Abstract
During collision induced dissociation phosphoserine- and phosphothreonine-containing peptides frequently undergo neutral loss of phosphoric acid. Subsequent amide bond cleavage N-terminal to the site of phosphorylation results in a y ion with a mass 18 Da lower than the corresponding unmodified y fragment. We report here that when the phosphoserine or phosphothreonine is directly preceded by a proline, an unusual fragment with a mass 10 Da higher than the corresponding unmodified y ion is frequently observed. Accurate mass measurements are consistent with elimination of the phosphoric acid followed by fragmentation between the α carbon and the carbonyl group of the proline residue. We propose a cyclic oxazoline structure for this fragment. Our observation may be explained by the charge-directed SN2 neighboring group participation reaction proposed for the phosphoric acid elimination by Palumbo et al. [Palumbo, A.M., Tepe, J.J., Reid, G.E.: Mechanistic Insights into the Multistage Gas-Phase Fragmentation Behavior of Phosphoserine- and Phosphothreonine-Containing Peptides. J. Prot. Res. 7(2), 771-779 (2008).]. Considering such specific fragment ions for confirmation purposes after regular database searches may boost the confidence of peptide identifications as well as phosphorylation site assignments.
Keywords: CID, HCD, fragmentation, phosphorylation, database search
Introduction
High throughput proteomics experiments have been used for qualitative and quantitative characterization of protein complexes, cell organelles as well as cell lysates. With the advent of ever faster and more sensitive new instrumentation, over 10,000 MS/MS spectra can be acquired in a 90 minutes LC-MS/MS run. Thus, the identification of these spectra has to be performed in an automated fashion and the reliability of the resulting peptide assignments needs to be addressed. Collision-induced dissociation (CID) of peptides may yield a wide variety of fragment ions [nomenclature: 1; review: 2]. Despite the abundance of fragment ion types, search engines usually consider only a few “favorites” when matching spectra against a database. The products of the peptide bond cleavage, i.e. the N-terminal b and C-terminal y fragments are considered rightfully the most important for CID spectra. In addition, some neutral losses from these fragments are permitted, such as CO losses from b ions (a fragment formation), as well as ammonia- and water losses from both b and y fragments. Such losses are usually linked to the amino acid composition of the fragment ion: the presence of Asn, Gln, Lys and Arg promotes NH3 loss, while Asp, Glu, Ser and Thr-containing fragments may lose H2O.
Certain search engines, such as Mascot, Spectrum Mill and Protein Prospector will consider instrument-dependent fragmentation. For example, they may consider internal ion formation in CID-experiments performed in quadrupole collision cells (such as triple quadrupole and QTOF instruments, as well as HCD in Orbitraps). However, permitting too many fragment-types during the database searches may slightly increase the search time and increase the rate of false positive identifications [3]. At the same time, judiciously considering “minor” fragment-types, as well as unusual, sequence-specific fragment ions may increase the confidence in peptide assignments and eliminate false positives, if applied as a confirmation measure of the initial database search results.
While they share many common features, ion trap CID spectra are distinct from quadrupole collision cell CID data. However, the fact that they are both referred to as “CID” spectra leads to some confusion. Resonance excitation in ion traps generally leads to excitation and fragmentation of a specific m/z range. Therefore, fragment ions (which typically have m/z values distinct from the precursor) do not undergo secondary fragmentation. In terms of gas pressure and peptide collision energy, the design of quadrupole collision cells promotes multiple fragmentation events. Thus, one would expect more ions derived from secondary fragmentation using a quadrupole collision cell relative to an ion trap. Internal ion formation is a well known, though frequently ignored phenomenon. Other unusual fragments also have been reported. For example, it has been described that sequences featuring a Gln residue in their 2nd position usually produce an abundant c1 fragment [4]. Rearrangement reaction of b-type fragments also has been documented [5].
Software such as Peptide Prophet or Scaffold statistically reevaluate peptide assignments after the database search is finished, however neither of them utilize any new fragments in the confirmation process [6]. Protein Prospector may be manually prompted to assign fragment ion types that were not considered during the database search, for example, ions formed via multiple neutral losses. However, we are not aware of any software that will consider specific minor fragments during the confirmation/evaluation process.
In this paper we report an unusual fragment formation observed prominently from phosphoserine or phosphothreonine-containing peptides that contain a proline immediately preceding the site of modification. This fragmentation is observed, albeit less intensely, when the ProSer or ProThr sequence is not phosphorylated. Such fragmentation was detected in CID acquired in QTOF instruments as well as in HCD on an LTQ-Orbitrap, but was not observed in ion trap CID.
Experimental
Samples were tryptic digests from different projects, with or without iTRAQ-derivatization, unmodified and phosphorylated peptides separated by an enrichment step.
The synthetic peptide, AGSPS(phospho)ADPFR was a gift from Nick Agard (UCSF, School of Pharmacy), and was analyzed from the raw synthetic mixture without further purification. All MS/MS data were acquired in LC-MS experiments in a data dependent fashion, when multiply charged precursor ions were computer-selected from the MS survey scan, and the activation conditions were adjusted to the m/z and z of the precursor ion. Mass spectrometers used for data acquisition were a QSTAR Pulsar, QSTAR Elite (MDS SCIEX, Concorde, Canada) well as an LTQ-Orbitrap Velos (Thermo Fisher, San Jose, CA) for the synthetic peptide. Database searches were performed by Protein Prospector adjusting the search parameters according to the experiments, and the search results were manually inspected. Instrument-specific fragment ion lists were obtained using MS-Product of Protein Prospector. http://prospector.ucsf.edu/prospector/mshome.htm
Results and Discussion
Manual inspection of the quadrupole collision cell CID data of phosphopeptides from various enrichment experiments revealed a novel, frequently abundant ion for peptides containing a phosphorylated ProSer or ProThr sequence. The ProSer sequence is very common in phosphorylation motifs. Thus, it was first noticed that in ProSer(Phospho)-containing peptides fragmentation between the proline and phosphorylated serine residues resulted in a “y” ion where the serine had an apparent mass of 97 Da (rather than 87) (Figure 1). In a small dataset of 176 biologically “interesting” phosphopeptides 16 unique, unrelated sequences featured the Pro followed by a phosphorylated Ser residue, and 8 of the CID spectra displayed this fragment (Data not shown). However, the expected y ion that lost the phosphoric acid was also detected in each instance. We further investigated the phenomenon by examining other phosphopeptide datasets generated in our laboratory. Our observation was further supported by numerous quadrupole collision cell CID spectra featuring this unusual fragment ion (Table 1). In addition, the same phenomenon was detected in some ProThr(Phospho)-containing peptides: the apparent mass of the Thr was 111 Da, i.e. an unusual fragment was detected at an m/z value 10 Da higher than a y fragment without any modification (Table 1). Observation of this unusual fragment ion seemed to require an intense y ion series. In general, y ion intensity decreases with distance from the carboxy-terminus, and observation of this fragment ion was mostly limited to peptides where the ProSer/ProThr pair was within 12 amino acids of the C-terminus. Accurate mass measurements revealed that the 97 Da mass increment corresponds to an elemental composition of C4H3NO2 (the elemental composition of proline is C5H7NO) (Figure 2, Table 2). After neutral loss of the phosphoric acid, cleavage between the proline’s α carbon and its carbonyl group (similar to x fragment formation) would result in the observation of a new fragment ion 10 Da higher in mass than a y fragment corresponding to an unmodified serine. The potential pathways for the elimination of phosphoric acid from Ser/Thr-phosphorylated peptides have been studied by Palumbo et al. using a linear ion trap [7]. Three different pathways were suggested. Pathway A is the canonical (charge-remote) β-elimination pathway, while pathways B and C represent alternative, charge-directed reaction mechanisms: E2 elimination reaction and SN2 neighboring group participation reaction. They found that the charge-directed reactions represent the dominant process, and they provided experimental evidence for the phosphoric acid elimination via the SN2 neighboring group participation reaction, i.e. pathway C (Scheme 1). Pathway C results in a structure that makes normal y ion formation unlikely because three bonds would need to be broken to observe that y ion. Since in most quadrupole CID spectra, the y ion resulting from fragmentation of the peptide bond at the N-terminal side of the phosphoSer/Thr residue is observed, we speculate that the other pathways must be preferred during quadrupole CID of multiply charged peptides. However, phosphoric acid elimination via pathway C would explain the formation of the novel fragment described here (Scheme 1). While this fragmentation was originally identified from QSTAR data, examination of LTQ-Orbitrap data of such peptides revealed that the phenomenon occurs in HCD analysis, but was not observed in ion trap CID. We confirmed this observation using a synthetic peptide containing the Pro-Ser motif (AGSPS(phospho)ADPFR). This peptide was analyzed in LC-MS/MS using an LTQ-Orbitrap Velos instrument, subjecting the peptide to both ion trap CID and HCD analysis. The HCD spectrum of the phosphopeptide indeed showed the presence of the above described unusual fragment ion (Figure 3a), the corresponding ion trap CID did not display such fragmentation (Figure 3b). To our surprise, the unmodified version of the peptide also formed this fragment ion, albeit to a lesser extent (Figure 3c). Further investigation revealed that indeed non-modified Pro-Ser containing peptides also may undergo such fragmentation (Table 3).
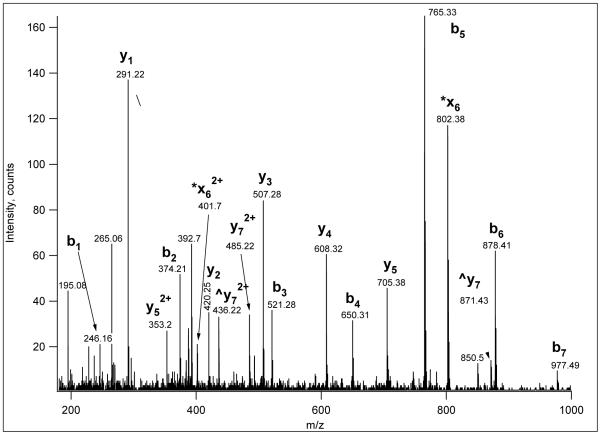
Figure 1.
CID spectrum of an iTRAQ-labeled phosphopeptide acquired in LC/MS mode on a QSTAR mass spectrometer from precursor ion m/z 649.3(3+). The peptide was identified as human Dystrobrevin peptide TQFEDLVPS(phospho)PTSEK. The fragment designated ‘*x6’ is 97 Da higher than y5. In this spectrum, it was detected both singly and doubly charged. ^indicates elimination of H3PO4.
Table 1.
List of phosphopeptides featuring the unusual fragment ion (From refs. 8-10)
| Gene | Protein | Peptide Sequence | aBP % | bx/y-98 |
|---|---|---|---|---|
| PS(Phospho)-containing sequences | ||||
| A3KGU4_MOUSE | Spectrin alpha 2 | TSSKE(ss)PVPsPTLDRc | 50 | 4.4 |
| A3KGU4_MOUSE | Spectrin alpha 2 | E(ss)PVPsPTLDR | 28 | 3.5 |
| A3KGU4_MOUSE | Spectrin alpha 2 | RPPsPDPNTK | 5 | 6.7 |
| SPTB2_MOUSE | Spectrin beta chain, brain 1 | (tssKEss)PVPsPTLDR | 80 | 4 |
| ANK2_MOUSE | Ankyrin-2 | TEWQPsPTDIPLQK | 20 | 4.7 |
| B2RQQ5_MOUSE | Microtubule-associated protein 1B | SPSLSPSPPsPIEK | 30 | 0.3 |
| MAP1A_MOUSE | Microtubule-associated protein 1A | AELEEMEEVHPsDEEEEETK | 9 | 1.8 |
| NFH_MOUSE | Neurofilament heavy polypeptide | IGFGPsPFSLTEGLPK | 14 | 1.8 |
| B1ART6_MOUSE | Microtubule-actin crosslinking factor 1 | LLDAEDVDVPsPDEK | BP | 8.9 |
| SYGP1_RAT | Ras GTPase-activating protein SynGAP | LPsPTK | BP | 6.4 |
| LRRC7_MOUSE | Leucine-rich repeat-containing protein 7 | TPsPFEDR | 43 | 2 |
| KPCG_MOUSE | Protein kinase C gamma | MGP(sss)PIPSPSPsPTDSKR | (2+)BP (+) 69 |
3.3 3 |
| LMNA_MOUSE | Prelamin-A/C | LSPsPTSQR | BP | 4.6 |
| STE20_CANAL | Serine/threonine-protein kinase CST20 | IPsNPTSTR | 52 | 1.8 |
| STE20_CANAL | Serine/threonine-protein kinase CST20 | TPsSQSLPR | 18 | 0.4 |
| PMA1_CANAL | Plasma membrane ATPase 1 | GYLVAMTGDGVNDAPsLK | 29 | 7.3 |
| ACT_CANAL | Actin | EITALAPsSMK | 18 | 2.5 |
| TSA1_CANAL | Peroxiredoxin TSA1 | YGEVCPANWHPGDETIKPsPEASK | 7 | y-98 not detected |
| TSA1_CANAL | Peroxiredoxin TSA1 | APVVQQPAPsFKK | 16 | 5 |
| RS13_CANMA | 40S ribosomal protein S13 | NAPsWFK | 9 | 3 |
| RAD18_CANAL | Postreplication repair E3 ubiquitin- protein ligase RAD18 |
ANVASPsPVAQSTVHK | 12 | y-98 not detected |
| PT(Phospho)-containing sequences | ||||
| B2RQQ5_MOUSE | Microtubule-associated protein 1B | DLTGQVPtPPVK | 52 | 2.8 |
| RBM14_MOUSE | RNA-binding protein 14 | QPtPPFFGR | 5 | 0.3 |
| TBA1B_MOUSE | Tubulin alpha-1B chain | sIQFVDWCPtGFKd | 13 | 2.8 |
| LRFN6_MOUSE | Leucine-rich repeat and fibronectin type- III domain-containing protein 6 |
VFSLDVPDHPtPTGLAKd | 16 | 1.5 |
| SSN2_CANAL | Mediator of RNA polymerase II transcription subunit 13 |
SNDYAPtPMIQDK | 5 | y-98 not detected |
| CHS5_CANAL | Chitin biosynthesis protein CHS5 | IESVPtDEIDTK | 4 | 3 |
| STE7_CANAL | Serine/threonine-protein kinase STE7 homolog |
ILHIPtQK | 23 | 1 |
the relative intensity of the ‘x’ fragment within the spectrum; BP = base peak
the intensity ratio of the ‘x’ fragment and the normal y-H3PO4 fragment
amino acids in lower case indicate phosphorylation sites; multiple residues in parentheses indicate ambiguity of the site assignment.
iTRAQ-labeled peptides
Figure 2.
From the CID spectrum of an iTRAQ-labeled phosphopeptide, KDYLIENEELPS(phospho)P displaying the PS-specific fragment ion, ‘*x2’. Data were acquired on a QSTAR Elite mass spectrometer. The spectrum was recalibrated using m/z 185 and 282 as internal standards (See Table 2). There is only one potential elemental composition for ‘*x2’within 20 ppm that also may fit a peptide fragment: C9 H13 N2 O4. Once the elemental composition of the C-terminal Pro is subtracted, C4 H3 N1 O2 is left.
Table 2.
Mass accuracy for the ions in Figure 2. Fragments m/z 185 and 282 served as internal standards.
| m/z exp. | m/z calc. | dev [ppm] | assignment |
|---|---|---|---|
| 185.0921 | 185.0921 | Ref1 | y2-H3PO4 |
| 213.0909 | 213.0870 | 18 | y2 |
| 226.0886 | 226.0822 | 28 | EN/NE-H2O |
| 244.0947 | 244.0928 | 8 | EN/NE |
| 259.0963 | 259.0925 | 15 | EE |
| 265.0605 | 265.0584 | 8 | PS(phospho) |
| 282.1448 | 282.1448 | Ref2 | y3-H3PO4 |
| 348.2084 | 348.2020 | 18 | b3(2+) |
| 355.1240 | 355.1248 | −2 | ENE/NEE-H2O |
| 380.1256 | 380.1217 | 10 | y3 |
| 404.7454 | 404.7440 | 3 | b4(2+) |
Scheme 1.
Fragmentation mechanism proposed. The charge-directed SN2 neighboring group participation reaction for phosphoric acid elimination was presented as Pathway C in ref.7.
Figure 3a.
HCD spectrum of synthetic peptide, AGSPS(phospho)ADPFR-amide acquired from m/z 542.2375(2+) in LC/MS/MS mode using an LTQ-Orbitrap Velos. ^ indicates phosphoric acid elimination, ‘*x6’ stands for the Pro-Ser-sequence-specific fragment. Its calculated mass is 701.3366 that is 4 ppm lower than the experimental value. Fragments y5 and y7-H3PO4 were also measured with the same mass accuracy.
Figure 3b.
Ion trap CID of the same precursor ion acquired in the next scan. No Pro-Ser-sequence-specific fragment ion was detected in this experiment. The insert shows the region of interest magnified tenfold.
Figure 3c.
HCD spectrum acquired from the doubly charged ion (m/z 502.25) of the unmodified peptide, AGSPSADPFR-amide. Both the expected y6 fragment and the Pro-Ser-sequence-dependent ‘*x6’ ion was observed in the experiment.
Table 3.
Some unmodified peptides yielding the unusual fragment ion a
| Sequence | ‘x’ m/z exp.b |
‘x’ m/z calc. |
Δ ppm |
|---|---|---|---|
| VLALPEPSPAAPTLR | 822.4538 | 822.4464 | 9 |
| DTHDQLSEPSEVR | 500.2472 | 500.2463 | 2 |
| <QLMHNGHPSEK | 373.1753 | 373.1717 | 9 |
| TPDSLEPSPLK | 454.2625 | 454.2659 | 7 |
| <QTPSGTASR | 588.2673 | 588.2736 | 11 |
Data were acquired on a QSTAR Pulsar mass spectrometer.
after internal calibration
Conclusions
In quadrupole collision-cell CID experiments, a novel fragment ion may be formed from peptides containing Pro-Ser/Thr sequences. The novel fragment is most likely formed in two consecutive steps: first the phosphoric acid/or water is eliminated via a charge-directed neighboring group assisted SN2 reaction, and a cyclic oxazoline structure is formed [7]. Then a cleavage occurs between this ring and the α carbon of the proline residue. When the hydroxy amino acid is not phosphorylated, this fragment ion is less intense than the expected y ion. However, in the case of a phosphorylated serine or threonine, this fragment may be very abundant. Thus, this fragment ion could be utilized after regular database searching for peptide assignment confirmation as well as aiding phosphorylation site assignment. In general, we believe that considering sequence specific “minor” fragmentation may improve the reliability of peptide identifications in large scale proteomics experiments.
Acknowledgements
This work was supported by NIH grants NCRR P41RR001614 and 1S10RR012961, and the Howard Hughes Medical Institute (to the Bio-organic Biomedical Mass Spectrometry Resource at UCSF, director A.L. Burlingame).
References
- 1.Biemann K. Nomenclature for peptide fragment ions (positive-ions) Meth. Enzymol. 1990;193:886–887. doi: 10.1016/0076-6879(90)93460-3. [DOI] [PubMed] [Google Scholar]
- 2.Medzihradszky KF. Peptide Sequence Analysis. Meth. Enzymol. 2005;402:209–44. doi: 10.1016/S0076-6879(05)02007-0. [DOI] [PubMed] [Google Scholar]
- 3.Baker PR, Medzihradszky KF, Chalkley RJ. Improving software performance for peptide electron transfer dissociation data analysis by implementation of charge state- and sequence-dependent scoring. Mol Cell Proteomics. 2010;9(9):1795–1803. doi: 10.1074/mcp.M110.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee YJ, Lee YM. Formation of c1 fragment ions in collision-induced dissociation of glutamine-containing peptide ions: a tip for de novo sequencing. Rapid Commun Mass Spectrom. 2004;18(18):2069–2076. doi: 10.1002/rcm.1593. [DOI] [PubMed] [Google Scholar]
- 5.Erlekam U, Bythell BJ, Scuderi D, Van Stipdonk M, Paizs B, Maître P. Infrared spectroscopy of fragments of protonated peptides: direct evidence for macrocyclic structures of b5 ions. J. Am Chem Soc. 2009;131(32):11503–11508. doi: 10.1021/ja903390r. [DOI] [PubMed] [Google Scholar]
- 6.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. http://www.proteomesoftware.com/ [DOI] [PubMed] [Google Scholar]
- 7.Palumbo AM, Tepe JJ, Reid GE. Mechanistic Insights into the Multistage Gas-Phase Fragmentation Behavior of Phosphoserine- and Phosphothreonine-Containing Peptides. J. Prot. Res. 2008;7(2):771–779. doi: 10.1021/pr0705136. [DOI] [PubMed] [Google Scholar]
- 8.Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL. Quantitative Analysis of Synaptic Phosphorylation and Protein Expression. Mol Cell Proteomics. 2008;7(4):684–696. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Trinidad JC, Thalhammer A, Specht CG, Schoepfer R, Burlingame AL. Comprehensive Identification of Phosphorylation Sites in Postsynaptic Density Preparations. Mol Cell Proteomics. 2006;5(5):914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Beltrao P, Trinidad JC, Fiedler D, Toguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of Phosphoregulation: Comparison of Phosphorylation Patterns across Yeast Species. PLoS Biology. 2009;7(6):e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]