA = bovine growth hormone polyadenylation signal. B, In each panel, UBC-HS-GRtg/+ (left) and wild type (right) E11.5 embryos viewed under a stereofluorescence microscope. C, Confocal images of embryonic tissues from UBC-HS-GRtg/+ mice. Left panels, H2B-EGFP; center panels, mCherry-GPI; right panels, composite images. Arrows indicate mitotic cells. Scale bar = 30 μm.
We obtained UBC-HS-GR founder animals with similar efficiency to that of other transgenes and most founders expressed detectable levels of the fluorescent proteins (13 of 14 founders expressed the transgene in ear punches); however, most of the male and female founders were infertile. Three of the founders transmitted the transgene to F1 progeny; however, the F1 transgenic animals were infertile. We were only able to maintain one UBC-HS-GR transgenic line with ubiquitous HS-GR expression on an outbred genetic background sufficiently long enough to collect data.
The UBC-HS-GR transgene appears to be widely expressed. UBC-HS-GR male mice were bred to Swiss Webster females and embryos were collected at embryonic day (E) 8.5 and E12.5. Whole E12.5 wild-type and transgenic embryos were observed under a stereofluorescence microscope. The entire transgenic embryo fluoresced in red and green channels. No fluorescence was observed in wild-type embryos (Fig. 2B).
We examined transgene expression in various tissues at E8.5 and E12.5. At both embryonic stages, H2B-EGFP labeled cell nuclei in non-dividing cells and chromosomes could be discerned in mitotic cells. The mCherry-GPI labeled cell membranes and also intracellular vesicular structures (Fig. 2C).
Our laboratory has a specific interest in mammalian sexual development; therefore we sought to test our bicolor transgenic system to study the development of the embryonic gonads. Both XX and XY mammals initiate gonadal development with the formation of a bipotential gonad which is capable of giving rise to either a testis or ovary (Swain and Lovell-Badge, 1999). In mice, this occurs between E10.5 and E11.5. The bipotential gonad consists predominantly of two cell types: primordial germ cells (PGCs) and supporting somatic cells. In XX gonads the germ cells and supporting cells give rise to oogonia and granulosa cells, respectively. In XY gonads, germ cells become spermatogonia while the supporting cells differentiate into Sertoli cells.
PGCs, like certain stem cells, express Oct4/Pou5f1, a well-characterized stem cell transcription factor gene. In preimplantation embryos, the Oct4 gene is expressed in all cells until the blastocyst stage, when it restricts expression to the ICM. However, Oct4 protein can be detected in both the ICM and trophectoderm until late blastocyst stages (Palmieri et al., 1994). Postimplatation Oct4 expression is further restricted to the epiblast. It is expressed by PGCs during and after their migration to the genital ridge (Rosner et al., 1990; Scholer et al., 1990). In XY gonads, germ cells continually express Oct4 throughout development and Oct4 remains expressed in type A spermatogonia after birth. In XX gonads, germ cells express Oct4 until it is silenced upon entry into meiosis at E13.5. The Oct4 gene remains silenced until it is reactivated during oocyte maturation (Pesce et al., 1998).
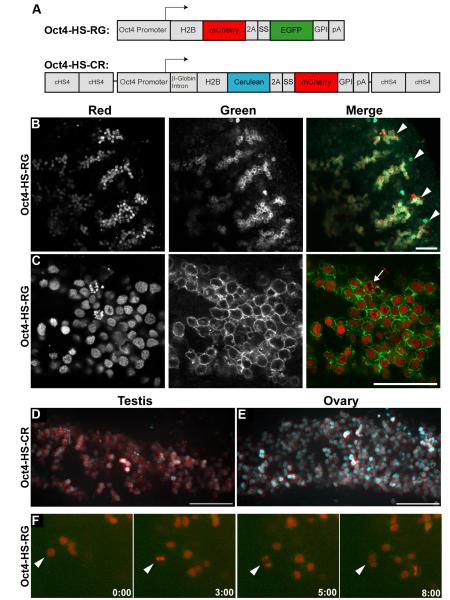
We generated two gene constructs utilizing 2.8 kb of 5′ flanking sequence, containing both the proximal and distal promoters, of the mouse Oct4/Pou5f1 gene (Nordhoff et al., 2001). In the first construct, the Oct4 promoter directs expression of H2B-mCherry-2A-EGFP-GPI (Oct4-HS-RG). This construct labels chromatin with mCherry FP and the plasma membrane with EGFP. In the second construct, the Oct4 promoter directs expression of H2B-Cerulean-2A-mCherry-GPI (Oct4-HS-CR). This construct labels chromatin with Cerulean FP and the plasma membrane with mCherry FP. A schematic of the two gene constructs is illustrated in Fig. 3A. Of note, the Oct4-HS-CR construct is flanked by four cHS4 transcriptional insulator sequences, whereas the Oct4-HS-RG construct is not. Founder mice were screened for transgene expression by crossing to outbred Swiss Webster female mice, collecting embryos on E12.5, and analyzing the gonads for fluorescence. Interestingly, all five Oct4-HS-CR founders expressed the transgene, whereas only two of ten Oct4-HS-RG founders expressed. Thus, it appears that the addition of insulators facilitated Oct4-directed transgene expression.
Fig. 3.
Generation and characterization of Oct4-HS-RG and Oct4-HS-CR transgenic mice. A, Schematic of the Oct4-HS-RG and Oct4-HS-CR gene constructs. cHS4 = chicken hypersensitive site 4; SS = endoplasmic reticulum signal peptide; pA = bovine growth hormone polyadenylation signal. B, C, Oct4-HS-RGtg/+ E13.5 testis. Left panels, H2B-mCherry; center panels, EGFP-GPI; right panels, composite images. B, Four cords with germ cells (arrowheads). Scale bar = 100 μm. C, Arrow indicates a germ cell in anaphase. Scale bar = 50 μm. D, Oct4-HS-CRtg/+ E12.5 testis; composite. Scale bar = 100 μm. E, Oct4-HS-CRtg/+ E12.5 ovary; composite. Scale bar = 100 μm. F, Frames at different hours from a time-lapse video of an Oct4-HS-RGtg/+ E11.5 gonad. Arrowhead indicates a cell that undergoes mitosis during the video.
Gonads from Oct4-HS-RGtg/+ (Line 8) and Oct4-HS-CRtg/+ (Line 1) mice were collected between E11.5 and birth, postnatal stages and adult (>8 wk). Transgene expression was determined by viewing under a stereofluoresence microscope. Embryonic gonads were imaged as fresh whole tissue by confocal microscopy. Postnatal gonads were cryosectioned before confocal imaging.
In XY gonads, both transgenes were expressed from E11.5 through E17.5; however, fluorescence levels at E15.5 and E17.5 were significantly lower than at earlier stages. In XX gonads, both transgenes were specifically expressed in the germ cells at E11.5 and E13.5, but were undetectable at later stages (data not shown). This transgene expression pattern correlates with the endogenous expression of the Oct4 gene. In XY gonads, the Oct4 gene is continually expressed throughout development, whereas in XX gonads, the Oct4 gene is only expressed until the germ cells enter meiosis on E13.5. Representative images of E13.5 Oct-RG testes are shown in Figs. 3B, C. Under low magnification, the germ cells of the E13.5 testis are arranged in cords (Fig. 3B). In higher magnification images mitotic events can be discerned as well as the shape of the germ cells and their spatial relationships to each other (Fig. 3C). Representative images of an E12.5 Oct4-HS-CR testis and ovary are shown in Fig. 3D, E, respectively. The H2B-Cerulean labeled the germ cell nuclei and the mCherry-GPI labeled the plasma membrane. Gonads from Oct4-HS-RGtg/+ embryos at E11.5 were cultured ex vivo and imaged for 24 h using a spinning disc confocal microscope (Video S2, S3). Germ cells rapidly proliferate during this period of development. Indeed, we observed many mitotic germ cells in these organ culture experiments. Representative frames are presented in Fig. 3F.
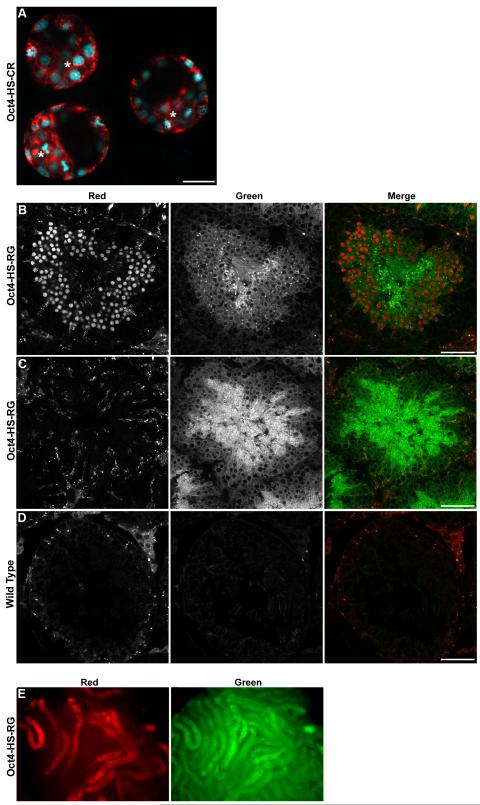
Both Oct4-HS-RG and Oct4-HS-CR transgenes were active in the blastocyst. Representative confocal images of Oct4-HS-CR E3.5 blastocysts are presented in Fig. 4A. The dual fluorescent proteins were detectable in both the inner cell mass and trophoblast. This is in contrast to the mRNA expression pattern for Oct4, which is restricted to the ICM. However, it is in agreement with immunohistochemical analysis of the Oct4 protein, which can be found in both the ICM and trophectoderm until late blastocyst development (Palmieri et al., 1994).
Fig. 4.
A, Oct4-HS-CRtg/+ blastocysts. Composite confocal image. Asterisks indicate ICM. Scale bar = 100 μm. B, C, Oct4-HS-RGtg/+ adult testis. Left panels, H2B-mCherry; center panels, EGFP-GPI; right panels, composite images. Scale bar = 50 μm. B, Seminiferous tubule containing round spermatids, expressing H2B-mCherry. C, Seminiferous tubule containing no spermatids. D, Seminiferous tubule from a non-transgenic adult male. E, Oct4-HS-RGtg/+ testis viewed under a stereofluorescence microscope. Left panel, H2B-mCherry; right panel, EGFP-GPI. Subregions of the seminiferous tubules express mCherry, whereas all regions express EGFP.
Interestingly, in the seminiferous tubules of the adult testis of Oct4-HS-RG and Oct4-HS-CR transgenic mice, the nuclear label was not detectable in spermatogonia or spermatocytes, but was observed in round spermatids (Fig. 4B) and elongating spermatids (data not shown). The membrane label (EGFP-GPI or mCherry-GPI) filled the germ cell cytoplasm and was observed in all seminiferous tubules regardless of the presence of round/elongating spermatids (Fig. 4B, C and not shown). The detection of the membrane label throughout all seminiferous tubules was not due to tissue autofluorescence because little to no fluorescence was observed in seminiferous tubules from non-transgenic mice (Fig. 4D). Similarly, under the stereofluorescence microscope, all seminiferous tubules are positive for the membrane FP; however, only those tubules that contain spermatids are positive for the nuclear FP (Fig. 4E). Thus, although the nuclear expression pattern does not match that of endogenous Oct4, these mice may be useful for determining the approximate stage of spermatogenesis in individual seminiferous tubules.
Fluorescent protein reporters are important tools for visualizing live cells and their behaviors during tissue differentiation and morphogenesis (Hadjantonakis et al., 2003). Using only one color of FP limits the number of different cell types that can be imaged in a complex tissue. In addition, using traditional cytoplasmically-localized FPs, although useful, sometimes provides limited information about cell characteristics. FPs fused with other proteins or protein domains that direct subcellular localization provide more information about cell structure, morphology, and behavior (Hadjantonakis et al., 2003). Recently, Trichas et al. (2008) generated a gene construct to simultaneously express, in tissue culture cells and mouse embryos, a Tomato FP localized to plasma membranes with a myristoylation signal (myr-Tomato) and H2B-EGFP using the 2A system described here. In their case, they placed the membrane-localized FP 5′ of the H2B-localized FP, whereas ours is reversed. Both 2A strategies appeared to work well. In addition, Nowotschin et al. (2009) co-electroporated separate constructs to generate embryonic stem cell lines that co-express H2B-Cherry and GPI-GFP or H2B-GFP and myr-mRFP1. Mouse embryos generated from the H2B-Cherry; GPI-GFP ES cell lines labeled nuclei with Cherry FP and plasma membranes with GFP. Mixing the RG and GR ES cell lines resulted in colonies in which individual ES cells could be recognized by the two distinct color codes. We extend the above studies by creating a binary color-coding system using four distinct FPs that generates a set of 16 different fluorescent protein gene cassettes that simultaneously label the nucleus/chromosomes and plasma membrane. This binary color-coding system provides a means to unequivocally label different cells types in complex tissues for static and real time studies of tissue and organ formation. In the future, transgenic mouse lines that express our different color codes in different cell types can be bred together to generate bi-, tri- or perhaps quadri-genic mice whose developing tissues can be analyzed by static or time-lapse imaging to determine how cell types interact during organogenesis.
While our system labels most embryonic cell types correctly, some tissues do not exhibit ideal cell membrane labeling. In addition, EGFP expression in Oct4-GR in the seminiferous tubules of the adult testes was distributed throughout the cytoplasm. This may be a post-translational problem specifically affecting the FP fused to the GPI signal sequence, because the H2B-FP was always localized to the nucleus. It is possible that over-expression of the GPI-anchored FP may exceed the capacity of the processing enzymes for GPI addition or the GPI-anchored protein sorting system in some cells and thus results in mislocalization of the FP, which is trapped in cytoplasmic vesicles. There are also phospholipase enzymes expressed in many tissues which may cleave the GPI anchor and release the attached protein into the extracellular environment (Fouchier et al., 1990; Hooper et al., 1997; Littlewood et al., 1989). GPI-anchored endogenous proteins are usually sorted into specific membrane domains or “rafts” (Mayor and Riezman, 2004), which may explain the uneven distribution of membrane fluorescent signal observed in some tissues. Very different results were presented in the two previous reports on in vivo GPI-anchored FP distribution. Kondoh et al. (1999) described uneven distribution in most tissues, whereas Rhee et al. (2006) reported even labeling in all the tissues they examined. As an alternative, FPs with myristoylation domains could be incorporated into our binary color code system (Larina et al., 2009).
Transgenes flanked by cHS4 insulators and utilizing the human UBC promoter direct expression of FPs at levels that are easily detectable by fluorescence microscopy in almost all of the transgenic founders we examined. This suggests that this gene construct design is less susceptible to position effects and is useful for transgene expression in most founders (Hsiao et al., 2004). However, we observed a fertility problem in our UBC-HS-GR lines. This could be a consequence of high levels of H2B- or GPI-tagged FPs in germ cells or the supporting niche. It may be better to generate UBC-HS-XY transgenic mice without the cHS4 insulators to create lines with lower but sufficient levels of FP expression that might be compatible with fertility. This should be achievable because CAG-myr-tomato-2A-H2B-EGFP transgenic mouse lines express sufficient levels of FPs for fluorescent imaging but not such high levels to compromise fertility (Trichas et al., 2008). Our Oct4-HS-CR construct included cHS4 insulators that appeared to increase the likelihood of PGC expression in transgenic mice. In these lines and the Oct4-GR lines, there was sufficient FP expression for static and time-lapse imaging but apparently the levels were low enough in the PGCs to be compatible with fertility. Indeed, we have also generated Gata4-HS-GR transgenic mouse lines that express in Sertoli cells of the testis and these mice are also fertile (Nel-Themaat and Behringer, unpublished observations). This suggests that tissue-specific regulatory elements combined with our color-code system may generally lead to the generation of fertile transgenic mice. It will be exciting to begin combining transgenes expressing distinct color codes in different cell types of forming organs to visualize their individual behaviors and interactions during organogenesis.
METHODS
Gene constructs
The pCAG-HS-XY plasmids for transient overexpression in HeLa cells are composed of a CAG promoter (Niwa et al., 1991), the individual HS cassettes and a bovine growth hormone polyadenylation signal. The plasmids were constructed from a pCMV (Boshart et al., 1985) backbone, and the chicken beta-actin promoter and intron was introduced by NdeI/XhoI digestion to reconstruct the CAG promoter. The human histone H2B sequence was subcloned from pCX-H2B/ECFP (a gift from Dr. Kat Hadjantonakis) by PCR into pCAG-HS-XY plasmids between the XhoI and BamHI sites. FPs in all constructs were PCR-amplified and subcloned from various sources (Rizzo et al., 2004; Shaner et al., 2004; Zapata-Hommer and Griesbeck, 2003). The 2A sequence and the acrosin secretion signal peptide sequence were reconstructed with oligonucleotides (Kondoh et al., 1999; Szymczak and Vignali, 2005). The GPI sequence was PCR-amplified from pCX-GFP-GPI (a gift from Dr. Kat Hadjantonakis) and subcloned between the BsrGI and XbaI restriction sites. The plasmid used for generating UBC-HS-GR mice utilizes the human Ubiquitin C promoter (Schorpp et al., 1996) (including the first intron) and the bovine growth hormone polyadenylation signal. The whole transgene is flanked by two copies of cHS4 insulators on both sides (Chung et al., 1993). The pXL-CAG-HS-GR(or-RG)-Neo constructs for stable expression in HeLa cells were made using a pXL-BacII piggyBac transposon (Li et al., 2005). CAG-driven HS-GR and -RG together with a Pgk-Neo-bpA selection marker were flanked by the transposon inverted repeats.
A plasmid containing the 2.8 kb Oct4 promoter/enhancer was generously provided by Dr. Paul Overbeek (Baylor College of Medicine). To generate the construct for creation of Oct4-HS-RG mice, the Oct4 promoter was amplified by PCR and subcloned into a previously made vector containing H2B-mCherry-2A-EGFP-GPI (HS-RG) by PCR followed by restriction digest and ligation. PCR primers were: 5′- GGA TCC TCG AGC CGG GGG CCT GGT GGA AAG (forward), 5′-GGT CAA GAT CTT GAC CGG ATT CGA GTA TG (reverse). To generate the construct for creation of Oct4-HS-CR mice, the same Oct4 promoter was used to drive expression of an HS-CR cassette. Of note, the Oct4-HS-CR construct is flanked by cHS4 insulators and includes a β-globin intron in front of the coding sequence, whereas the Oct4-HS-RG construct does not.
Cell culture and transfection
HeLa cells were cultured in DMEM medium supplemented with 10% fetal bovine serum in a 37°C incubator with 5% CO2. HeLa cells were transfected using Lipofectamine (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. For stable transfections, the transposase expressing plasmid, pcDNA3-mPB (Cadinanos and Bradley, 2007), was cotransfected into HeLa cells, and transfected cells were selected with G418 (500 μg/ml, beginning 2 days after transfection) for four passages as mixed cultures.
Generation of transgenic mice
UBC-HS-GR and Oct4-HS-CR mice were generated in our laboratory according to standard procedures (Nagy, 2003). Briefly, gene constructs were released from their plasmid backbones by restriction enzyme digestion and isolated by electrophoresis and gel purification using a Gel Extraction kit (Qiagen). Purified DNA was eluted in injection buffer (10 mM Tris pH 7.4, EDTA 0.1 mM) and subsequently diluted with injection buffer to a working concentration of 2 ng/μl. Zygotes for microinjection were collected from matings between superovulated B6SJLF1/J females and B6SJLF1/J males. Pseudopregnant recipient females were obtained from crosses of Swiss Webster (Taconic) female and vasectomized Swiss Webster male mice. Oct4-HS-RG mice were generated by the Genetically Engineered Mouse Facility at U.T. M.D. Anderson Cancer Center using B6D2F1 zygotes. All experimental animals were maintained in accordance with the Institutional Animal Care and Use Committee of the M.D. Anderson Cancer Center and the NIH Guide for the Care and Use of Laboratory animals.
For UBC-HS-GR, progeny were genotyped with primers: 5′-CTG AAG CTC CGG TTT TGA ACT ATG CG (forward) and 5′-CCT TAG CGC TGG TGT ACT TGG TG (reverse), yielding a 550 bp fragment or by examining fluorescence of clipped toes. Oct4-HS-RG and Oct4-HS-CR transgenic mice were genotyped with the following primers: 5′-TAA GGG TTG TCC TGT CCA GAC G (forward) and 5′-TTT CTT CTG CGC CTT AGT CAC C (reverse), yielding a ~250 bp (Oct4-HS-RG) or ~750 bp (Oct4-HS-CR) fragment. The UBC-HS-GR transgenic line was maintained as hemizygotes on a Swiss Webster outbred background. Oct4-HS-RG transgenic mice were maintained on a B6D2 hybrid background. Oct4-HS-CR transgenic mice were maintained on a B6SJL hybrid background.
The progeny of Oct4-HS-RG and Oct4-HS-CR transgene positive founders were screened for expression by collecting gonads at E12.5 and viewing them with a stereofluorescence microscope. E12.5 was chosen for screening because the Oct4 gene is expressed in both XY and XX germ cells on this day and this is the first day in which XY and XX gonads can be visually differentiated by the presence or absence of testis cords, respectively.
Tissue collection and microscopy
UBC-HS-GRtg/+, Oct4-HS-RGtg/+ or Oct4-HS-CRtg/+ male mice were mated with outbred Swiss Webster females. For our studies, 12 pm on the day of the vaginal plug was considered E0.5. Blastocysts were collected on E3.5.
Blastocysts were flushed from uteri with DMEM supplemented with 10% FBS, 2 mM glutamine, 20 mM HEPES and antibiotics. Healthy blastocysts were selected and placed in a drop culture using the same medium under oil and imaged using a Perkin Elmer spinning disc confocal microscope. Embryonic gonads were imaged as live tissue using either a Zeiss LSM 510 confocal or a Perkin Elmer spinning disc confocal microscope. Stereofluorescence images were taken on a Zeiss Lumar V12 stereofluorescence microscope. Live HeLa cells were also imaged on a Perkin Elmer spinning disc confocal microscope after plating on glass bottom dishes.
Supplementary Material
Video S1. Movie of HeLa cells. HeLa cells stably expressing CAG-HS-GR and -RG constructs were pooled together and cultured for 48 h. Confocal images were captured every 30 min.
Video S2. Movie of Oct4-HS-RG XX organ culture. The genital ridge and accompanying mesonephros were dissected from XX embryos at E11.5 and cultured for 13.5 h. Confocal images were captured every 30 min.
Video S3. Movie of XY Oct4-HS-RG organ culture. The genital ridge and accompanying mesonephros were dissected from XY embryos at E11.5 and cultured for 17.5 h. Confocal images were captured every 30 min.
ACKNOWLEDGEMENTS
We thank Hank Adams for advice with microscopy, the Genetically Engineered Mouse Facility at U.T. M.D. Anderson Cancer Center for the generation of Oct4-HS-RG founder mice. These studies were supported by NCI training grant CA009299 to M.D.S. and NIH grant HD30284 and the Ben F. Love Endowment to R.R.B. DNA sequencing and veterinary resources were supported by the National Institutes of Health (NIH) Cancer Center Support Grant CA16672.
REFERENCES
- Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Fouchier F, Baltz T, Rougon G. Identification of glycosylphosphatidylinositol-specific phospholipases C in mouse brain membranes. Biochem J. 1990;269:321–327. doi: 10.1042/bj2690321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Papaioannou VE. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM, Cook S, Laine J, Lebel D. Identification of membrane dipeptidase as a major glycosyl-phosphatidylinositol-anchored protein of the pancreatic zymogen granule membrane, and evidence for its release by phospholipase A. Biochem J. 1997;324(Pt 1):151–157. doi: 10.1042/bj3240151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YC, Chang HH, Tsai CY, Jong YJ, Horng LS, Lin SF, Tsai TF. Coat color-tagged green mouse with EGFP expressed from the RNA polymerase II promoter. Genesis. 2004;39:122–129. doi: 10.1002/gene.20038. [DOI] [PubMed] [Google Scholar]
- Kondoh G, Gao XH, Nakano Y, Koike H, Yamada S, Okabe M, Takeda J. Tissue-inherent fate of GPI revealed by GPI-anchored GFP transgenesis. FEBS Lett. 1999;458:299–303. doi: 10.1016/s0014-5793(99)01172-2. [DOI] [PubMed] [Google Scholar]
- Larina IV, Shen W, Kelly OG, Hadjantonakis AK, Baron MH, Dickinson ME. A membrane associated mCherry fluorescent reporter line for studying vascular remodeling and cardiac function during murine embryonic development. Anat Rec (Hoboken) 2009;292:333–341. doi: 10.1002/ar.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Harrell RA, Handler AM, Beam T, Hennessy K, Fraser MJ., Jr. piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol. 2005;14:17–30. doi: 10.1111/j.1365-2583.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- Littlewood GM, Hooper NM, Turner AJ. Ectoenzymes of the kidney microvillar membrane. Affinity purification, characterization and localization of the phospholipase C-solubilized form of renal dipeptidase. Biochem J. 1989;257:361–367. doi: 10.1042/bj2570361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo : a laboratory manual. 3rd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2003. p. x.p. 764. [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Nordhoff V, Hubner K, Bauer A, Orlova I, Malapetsa A, Scholer HR. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm Genome. 2001;12:309–317. doi: 10.1007/s003350010279. [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Eakin GS, Hadjantonakis AK. Dual transgene strategy for live visualization of chromatin and plasma membrane dynamics in murine embryonic stem cells and embryonic tissues. Genesis. 2009;47:330–336. doi: 10.1002/dvg.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Pesce M, Wang X, Wolgemuth DJ, Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Potts W, Tucker D, Wood H, Martin C. Chicken beta-globin 5′HS4 insulators function to reduce variability in transgenic founder mice. Biochem Biophys Res Commun. 2000;273:1015–1018. doi: 10.1006/bbrc.2000.3013. [DOI] [PubMed] [Google Scholar]
- Rhee JM, Pirity MK, Lackan CS, Long JZ, Kondoh G, Takeda J, Hadjantonakis AK. In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis. 2006;44:202–218. doi: 10.1002/dvg.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. Embo J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorpp M, Jager R, Schellander K, Schenkel J, Wagner EF, Weiher H, Angel P. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 1996;24:1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Swain A, Lovell-Badge R. Mammalian sex determination: a molecular drama. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Vignali DA. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin Biol Ther. 2005;5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- Trichas G, Begbie J, Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata-Hommer O, Griesbeck O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Movie of HeLa cells. HeLa cells stably expressing CAG-HS-GR and -RG constructs were pooled together and cultured for 48 h. Confocal images were captured every 30 min.
Video S2. Movie of Oct4-HS-RG XX organ culture. The genital ridge and accompanying mesonephros were dissected from XX embryos at E11.5 and cultured for 13.5 h. Confocal images were captured every 30 min.
Video S3. Movie of XY Oct4-HS-RG organ culture. The genital ridge and accompanying mesonephros were dissected from XY embryos at E11.5 and cultured for 17.5 h. Confocal images were captured every 30 min.