Abstract
How does the sequence of a single Small Heat Shock Protein (sHSP) assemble into oligomers of different sizes? To gain insight into the underlying structural mechanism, we determined the crystal structure of an engineered variant of Methanocaldococcus jannaschii Hsp16.5 wherein a 14 amino acid peptide from human heat shock protein 27 (Hsp27) was inserted at the junction of the N-terminal region and the α-crystallin domain. In response to this insertion, the oligomer shell expands from 24 to 48 subunits while maintaining octahedral symmetry. Oligomer rearrangement does not alter the fold of the conserved α-crystallin domain nor does it disturb the interface holding the dimeric building block together. Rather, the flexible C-terminal tail of Hsp16.5 changes its orientation relative to the α-crystallin domain which enables alternative packing of dimers. This change in orientation preserves a peptide-in-groove interaction of the C-terminal tail with an adjacent β-sandwich thereby holding the assembly together. The interior of the expanded oligomer, where substrates presumably bind, retains its predominantly non-polar character relative to the outside surface. New large windows in the outer shell provide increased access to these substrate-binding regions, thus accounting for the higher affinity of this variant to substrates. Oligomer polydispersity regulates sHSPs chaperone activity in vitro and has been implicated in their physiological roles. The structural mechanism of Hsp16.5 oligomer flexibility revealed here, which is likely to be highly conserved across the sHSP superfamily, explains the relationship between oligomer expansion observed in disease-linked mutants and changes in chaperone activity.
Keywords: Small heat shock proteins, Hsp16.5, Chaperone, crystal structure
Cells face the continual challenge of preventing or reversing protein aggregation (1). Driven by stress, mutations and inefficient folding, the accumulation of stable aggregates is associated with a range of protein deposition maladies (2, 3). Five superfamilies of heat shock proteins function as molecular chaperones binding non-native states of proteins thus providing a competing pathway for these toxic intermediates (4). Among heat shock proteins, the ubiquitously expressed small heat shock proteins (sHSPs) (5-7) sequester unfolded or misfolded proteins without direct ATP hydrolysis. This function is exemplified by their ability to buffer rapid and persistent increases in aggregation-prone intermediates, thereby preventing futile refolding cycles by other HSPs (8).
While mechanistic elements of sHSP chaperone activity appear to be conserved, the structural scaffold has undergone significant divergence (6). Expressed as subunits of 12-42 kDa (9), sHSPs assemble into large oligomers (12-40 subunits) with a remarkable spectrum of symmetries and size across the superfamily, ranging from ordered (10, 11) to heterogenous and dynamic ensembles of interconverting oligomers (9, 12-15). This architectural diversity stands in stark contrast to the conservation of tertiary and quaternary structures in other heat shock protein families such as Hsp60, 70, 90 and 100. The property of forming multiple oligomers with dynamic assembly and disassembly, prevalent in metazoan sHSPs, has been linked to the regulation of substrate recognition and binding (6, 16-18). Disease-linked mutations in human sHSPs lead to changes in their polydisperisty often inducing the formation of larger oligomers (19-21).
An outstanding question in the chaperone field is how are sHSPs able to form a diverse group of oligomers using a single sequence. Sequence identity is highest in a 90-100 amino acid module referred to as the α-crystallin domain and considered the defining characteristic for membership in the superfamily (9, 22). This domain has a conserved core fold consisting of a 7-stranded β-sandwich (10, 11, 14, 23-25). Furthermore, dimers of the α-crystallin domain form the building blocks of sHSP oligomers. C-terminal to the α-crystallin domain is a stretch of 10-20 amino acids, the C-terminal tail or extension, containing a conserved 4-residue stretch critical for oligomer assembly (26). The most divergent region in size and sequence is the N-terminal extension which ranges from short amino acid sequences to entire domains in human sHSPs (9, 27). Sequence similarity in this domain is only observed between closely related species.
In contrast to the wealth of biochemical data on sHSP chaperone activity, there is a dearth of high resolution structures of their assemblies. Indeed, the extreme polydispersity of metazoan sHSP has frustrated crystallographic analysis, and structures of the native oligomer are limited to the ordered archeal Methanocaldococcus jannaschii Hsp16.5 (10) and eukaryotic Triticum aestivum (wheat) Hsp16.9 (11). Although both Hsp16.5 and Hsp16.9 are built from similar dimers of the α-crystallin domain, the former assembles into a spherical oligomer of 24 subunits with octahedral symmetry while the Hsp16.9 oligomer consists of two stacked hexameric rings. The two structures provide an example of the interplay between the three sequence modules in determining higher order assemblies. A conserved motif in the C-terminal tail straps α-crystallin dimers through interaction with the edge of the α-crystallin domain β-sandwich. The different oligomer architectures of Hsp16.5 and Hsp16.9 arise from different orientations of the C-terminal tail relative to the α-crystallin domain.
Extension of these architectural principles to polydisperse sHSP oligomers is complicated by sequence divergence particularly the expansion of the N-terminal region. In polydisperse sHSP, the N-terminal domain plays an important role in stabilizing the assembly and conferring its dynamic properties. Truncation of this domain in α-crystallin and Hsp27 decreases the average oligomer size to dimers and tetramers and inhibits the phenomenon of subunit exchange (24, 28-31). Furthermore, phosphorylation of αB-crystallin and Hsp27 at sites in the N-terminal domain shifts the oligomer equilibrium towards monomers and dimers respectively (17, 32). This shift is tightly coupled to substrate binding in vitro (17, 33), and is physiologically important (34, 35). For instance Hsp27 phosphorylation increases substrate affinity through the exposure of the otherwise buried N-terminal domain upon dissociation to a dimer. Hsp27 dissociation regulates its ability to rescue neuronal plasticity in tau transgenic mice (35).
In contrast to α-crystallins and Hsp27, deletion of the entire N-terminal region of Hsp16.5 does not affect the overall size and symmetry of the oligomeric assembly (36). Previous EPR and cryoEM analyses have shown that the construct consisting of the α-crystallin domain and C-terminal tail encodes all the interactions necessary to assemble the Hsp16.5 oligomer (37). However, this region is functionally important and sequence modifications in the N-terminal region alter the size and order of the assembly. Shi et al. constructed Hsp16.5 variants that recapitulate aspects of mammalian sHSP oligomers such as size expansion and polydispersity (38). A 14 amino acid peptide, referred to as P1, unique to Hsp27 was inserted into the N-terminal region of Hsp16.5. Deletion of P1 inhibits Hsp27 equilibrium dissociation to dimers by stabilizing its ensemble of large oligomers. When P1 was inserted at the junction of the N-terminal region and C-terminal domain of Hsp16.5 (this construct is referred to hereafter as Hsp16.5-P1), the oligomer size increased from 24 to 48 subunits. In contrast, P1 insertion in the middle of a helix in the N-terminal domain yielded an ensemble of polydisperse oligomers. Both constructs displayed higher affinity toward substrate suggesting oligomer expansion is required for substrate binding.
To understand the molecular determinants that enable Hsp16.5 to form oligomeric shells of both 24 and 48 subunits, and to inform molecular mechanisms for polydispersity in other sHSPs, we determined the crystal structure of Hsp16.5-P1 at 2.7Å resolution (Table 1, Figure S1). The structure of the 48 subunit octahedral shell reveals that oligomer expansion is mediated by reorientation of the C-terminal tail rather than changes in the α-crystallin dimer architecture in agreement with predictions based on low resolution cryoEM models (38). Comparison with the WT Hsp16.5 structure reveals the molecular strategies used by sHSPs to form multiple oligomers. Changes in the N-terminal region are propagated to the C-terminal tail enabling alternative assemblies. Analysis of the structure rationalizes the increase in substrate affinity that has been reported to accompany oligomer expansion induced by disease-associated mutations (19).
Table 1. Data collection and refinement statistics.
| Data Collection | |
|---|---|
| Space Group | I432 |
| Cell Dimensions (Å) | |
| a, b, c (Å) | 184.58 |
| α, β, γ (°) | 90.00 |
| Resolution (Å) | 2.7 |
| Rsym (%) | 4.3(60) |
| I/σI | 16.5(2.1) |
| Completeness (%)* | 98.9(94.4), 98.9(100) |
| Redundancy | 4.7(4.7) |
| Refinement | |
| Resolution range (Å) | 50-2.7 |
| No. of reflections | 14,902 |
| Rfactor (%) | 21.4 |
| Rfree (%) | 23.7 |
| Total Protein Atoms | 1,791 |
| Total Water Molecules | 10 |
| RMSD | |
| Bond lengths | 0.009 |
| Bond angles | 1.089 |
| Ramachandran (%) | |
| Favored | 98.7 |
| Allowed | 1.3 |
| Outliers | 0 |
| B-factors | |
| All | 92 |
| Main Chain | 87 |
| Side Chain | 96 |
| Solvent | 70 |
Numbers in parenthesis describe data in the highest resolution bin (2.79-2.70 Å)
For completeness two sets of numbers are given. The first are completeness of data used in refinement (including the test set). The second are completeness from data reduction.
Materials and Methods
Expression, purification, crystallization and data collection
The construction of Hsp16.5-P1 was previously described (38). Briefly, a sequence encoding human Hsp27 amino acids 57-70 (PLPPAAIESPAVAA) was inserted between residues 33 and 34 of Hsp16.5. Hsp16.5-P1 was expressed in E. coli BL21 (DE3) cells containing a pET20b+ derived plasmid (38). Cell disruption was followed by addition of polyethyleneimine to precipitate the DNA. Following high speed centrifugation, the supernatant was loaded on an anion exchange chromatography column. Fractions containing Hsp16.5-P1 were pooled and further purified by gel filtration in 20 mM HEPES pH 7.5 and 100 mM NaCl on a Superose 6 column.
Freshly purified protein was concentrated to 40 mg/mL before crystallization. Hanging drop vapor diffusion was used in crystallization trials. Crystals grew from 1.8 M (NH4)2SO4, 20 mM Tris pH 8 and 10% glycerol at 25 °C. Diffraction data were collected at SER-CAT ID-22 at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL.
Structure determination and analysis
Data were indexed, integrated, and scaled with HKL2000 (39). The space group of engineered Hsp16.5-P1 crystal was I432. There are two non-equivalent monomers forming a dimer in an asymmetric unit. Molecular replacement solutions were obtained using program PHASER (40) and were evaluated with omit maps calculated in CCP4 (41) and displayed in COOT (42). The structure was refined in CNS (43) and Phenix (44) and manually rebuilt into composite-omit maps (calculated initially in CNS and in later stages using Phenix) using COOT (42). The overall Rwork was 21.4 % and Rfree was 23.7 %. Rms of bond lengths is 0.009 Å. Rms of bond angles is 1.1°. The final model contains 10 water molecules. There are zero Ramachandran outliers with 98.7% of residues in favored and 1.3% in allowed regions as determined by Phenix/Molprobity (44, 45). Data collection and refinement statistics are summarized in Table 1. Figures were produced in Pymol (46). Solvent accessible surface areas were calculated in Pymol using a solvent sphere with a radius of 1.4 Å (47, 48).
Results
Hsp16.5-P1 oligomer assembly
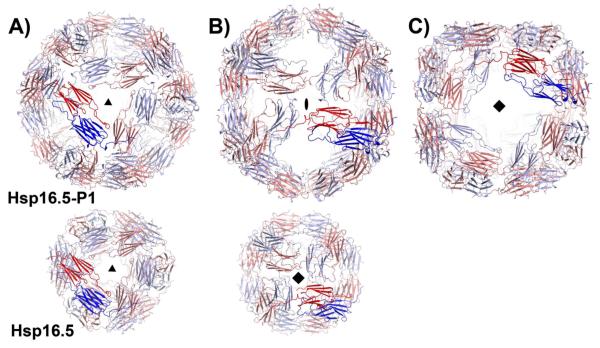
HSP16.5-P1 crystallized in the cubic space group I432 with 2 subunits in the crystallographic asymmetric unit. The octahedral shell is formed by crystallographic symmetry operators that allow formation of a spherical oligomer of 48 subunits in agreement with the oligomer size determined by light scattering and low resolution electron microscopy reconstructions (38). Figure 1A, 1B and 1C shows the views along the three windows of the outer shell capturing three symmetry axes that relate dimers of Hsp16.5-P1 (shown in dark red and blue). Thus, 24 Hsp16.5-P1 dimers are related by octahedral symmetry while the same symmetry operations relate 24 monomers in the WT (10). Hsp16.5-P1 is expanded with an outer diameter of 163Å compared to 115Å for the WT. The expansion is manifested by a new large square opening in the outer shell (Figure 1C). The sides of this window consist of dimers of the α-crystallin domain related by four-fold symmetry. The triangular three-fold window is similar to that of the WT (Figure 1A). A two-fold window between dimers replaces the four-fold window relating monomers in the WT structure (Figure 1B).
Figure 1.
Views of Hsp16.5-P1(upper row) and Hsp16.5-WT assemblies (PDBID 1SHS, lower row) along their respective symmetry axes. The two shells are scaled to highlight the expansion of the P1 variant. Hsp16.5-P1 has A) three-, B) two- and C) four-fold symmetry axes that relate dimers of the α-crystallin domain (blue and red). (B) By comparison, WT dimers of the α-crystallin domain are related by A) three- and B) four-fold symmetry.
In contrast to the α-crystallin domain and the C-terminal extension, the 32 amino acid N-terminal segment along with the 14 amino acid peptide insertion (P1) are crystallographically disordered, and therefore not visible in the structure. Previous site-directed spin labeling analysis assigned internal density observed in cryoEM images of the WT to the N-terminal region (37). Although weak internal density, arising from the N-terminal segment and P1, was observed in the cryoEM model of Hsp16.5-P1, its poor reconstruction was interpreted as evidence of P1 flexibility (38). The absence of density for this region, even in electron density maps calculated using only low-resolution terms (50-15Å), indicates that these residues do not obey octahedral symmetry. The mechanism by which the inserted peptide P1 induces a gross oligomer reorganization is thus not directly observable. However, the structural consequences of this reorganization have herein been observed at 2.7 Å resolution, resulting in an atomic model describing a structural strategy used by sHSPs to expand the oligomer shell.
The α-crystallin dimer
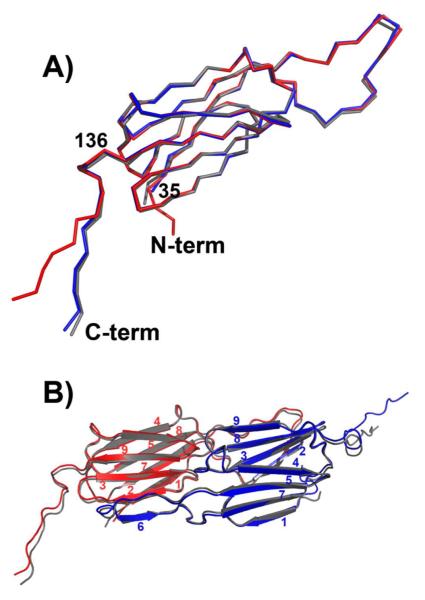
Despite the extensive reorganization of the oligomer as a result of P1 insertion, the overall fold of the α-crystallin domain is unchanged. Nine β-strands are arranged in two sheets forming a β-sandwich (Figure 2A). A structure-based alignment with Hsp27 (Figure S2) highlights the conservation of the seven-strand core despite marginal sequence similarity between the archeal and human sHSPs. Superposition of the β-sandwich from Hsp16.5-WT with that of the P1 variant yielded a root mean square deviation of 0.3 Å for main-chain atoms of residues 45 to 148 demonstrating the similarity of the two structures. A long loop extends from the sandwich enabling a short β-strand (strand 6) to hydrogen bond to an edge strand (strand 2) of another α-crystallin domain to form a composite β-sheet (Figure 2B). In addition to strand swapping, the α-crystallin domain dimer in Hsp16.5-P1 is stabilized by extensive hydrophobic contacts, hydrogen bonds and ionic interactions along loop regions that connect the two sheets of the two β-sandwiches. The dimer interface which buries ~3400 Å2 of solvent accessible surface area is identical to that found in the WT assembly (Figure 2B). The β-sandwich and the loops at the dimer interface have the lowest B factors in the structure (Figure S3).
Figure 2.
A) Superposition of subunits from Hsp16.5-P1 (red and blue) and Hsp16.5-WT (gray) demonstrates that the α-crystallin domain fold is conserved and highlights the change in orientation of the C-terminal tail in one of the Hsp16.5-P1 subunits. B) Ribbon diagram of the dimer from Hsp16.5-P1 (red and blue) superimposed on the dimer from Hsp16.5-WT (gray) shows that that oligomer expansion does not perturb the interface of the dimer involving stand swapping (strand 6) and extensive networks of contacts between loops.
Thus, the expansion of the oligomer does not involve repacking at the interface of the dimer or changes in the relative orientations of dimer subunits. Instead, the two subunits in the Hsp16.5-P1 dimer are rendered structurally inequivalent by the distinct orientation of the C-terminal tail relative to the β-sandwich. Figure 2B shows that while the β-sheet core of the α-crystallin domain is superimposable, the C-terminal tail of one of the two subunits in the dimeric building block of Hsp16.5-P1 undergoes a change of orientation relative to the β-sandwich. Whereas subunit A is overly similar to the WT structure, there is a clear displacement of the tail of subunit B. The change in orientation of this region is enabled by a reorientation at Ile140 such that there is an ~30° difference in the orientation of residues 141-147 between the two subunits (Figure 2A).
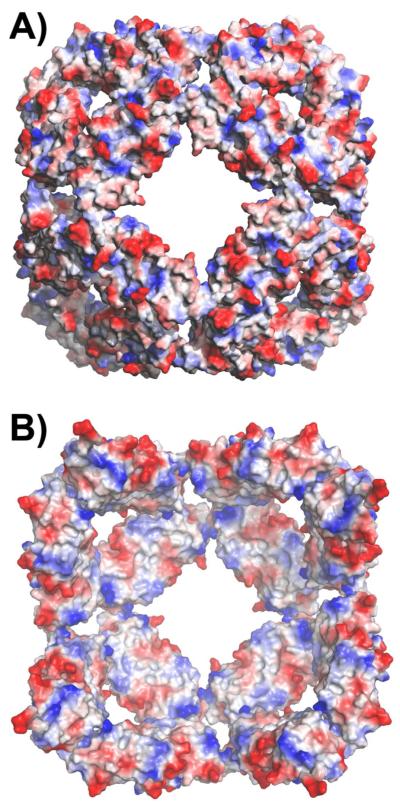
Similar dimer structures and octahedral symmetry of the assembly preserve the electrostatic contrast between the outside and inside of the sphere observed in the WT structure (Figure 3A and B) (10). The accessible surface on the inside is largely non-polar, whereas the three- and four-fold windows on the surface are decorated by charged residues. This observation is consistent with recent cryoEM analysis demonstrating that T4 Lysozyme binds to the inside of the Hsp16.5-P1 particle (Shi, J., Koteiche, H.A., Stewart, P.L., Mchaourab, H.S., unpublished results) and supports a binding mode in which unfolded proteins can interact with the largely hydrophobic interior surface.
Figure 3.
A) Exterior and B) interior of the Hsp16.5-P1 sphere viewed along the 4-fold symmetry axis and colored by residue polarity. The predominant non-polar character of the internal surface provides plausible contact sites for bound substrates.
Two C-terminal tail orientations enable distinct dimer-dimer packing
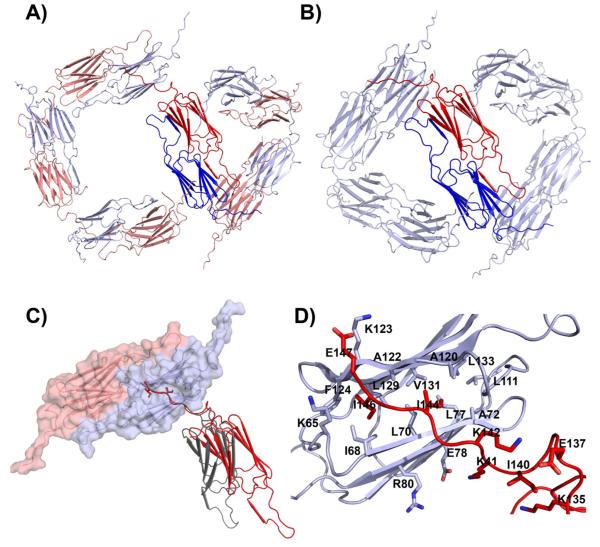
To assemble the expanded Hsp16.5-P1 shell, the two subunits in a dimer, inequivalent by virtue of the C-terminal tail conformations, interact along 2-, 3- and 4-fold symmetry interfaces. At the corner of the three-fold window, the dimer-dimer interface is very similar to that of the WT (Figure 4 A and B) (10). This interface has the least extensive contacts in the assembly burying ~400 Å2 of solvent accessible surface area whereas ~530 Å2 are buried in the WT structure. In both instances, this interaction is stabilized by the C-terminal tail, which extends out and interacts with the hydrophobic groove at the edge of the neighboring β-sandwich. Ile 144 and Ile 146 from the C-terminal tail of one subunit make contacts with hydrophobic pockets on the edge of the β-sandwich of an adjacent subunit. Ile 144 is buried in a pocket formed by L70, L77, A72, L133, A120, V131, A122, and Ile 146 is buried in a pocket formed by F124, L129, A122, I68, L70 (Figure 4C,D). This peptide-in-groove interaction is quite extensive and buries ~1,100 Å2 of solvent accessible surface area, allowing for flexibility at this interface. The conformation of the C-terminal tail along the three-fold window is identical to that observed in the WT subunit.
Figure 4.
A) Dimer packing around three- and four-fold windows of Hsp16.5-P1. The different angles between α-crystallin domain in the two windows are enabled by two distinct orientations of the C-terminal tail in the blue and red subunits. B) By comparison, the angle between dimers is uniform in the WT assembly where a single orientation of the C-terminal tail is observed. C) Peptide-in-groove interaction stabilizing contacts between subunits in the Hsp16.5-P1 (red and Hsp16.5-WT (gray). Note the change in relative orientation between the gray and red subunits. The gray orientation is seen in Hsp16.5-WT and in three-fold window of Hsp16.5-P1. The red orientation is seen only in the four-fold window of Hsp16.5-P1. D) Close-up view of C) showing the side chains stabilizing the peptide-in-groove interaction.
The large four-fold window, which distinguishes the structure of Hsp16.5-P1, is assembled using the same dimer-dimer interactions observed at the corners of the three-fold window. However, the angle between α-crystallin domain dimers is closer to 90° whereas it is closer to 60° at the three-fold window (Figure 4A). This interface highlights the significance of the assembly strategy utilized by sHSPs. While similar to the interfaces at the corners of the three-fold window, the wider angle between α-crystallin domain dimers results in a complete loss of contacts between these domains. No buried surface area on the α-crystallin domain is associated with this interaction, and the hydrophobic peptide-in-grove interaction described in the previous paragraph (Figure 4C,D) suffices to stabilize this interface. The combination of the strong interaction between the C-terminal tail and the hydrophobic groove at the edge of the β-sandwich and the relatively weak interaction between adjacent α-crystallin domains gives these interfaces both stability and flexibility
Discussion
This paper reports the first structural insight into the basis of oligomer flexibility in a single sHSP. Oligomer polydispersity has been reported for evolutionary distant sHSPs and is induced by an increase in temperature, disease-causing mutations or substrate binding. Our findings reveal how sequence modifications in the N-terminal domain can remodel the overall assembly without altering the α-crystallin domain fold or the dimeric building block. Repacking of the N-terminal domain, here as a consequence of sequence insertion, is propagated to the C-terminal tail which can adopt two conformations compatible with a different assembly size built by the same symmetry elements. In addition to the 48-subunit assemblies reported here, Hsp16.5 can be induced to form heterogeneous ensembles of structures presumably mediated by the same elements of flexibility (38). The linkage between shell size and C-terminal tail conformation uncovered by the structure of Hsp16.5-P1, illustrates how larger N-terminal domain sequences, characteristic of eukaryotic sHSP, can modulate the size and stability of the oligomeric assembly. Consistent with this notion, two different conformations of the C-terminal tail appear to be associated with different conformations of the N-terminal domain in Hsp16.9 (26). Furthermore, recent NMR and mass spectrometry based modeling of αB-crystallin invoked fluctuations of the C-terminal tail as a determinant of its oligomer polydispersity (49).
The expansion of Hsp16.5-P1 provides insight into sHSP activation which is a critical element of their chaperone mechanism. There is a mounting body of evidence implicating sequences in the N-terminal regions of sHSPs in high affinity binding to client proteins (7). Two models have been advanced to describe the basis of substrate recognition and binding. For the polydisperse and dynamic oligomers, substrate binding is mediated by a dimeric unit that is in equilibrium with the ensemble of large oligomers (17). Dissociation to dimers exposes the N-terminal domain in an unstructured conformation (50). In contrast, a dissociation-independent model (51), proposed for ordered sHSPs, invokes conformational changes in the oligomer as the mechanism for access to the otherwise buried N-terminal domain. The Hsp16.5-P1 structure is the first direct view of how access to binding sites in the N-terminal domain can be accomplished without subunit dissociation. Indeed, Hsp16.5-P1 has an order of magnitude higher apparent affinity for substrates than does the WT (38). Because assembly rearrangement does not alter the hydrophobic nature of the internal surface of the oligomer, the higher apparent substrate affinity of Hsp16.5-P1 likely arises from increased access enabled by the large windows in its shell.
Considering the binding capacity of sHSPs which can reach one subunit of equal molecular mass, loss of regulation of their interaction with client proteins can have deleterious consequences on proteostasis as evident from the association of sHSP point mutations with multiple inherited diseases in mouse models (52-54) and in humans (55-58). Remarkably, many of these mutants have higher affinity to model substrates (19, 59). Consistent with the two activation mechanisms described above, these mutations either induce expansion of the oligomer or increase dissociation. For example, αA-crystallin mutant R116C (20, 60), which causes a dominant human cataract phenotype, and αB-crystallin R120G (21), which is a dominant human mutation linked to cardiomyopathy, show increased average molecular mass. Determination of modes of sHSP oligomer flexibility is thus critical to understanding the molecular basis of these pathologies.
Supplementary Material
Acknowledgements
The authors thank Dr. Hanane Koteiche for critical reading of the manuscript.
This work was supported by the National Eye Institute, National Institute of Health grant R01-EY12018 to Hassane S. Mchaourab. The structure and data files have been deposited at the protein data base with the accession code 4ELD.
Abreviations
- sHSP
small heat shock protein
- Hsp16.5
Methanocaldococcus jannaschii heat shock protein 16.5
- Hsp16.9
Triticum aestivum heat shock protein 16.9
- Hsp27
heat shock protein 27
- WT
wild-type
References
- 1.Dobson CM. Principles of protein folding, misfolding and aggregation. Seminars in Cell & Developmental Biology. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Gidalevitz T, Kikis EA, Morimoto RI. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Current Opinion in Structural Biology. 2010;20:23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 4.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 5.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 6.McHaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry. 2009;48:3828–3837. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basha E, O’Neill H, Vierling E. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO Journal. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong WW, Caspers GJ, Leunissen JA. Genealogy of the α-crystallin--small heat-shock protein superfamily. International Journal of Biological Macromolecules. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 10.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 11.van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nature Structural Biology. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 12.Haley DA, Bova MP, Huang QL, Mchaourab HS, Stewart PL. Small heat-shock protein structures reveal a continuum from symmetric to variable assemblies. Journal of Molecular Biology. 2000;298:261–272. doi: 10.1006/jmbi.2000.3657. [DOI] [PubMed] [Google Scholar]
- 13.Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J. Hsp26: a temperature-regulated chaperone. EMBO Journal. 1999;18:6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jehle S, van Rossum B, Stout JR, Noguchi SM, Falber K, Rehbein K, Oschkinat H, Klevit RE, Rajagopal P. alphaB-crystallin: a hybrid solid-state/solution-state NMR investigation reveals structural aspects of the heterogeneous oligomer. Journal of Molecular Biology. 2009;385:1481–1497. doi: 10.1016/j.jmb.2008.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haley DA, Horwitz J, Stewart PL. The small heat-shock protein, αB-crystallin, has a variable quaternary structure. Journal of Molecular Biology. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- 16.Koteiche HA, McHaourab HS. Mechanism of chaperone function in small heat-shock proteins. Phosphorylation-induced activation of two-mode binding in alphaB-crystallin. Journal of Biological Chemistry. 2003;278:10361–10367. doi: 10.1074/jbc.M211851200. [DOI] [PubMed] [Google Scholar]
- 17.Shashidharamurthy R, Koteiche HA, Dong J, McHaourab HS. Mechanism of chaperone function in small heat shock proteins: dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. Journal of Biological Chemistry. 2005;280:5281–5289. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- 18.Giese KC, Vierling E. Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. Journal of Biological Chemistry. 2002;277:46310–46318. doi: 10.1074/jbc.M208926200. [DOI] [PubMed] [Google Scholar]
- 19.Koteiche HA, McHaourab HS. Mechanism of a Hereditary Cataract Phenotype: Mutations in αA-crystallin activate substrate binding. Journal of Biological Chemistry. 2006;281:14273–14279. doi: 10.1074/jbc.M512938200. [DOI] [PubMed] [Google Scholar]
- 20.Cobb BA, Petrash JM. Structural and functional changes in the αA-crystallin R116C mutant in hereditary cataracts. Biochemistry. 2000;39:15791–15798. doi: 10.1021/bi001453j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bova MP, Yaron O, Huang Q, Ding L, Haley DA, Stewart PL, Horwitz J. Mutation R120G in αB-crystallin, which is linked to a desmin-related myopathy, results in an irregular structure and defective chaperone-like function. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6137–6142. doi: 10.1073/pnas.96.11.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulain P, Gelly JC, Flatters D. Detection and architecture of small heat shock protein monomers. PLoS ONE [Electronic Resource] 2010;5:e9990. doi: 10.1371/journal.pone.0009990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laganowsky A, Benesch JLP, Landau M, Ding L, Sawaya MR, Cascio D, Huang Q, Robinson CV, Horwitz J, Eisenberg D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Science. 2010;19:1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagneris C, Bateman OA, Naylor CE, Cronin N, Boelens WC, Keep NH, Slingsby C. Crystal Structures of α-Crystallin Domain Dimers of αB-Crystallin and Hsp20. Journal of Molecular Biology. 2009;392:1242–1252. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 25.Koteiche HA, Mchaourab HS. Folding pattern of the α-crystallin domain in αA-crystallin determined by site-directed spin labeling. Journal of Molecular Biology. 1999;294:561–577. doi: 10.1006/jmbi.1999.3242. [DOI] [PubMed] [Google Scholar]
- 26.Van Montfort R, Slingsby C, Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Advances in Protein Chemistry. 2001;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- 27.Jehle S, Vollmar BS, Bardiaux B, Dove KK, Rajagopal P, Gonen T, Oschkinat H, Klevit RE. N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6409–6414. doi: 10.1073/pnas.1014656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV. Three-Dimensional Structure of α-Crystallin Domain Dimers of Human Small Heat Shock Proteins HSPB1 and HSPB6. J. Mol. Biol. 2011;411:110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 29.Bova MP, Horwitz J, Fung BK. Subunit exchange of αA-crystallin. Journal of Biological Chemistry. 1997;272:29511–29517. doi: 10.1074/jbc.272.47.29511. [DOI] [PubMed] [Google Scholar]
- 30.Bova MP, Mchaourab HS, Han Y, Fung BK. Subunit exchange of small heat shock proteins. Analysis of oligomer formation of αA-crystallin and Hsp27 by fluorescence resonance energy transfer and site-directed truncations. Journal of Biological Chemistry. 2000;275:1035–1042. doi: 10.1074/jbc.275.2.1035. [DOI] [PubMed] [Google Scholar]
- 31.Berengian AR, Parfenova M, Mchaourab HS. Site-directed spin labeling study of subunit interactions in the α-crystallin domain of small heat-shock proteins. Comparison of the oligomer symmetry in αA-crystallin, HSP 27, and HSP 16.3. Journal of Biological Chemistry. 1999;274:6305–6314. doi: 10.1074/jbc.274.10.6305. [DOI] [PubMed] [Google Scholar]
- 32.Aquilina JA, Benesch JL, Ding LL, Yaron O, Horwitz J, Robinson CV. Phosphorylation of alphaB-crystallin alters chaperone function through loss of dimeric substructure. Journal of Biological Chemistry. 2004;279:28675–28680. doi: 10.1074/jbc.M403348200. [DOI] [PubMed] [Google Scholar]
- 33.Sathish HA, Stein RA, Yang G, Mchaourab HS. Mechanism of chaperone function in small heat-shock proteins. Fluorescence studies of the conformations of T4 lysozyme bound to alphaB-crystallin. J. Biol. Chem. 2003;278:44214–44221. doi: 10.1074/jbc.M307578200. [DOI] [PubMed] [Google Scholar]
- 34.Rogalla T, Ehrnsperger M, Preville X, Kotlyarov A, Lutsch G, Ducasse C, Paul C, Wieske M, Arrigo AP, Buchner J, Gaestel M. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. Journal of Biological Chemistry. 1999;274:18947–18956. doi: 10.1074/jbc.274.27.18947. [DOI] [PubMed] [Google Scholar]
- 35.Abisambra JF, Blair LJ, Hill SE, Jones JR, Kraft C, Rogers J, Koren J, Jinwal UK, Lawson L, Johnson AG, Wilcock D, O’Leary JC, Jansen-West K, Muschol M, Golde TE, Weeber EJ, Banko J, Dickey CA. Phosphorylation Dynamics Regulate Hsp27-Mediated Rescue of Neuronal Plasticity Deficits in Tau Transgenic Mice. The Journal of Neuroscience. 30:15374–15382. doi: 10.1523/JNEUROSCI.3155-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koteiche HA, McHaourab HS. The determinants of the oligomeric structure in Hsp16.5 are encoded in the alpha-crystallin domain. FEBS Letters. 2002;519:16–22. doi: 10.1016/s0014-5793(02)02688-1. [DOI] [PubMed] [Google Scholar]
- 37.Koteiche HA, Chiu S, Majdoch RL, Stewart PL, McHaourab HS. Atomic models by cryo-EM and site-directed spin labeling: application to the N-terminal region of Hsp16.5. Structure. 2005;13:1165–1171. doi: 10.1016/j.str.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Shi J, Koteiche HA, Mchaourab HS, Stewart PL. Cryoelectron Microscopy and EPR Analysis of Engineered Symmetric and Polydisperse Hsp16.5 Assemblies Reveals Determinants of Polydispersity and Substrate Binding. J. Biol. Chem. 2006;281:40420–40428. doi: 10.1074/jbc.M608322200. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, W M. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D-Biological Crystallography. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 41.Bailey S. The CCP4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;61:458–464. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Research. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 47.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 48.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. Journal of Molecular Biology. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 49.Baldwin AJ, Lioe H, Hilton GR, Baker LA, Rubinstein JL, Kay LE, Benesch JL. The polydispersity of alphaB-crystallin is rationalized by an interconverting polyhedral architecture. Structure. 2011;19:1855–1863. doi: 10.1016/j.str.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 50.McDonald ET, Bortolus M, Koteiche HA, Mchaourab HS. Sequence, structure, and dynamic determinants of Hsp27 (HspB1) equilibrium dissociation are encoded by the N-terminal domain. Biochemistry. 2012;51:1257–1268. doi: 10.1021/bi2017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franzmann TM, Wuhr M, Richter K, Walter S, Buchner J, Franzmann TM, Wuhr M, Richter K, Walter S, Buchner J. The activation mechanism of Hsp26 does not require dissociation of the oligomer. Journal of Molecular Biology. 2005;350:1083–1093. doi: 10.1016/j.jmb.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 52.Hsu C-D, Kymes S, Petrash JM. A transgenic mouse model for human autosomal dominant cataract. Investigative Ophthalmology & Visual Science. 2006;47:2036–2044. doi: 10.1167/iovs.05-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xi JH, Bai F, Gross J, Townsend RR, Menko AS, Andley UP, Xi J.-h., Bai F, Gross J, Townsend RR, Menko AS, Andley UP. Mechanism of small heat shock protein function in vivo: a knock-in mouse model demonstrates that the R49C mutation in alpha A-crystallin enhances protein insolubility and cell death. Journal of Biological Chemistry. 2008;283:5801–5814. doi: 10.1074/jbc.M708704200. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, Hewett T, Robbins J. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circulation Research. 2001;89:84–91. doi: 10.1161/hh1301.092688. comment. [DOI] [PubMed] [Google Scholar]
- 55.Graw J, Loster J, Soewarto D, Fuchs H, Meyer B, Reis A, Wolf E, Balling R, Hrabe de Angelis M. Characterization of a new, dominant V124E mutation in the mouse alphaA-crystallin-encoding gene. Investigative Ophthalmology & Visual Science. 2001;42:2909–2915. [PubMed] [Google Scholar]
- 56.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human α crystallin gene CRYAA. Human Molecular Genetics. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 57.Mackay DS, Boskovska OB, Knopf HLS, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. American Journal of Human Genetics. 2002;71:1216–1221. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, Chateau D, Chapon F, Tome F, Dupret JM, Paulin D, Fardeau M. A missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nature Genetics. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 59.Kumar MS, Koteiche HA, Claxton DP, McHaourab HS. Disulfide cross-links in the interaction of a cataract-linked alphaA-crystallin mutant with betaB1-crystallin. FEBS Letters. 2009;583:175–179. doi: 10.1016/j.febslet.2008.11.047. [DOI] [PubMed] [Google Scholar]
- 60.Berengian AR, Bova MP, Mchaourab HS. Structure and function of the conserved domain in αA-crystallin. Site-directed spin labeling identifies a β-strand located near a subunit interface. Biochemistry. 1997;36:9951–9957. doi: 10.1021/bi9712347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.