Abstract
The response and functions of proteasome regulators Pa28αβ (or 11S), Pa28γ, and Pa200 in oxidative-stress adaptation (also called hormesis) was studied in murine embryonic fibroblasts (MEF), using a well-characterized model of cellular adaptation to low concentrations (1.0 to 10.0μM) of hydrogen peroxide (H2O2), which alter gene expression profiles, increasing resistance to higher levels of oxidative-stress. Pa28αβ bound to 20S proteasomes immediately upon H2O2-treatment, whereas 26S proteasomes were disassembled at the same time. Over the next 24 hours, the levels of Pa28αβ, Pa28γ, and Pa200 proteasome regulators increased during H2O2-adaptation, whereas the 19S regulator was unchanged. Purified Pa28αβ, and to a lesser extent Pa28γ, significantly increased the ability of purified 20S proteasome to selectively degrade oxidized proteins; Pa28αβ also increased the capacity of purified immunoproteasome to selectively degrade oxidized proteins but Pa28γ did not. Pa200 regulator actually decreased 20S proteasome and immunoproteasome’s ability to degrade oxidized proteins but Pa200 and poly-ADP ribose polymerase may cooperate in enabling initiation of DNA repair. Our results indicate that cytoplasmic Pa28αβ and nuclear Pa28γ may both be important regulators of proteasome’s ability to degrade oxidatively-damaged proteins, and induced-expression of both 20S proteasome and immunoproteasome, and their Pa28αβ and Pa28γ regulators are important for oxidative-stress adaptation.
Keywords: Oxidative stress adaptation (hormesis), Ubiquitin-Proteasome System, Protein Degradation, Proteasome regulators, Pa28 (11S) regulator, Immunoproteasome
INTRODUCTION
Cells possess a complex and sophisticated system for the removal of oxidatively damaged proteins, which is vital for continued viability [1–3]. Many studies have shown a critical role for the Proteasome in the removal of such damaged proteins [2–8]. The core of the proteasome is a four ringed, barrel-shaped protein known as the 20S proteasome. The 20S proteasome is capable of functioning on its own or it can bind to a number of regulator complexes that modify its choice of substrates and its ATPase activity, as well as its proteolytic capacity; these include the 19S regulator, the Pa28αβ (or 11S) regulator, the Pa28γ regulator, and the Pa200 regulator. The 19S regulator can bind to both ends of the 20S proteasome cylinder, forming the ATP-stimulated 26S proteasome that is responsible for degrading ubiquitinated protein substrates. Previous work has demonstrated a relatively minor role for the 26S proteasome compared to the 20S proteasome in the proteolytic degradation of oxidized proteins [2–4, 9–18]. An alternative form of the proteasome known as the immunoproteasome also appears to be capable of the selective degradation of oxidized proteins [2] and to have a role in enhancing cellular resistance to oxidative stress [2, 19–21]. The role(s) that the Pa28αβ, Pa28γ, and Pa200 proteasome regulators may play in the removal of damaged proteins, and in cellular resistance to oxidative stress, is/are however largely unknown.
A process of transient adaptation to oxidative stress (sometimes also called hormesis) has previously been described. In this process, cells which have been exposed to a mild oxidative stress will become transiently more resistant to normally toxic oxidative stresses, via a combination of enzyme activation/deactivation steps and altered gene expression patterns [2, 3, 22, 23]. The 20S proteasome appears to be an important factor in increased cellular oxidative stress resistance. This occurs both through de novo synthesis and directly enhanced activity [2, 3, 23].
In addition to 20S proteasome and immunoproteasome, expression of the cytoplasmic proteasome regulator Pa28αβ is up-regulated in the process of adaptation, however, its function in this response is largely unknown [2, 23]. Two other activators of the 20S proteasome: Pa28γ and Pa200 are primarily located in the nucleus. Pa28γ is a genetic ortholog of Pa28αβ. Pa28γ forms a homoheptameric ring on the 20S proteasome and has been shown to weakly enhance the 20S proteasome’s ability to degrade peptide substrates [24]. The Pa200 regulator is a large 200kDa regulator which can bind to the 20S proteasome. Like the other regulators its attachment enhances the proteasome’s capacity to degrade short peptides. In addition, Pa200 expression is increased in response to ionizing radiation [25].
In this study we have investigated the roles of the three proteasome regulators Pa28αβ, Pa28γ, and Pa200 in the degradation of oxidized proteins and in the process of adaptation to the oxidative stress of hydrogen peroxide. Our data provides evidence of a highly complex set of interactions, between multiple forms of proteasome, with a variety of regulators. Our results also suggest that these regulators control a diverse range of proteasome functions in different parts of the cell.
EXPERIMENTAL
Materials
All materials were purchased from VWR unless otherwise stated. Murine Embryonic Fibroblasts (MEF), (catalog #CRL-2214) was purchased from ATCC (Manassas, VA, USA). Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM), (catalog #10-013-CV), from Mediatech (Manassas, VA) and supplemented with 10% Fetal Bovine Serum (catalog #SH30070.03) from Hyclone (Logan, UT, USA): henceforth referred to as ‘complete media.’ Cells were typically incubated at 37°C under 5% CO2 and ambient oxygen.
H2O2 Pretreatment
MEF cells were cultured to 10–20% confluence after which cell were pretreated with a mild dose of 1μM, 10μM, or 100μM H2O2 for 1 h. After this media was removed and fresh complete media added. Cells were then allowed 24 h to adapt before assays were performed.
H2O2 Challenge
MEF cells would be challenged with a toxic a toxic dose of H2O2 (1mM) 24 h after H2O2 pretreatment. Challenge would last 1 h after this media was removed and fresh complete media added. Cells were then allowed 24 h for cells to divide after which cell counts were taken.
Western Blot Analysis
MEF cells were harvested from 25–75 cm2 flasks by trypsinization. Cells were washed with PBS to remove trypsin and then lysed in RIPA buffer, (catalogue #89901) from Thermo Fisher (Waltham, MA, USA), supplemented with protease inhibitor cocktail (catalogue #11836170001) from Roche (Nutley, NJ, USA). Protein content was quantified with the BCA Protein Assay Kit (Pierce, Rockford, IL, USA) according to the manufacturer’s instructions. For Western analysis, 20 μg of protein was run on SDS–PAGE and transferred to PVDF membranes. Using standard Western blot techniques, membranes were treated with proteasome regulator subunit PA28α antibody (catalogue #PW8185-0100) from Enzo Life sciences, (Plymouth Meeting, PA, USA), anti-Pa200 (catalogue # ab5620) purchased from Abcam (Cambridge, MA, USA), anti-Pa28γ (catalogue # BML-PW8190-0025) from Enzo Life sciences, (Plymouth Meeting, PA, USA), immunoproteasome subunit anti-LMP2/β1i antibody (catalogue #ab3328) purchased from Abcam (Cambridge, MA, USA), 20S proteasome anti-β1 antibody (catalogue #sc-67345) and 3-tubulin (catalogue # 5661) purchased from Millipore (Billerica, Massachusetts, USA). The blocking buffer employed for Western blotting was Startingblock™ buffer (catalogue #37539) from Thermo Fisher (Waltham, MA, USA) and the wash buffer was 1x PBS containing 0.1% Tween 20. An enhanced chemiluminescence kit (Pierce, Rockford, IL) was used for chemiluminescent detection and membranes were analyzed using the biospectrum imaging system (UVP, Upland, CA, USA).
Immunoprecipitation
Protein A Sepharose CL-4B beads (catalogue # 17-0780-01) from GE Healthcare (Little Chalfont, UK), were washed twice with PBS and incubated in PBS + 1% BSA for 1hr then washed again with PBS, 24hr prior to assay. MEF cells were cultured to 50% confluence in 75–225cm2 flasks. Cells were washed twice with PBS and harvested through scraping in PBS. Cells were lysed with 3 freeze-thaw cycles. Cell debris was removed by centrifugation at 10,000g for 15 min. and all subsequent steps were performed at 4°C. Samples were incubated for 30 min with either anti-β-tubulin (catalogue # 5661) from Millipore (Billerica, Massachusetts, USA) or anti-porin (catalogue # ab15895) from Abcam (Cambridge, MA, USA), to remove non-specific binding. Samples were then pre-cleared by incubation with 100ul of sepharose bead slurry, for 30 min, followed by centrifugation and removal of beads. Samples were then incubated for 1hr either with PA28α antibody (catalogue #PW8185-0100) from Enzo Life sciences, (Plymouth Meeting, PA, USA), anti-Pa200 (catalogue # ab5620), anti-S3 (catalogue# ab110067) p purchased from Abcam (Cambridge, MA, USA), anti-Pa28γ (catalogue # BML-PW8190-0025) from Enzo Life sciences, (Plymouth Meeting, PA, USA), immunoproteasome subunit anti-LMP2/β1i antibody (catalogue #ab3328) purchased from Abcam (Cambridge, MA, USA), 20S proteasome anti-β1 antibody (catalogue #sc-67345) purchased from Santacruz biotechnology (Santa Cruz, CA, USA), anti-β-tubulin (catalogue # 5661) from Millipore (Billerica, Massachusetts, USA), or anti-porin (catalogue # ab15895) from Abcam (Cambridge, MA, USA). Next, 100ul of sepharose beads was added and samples were incubated for 4hr with agitation. Samples were washed 3 times with PBS and protein was then detached from beads using SDS-PAGE loading Buffer and boiling. Samples were run on then run on Western blots as described above.
Proteolytic Activity Assays with Purified Proteasome and Immunoproteasome
Purified 20S Proteasome (catalogue # PW8720), Immunoprteasome (catalogue # PW9645), Pa28αβ (catalogue # PW9420) and Pa28γ (catalogue #PW9875) were purchased from Enzo Life sciences (Plymouth Meeting, PA, USA). Purified Pa200 [25] was the kind gift of Dr Martin Rechsteiner of the University of Utah. All proteins were subjected to vigorous buffer exchange using centrifugal filter units (catalogue # UFC500308) from Millipore (Billerica, Massachusetts, USA), to exchange their storage buffer for proteolysis buffer 50 mM Tris, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 1 mM DTT (pH 7.5), shortly prior to use. Proteasome or immunoproteasome was incubated with a 4 fold molar excess of proteasome regulators for 30 min at 37°C. Peptidase activity of the 20S proteasome and of the immunoproteasome (± regulators) was determined by release of fluorescent 7-amino-4-methycoumarin (AMC) from the peptide substrate Suc-LLVY-AMC (catalogue #80053-860) purchased from VWR (Chester, PA, USA). Proteolytic activity of the 20S proteasome and of the immunoproteasome was measured by release of fluorescent AMC from, native or oxidatively modified, AMC-labeled hemoglobin, or histone proteins, as previously described [26]. Proteolytic substrates were added to the samples plates which were then incubated at 37°C and mixed at 300 rpm for 4 hr. Fluorescence readings were taken at 10 minute intervals using an excitation wavelength of 355nm and an emission of 444 nm. Following subtraction of background fluorescence, fluoresence units were converted to moles of free AMC, with reference to an AMC standard curve, constructed experimentally with known amounts of AMC (catalogue #164545) purchased from Merck (Whitehouse Station, NJ, USA). The substrates used were Suc-LLVY-AMC (catalogue #80053-860) purchased from VWR (Chester, PA, USA) or AMC labeled hemoglobin (Hb), oxidized hemoglobin (Hbox), histones, or oxidized histones. The Hb (catalogue #374834) was purchased from EMD Chemicals (Darmstadt, Germany) and the histones (catalogue # H5505) were purchased from Sigma Aldrich (St. Louis, MS, USA). Proteins were labeled with AMC and oxidized as described previously [26].
Proteolytic Activity Assays with Cell Lysates
MEF cells were cultured to 10% confluence, cells were then exposed to different doses of H2O2 for 1hr after which media was replaced, and cells were allowed to adapt for 24hr. Cells were then washed twice with PBS buffer and scraped from the flask and transferred into proteolysis buffer 50 mM Tris, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 1 mM DTT (pH 7.5). Cells were then lysed through 3, 5 minute freeze-thaw cycles and then centrifuged to remove non-lysed cells. Protein content was adjusted based on a BCA assay. Next, 5μM of the peptidase substrate Suc-LLVY-AMC, purchased from VWR (catalogue #80053-860, Chester, PA, USA) was added to each sample. Plates were then incubated at 37°C and mixed at 300 rpm for 4 hr. Fluorescence readings were taken at 10 minute intervals using an excitation wavelength of 355nm and an emission of 444 nm. Following subtraction of background fluorescence, fluorescence units were converted to moles of free AMC, with reference to an AMC standard curve, constructed experimentally with known amounts of AMC (catalogue #164545) purchased from Merck (Whitehouse Station, NJ, USA).
Immunocytochemistry
MEF cells were cultured to 50% confluence. Cells were fixed with 4% paraformaldehyde for 30mins then washed twice for 5mins with PBS. Exogenous proteinase activity was blocked through incubation with 10% methanol; 0.3% H2O2 in PBS for 20 min followed by two 5min PBS washes. Cells were permeabilized by incubation with 1% NP40 for 10 min followed by two 5 min washes with PBS. Samples were incubated in Startingblock™ buffer (catalogue #37539) from Thermo Fisher (Waltham, MA, USA) for 30 min, then incubated in blocking buffer containing a 1/1000 dilution of anti-Pa200 for 30mins. Samples were washed three times with PBS + 0.1% tween for 5 min. Samples were then incubated with Biotinylated -Secondary Rabbit antibody (catalogue PK-6101) purchased from Vector laboratories (Burlingame, CA, USA), in blocking buffer for 30 min. Samples were washed three times with PBS + 0.1% tween for 5 min then detected using a DAB detection kit (catalogue #SK-4100) purchased from Vector laboratories (Burlingame, CA, USA).
siRNA treatment
β1(catalog # sc-62865), β1i (Lmp2) (catalog # sc-35821), Pa28α (catalog # sc-151977), Pa200 (catalog # sc-151976) and Scrambled Control (catalog # sc-37007) siRNA were purchased from Santa Cruz biotechnology (Santa Cruz, CA). For experiments with these siRNAs, MEF were seeded at a density of 100,000 cells per well in 6 or 48 well plates and grown to 10% confluence. siRNA treatment was then performed as described in the Santa Cruz Biotechnology product manual.
RESULTS
Pa28αβ Regulator and 20S Proteasome
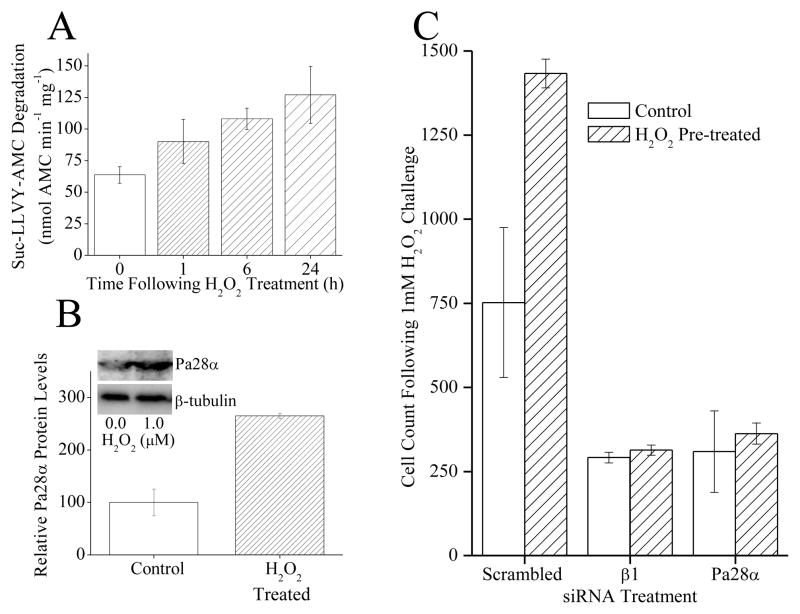
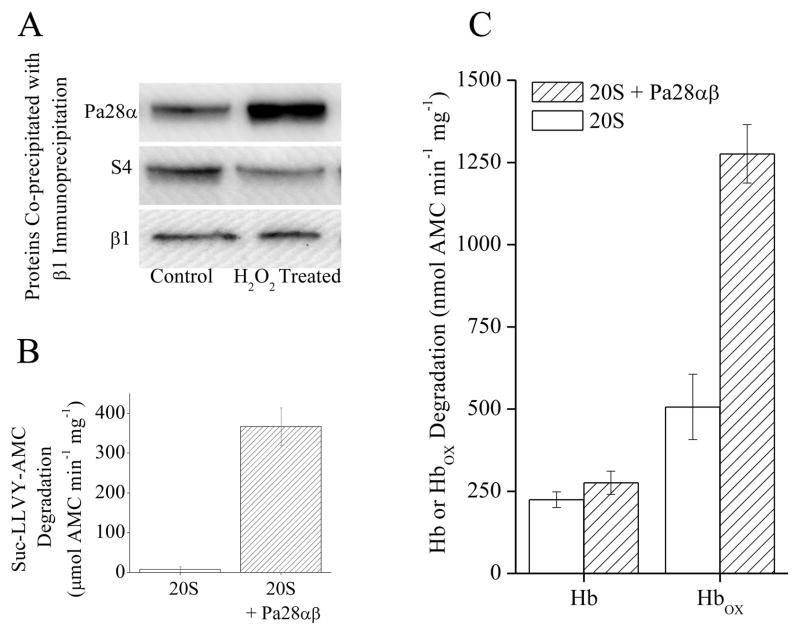
Exposure to a mild dose of hydrogen peroxide (H2O2), as well as other oxidants, has been shown to induce adaptation and produce an increase in proteolytic capacity in various mammalian cell types [2, 3, 23]. This increase was confirmed and extended in MEF cells under the conditions of the current experiments (Fig. 1A), revealing a progressive rise in proteolytic capacity over the 24 h following H2O2 exposure. This H2O2 adaptation also appears to produce an increase in the cellular concentration of Pa28αβ regulator over the same 24 h time period (Fig. 1B). It has been previously shown [2, 3, 23], and is now reaffirmed, that this increase is prevented by protein synthesis inhibitors and siRNA (data not shown). In addition it seems that treatment with siRNA directed against Pa28α or 20S proteasome β1 will decrease oxidative stress tolerance and block the H2O2 pre-treatment induced increase in oxidative stress tolerance (Fig. 1C). It has been observed that the initial response to H2O2 treatment produces a temporary (1–3 h) disassociation of 26S proteasome to form additional free 20S proteasomes, so increasing the proteolytic capacity of the cell, while the liberated 19S regulators are protected by HSP70 [2, 3]. This effect was then re-examined using new co-immunoprecipitation techniques. From this it appears that there is also a concurrent rise in association of free 20S proteasomes (β1 subunit) with Pa28αβ regulators occurring as rapidly as the first hour after H2O2 treatment (Fig. 2A). This rise in Pa28αβ regulator association occurred over the same time course as the dissociation of 19S regulators from 26S proteasomes, as shown by the diminished co-precipitation of the 19S S4 subunit by 20S proteasome β1(Fig. 2A). It should also be noted that, as previously [2, 3], There was no observed increase in 19S regulator levels (not shown). These results suggest that the Pa28αβ regulator perhaps might actually assist in the initial adaptive increase in proteolytic capacity following mild oxidative stress. It has been shown that Pa28αβ enhances 20S proteasome’s proteolytic capacity to degrade short peptide substrates [18, 24]. Under these conditions, it was found that if purified 20S proteasome was incubated with purified Pa28αβ, there was an ≈ 50 fold increase in proteolytic capacity to degrade the short peptide substrate Suc-LLVY-AMC (Fig. 2B).
Fig. 1.
(A) H2O2 exposure causes an increase in proteolytic capacity. MEF cells were pre-treated with 0, 1, 10 or 100μM H2O2. Then, 24 h later samples were harvested and lysed, and the capacity to degrade the short peptide Suc-LLVY-AMC was measured. Values are means ± SE where n = 3. (B) H2O2 exposure causes an increase in Pa28αβ protein levels. Samples were prepared as in panel A then run on Western blots which were screened with antibodies against either the α subunit of Pa28αβ or the loading control β-tubulin. (C) Blocking the induction of 20S proteasome β1 subunit, or Pa28ab proteasome regulator expression, with siRNAs, inhibited the H2O2 induced increase in oxidative stress tolerance, and blocked the increase in cell growth. siRNA knock-down with β1 siRNA, Pa28α siRNA, or scrambled siRNA was performed for 96 h. Cells were then pretreated with a mild dose of H2O2 then 24 h later challenged with a toxic dose of H2O2. Cell counts were taken 24 h later. Results are mean ± S.E. where n = 3.
Fig. 2.
(A), Following H2O2 treatment there is an increase in Pa28αβ association with 20S proteasome and a corresponding decrease in S4 (19S subunit) binding. MEF cells were treated with 1mM H2O2 for 1 h, after this immunoprecipitation of anti-β1 was performed. Samples were run on Western blots and screened for co-immunoprecipitation of anti-Pa28α, anti-S4, or anti-β1. (B) Pa28αβ increases the capacity of 20S proteasome to degrade the peptide substrate Suc-LLVY-AMC. 20S proteasome was incubated with a 4-fold molar excess of Pa28αβ for 30 minutes. Suc-LLVY-AMC was then added to samples and proteolytic capacity was measured. Values are means ± SE where n = 3. C. Pa28αβ increases the capacity of 20S proteasome to selectively degrade oxidized hemoglobin. 20S proteasome was incubated with a 4-fold molar excess of Pa28αβ for 30 minutes. Hb-AMC or HbOX-AMC was then added to samples and proteolytic capacity was measured. Values are means ± SE where n = 8.
An important aspect of cellular responses to oxidative stress is the capacity of cells to selectively degrade oxidized proteins. This represents the ability of cells to remove damaged proteins and permit normal cell function to return after an oxidative stress [1, 2, 4]. It has been shown that the capacity to degrade oxidized proteins increases under conditions of oxidative stress. It has also been shown that this increase is largely dependent on the synthesis of 20S proteasome [2, 3, 27] as well as the mitochondrial Lon protease [28–30]. If the Pa28αβ regulator plays an important role in oxidative stress adaptation, then it would be expected to enhance the ability of 20S proteasome to selectively degrade oxidized proteins. To test if this was the case purified 20S proteasome was incubated with or without purified Pa28αβ, and the enzyme’s capacity to degrade either oxidized or native hemoglobin was measured. Addition of Pa28αβ caused no change in 20S proteasome’s capacity to degrade native hemoglobin but more than doubled the capacity to degrade oxidized hemoglobin (Fig. 2C). This is highly supportive of the idea that Pa28αβ functions in oxidative stress adaptation through increasing the capacity of the 20S proteasome to degrade oxidized proteins.
Pa200 and 20S Proteasome
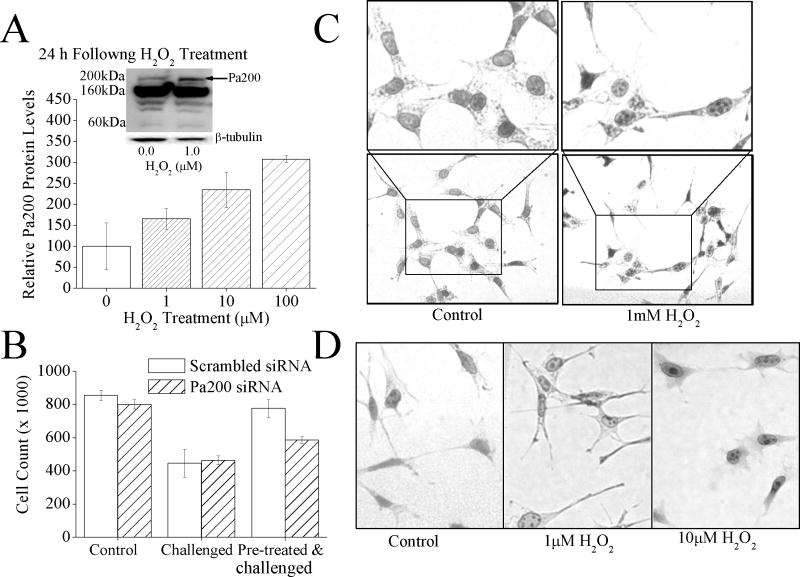
In addition to 19S and Pa28αβ, mammalian cells possess two other proteasome regulators: Pa28γ and Pa200; the functions of Pa28γ and Pa200, are however, largely unknown. As seen for Pa28αβ, it seems that exposure to 1μM H2O2 also produces a significant increase (t-test, p≤0.05) in Pa200 levels (Fig. 3A, inset). There are 3 isoforms of Pa200 (200kDa, 160kDa and 60kDa), A sharp increase in the levels of the 200kDa isoform was observed, which is thought to be the functional isoform, but no change in the level of the160kDa and 60kDa isoforms were observed. To further investigate this increase the levels of (200kDa) Pa200 was examined over a range of H2O2 concentrations. From this a progressive rise in Pa200 levels with H2O2 exposures was observed between 1μM and 100μM. In fact a 3 fold increase in Pa200 levels was observed at the peak H2O2 treatment of 100μM H2O2 (Fig. 3A). An H2O2 pre-treatment challenge assay was next performed, as done in fig. 1C. In this assay it was found that Pa200 levels increase under H2O2 exposure, and that this increase enhances oxidative stress resistance (Fig.3B). Pa200 is localized in the nucleus where it has been shown, to form punctuate foci under either strong ionizing radiation or toxic H2O2 levels of exposure [24, 25]. This has led to the hypothesis that Pa200 binds to chromatin in severe stress, and initiates some aspect of DNA repair [25]. To determine whether Pa200 behaved similarly under these experimental conditions, MEF cells were exposed to low adaptive doses of H2O2 (1–10μM), or to a 1.0mM toxic dose of H2O2 for 1 h then staining for Pa200. Interestingly, the formation of punctuate foci of Pa200 in the nuclei appeared both following toxic (1.0mM) H2O2 treatments and under the conditions of low 1.0 or 10.0μM adaptive H2O2 concentrations used throughout this paper; indeed, the formation of punctuate nuclear Pa200 foci seems to follow a fairly straightforward H2O2 concentration-curve profile (Fig. 3C and Fig. 3D).
Fig. 3.
(A) H2O2 exposure causes an increase in Pa200 protein levels. MEF cells were pretreated with 0, 1, 10, or 100μM H2O2, 24 h later samples were harvested and run on Western blots which were then screened with antibodies against either Pa200 or the loading control β-tubulin. Sample band intensity was measured and adjusted based on β-tubulin band intensity. Values are means ± SE where n = 3. The inset shows a representative blot. (B) By blocking the adaptive increase in Pa200, the H2O2 induced increase in oxidative stress tolerance is blunted. MEF cells treated with either Pa200 or (scrambled) siRNA for 24 h to block induction. Samples were then transiently adapted to oxidative stress by pre-treatment with 1μM of H2O2, for 1 h. Following a 24 h adaptation period, both adapted and non-adapted cells were challenged by incubation with a high dose of 100μM H2O2. Cell counts were then taken using a cell counter 24 h later. Results are means ± S.E. where n = 3. (C) Exposure to a toxic level of H2O2 causes the formation of Pa200 foci in the nucleus. MEF cells were exposed to 1.0mM H2O2 for 1hr. Immunocytochemistry was then performed and cells were stained with anti-Pa200. (D) Exposure to low, adaptive doses of H2O2 also results in the formation of nuclear Pa200 foci. Conditions were identical to those of panel C., except that 0, 1.0μM, or 10.0μM H2O2 exposures were used.
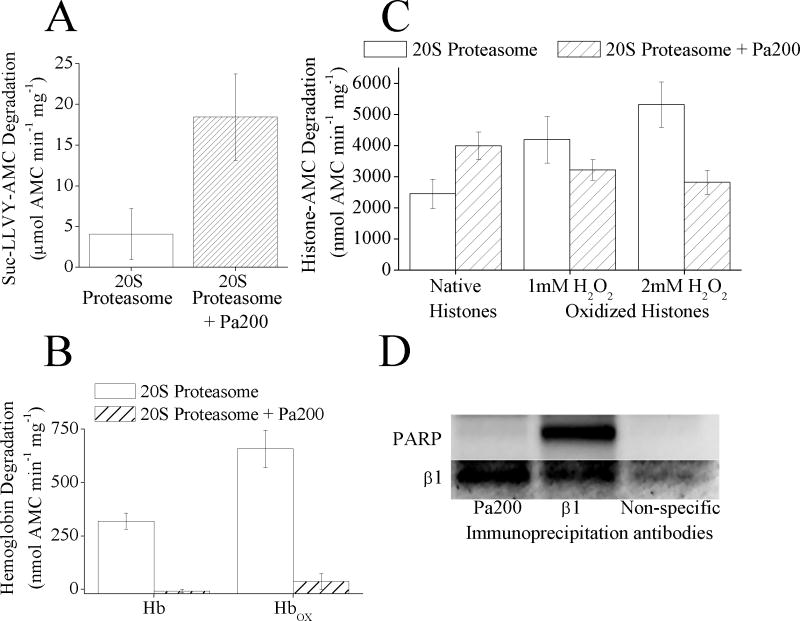
It is conceivable that Pa200 might have a role in enhancing the capacity of proteasome to degrade oxidized proteins in the nucleus. So like Pa28αβ the proteolytic activity of 20S proteasome with and without Pa200 was measured. First it was confirmed that Pa200 actually interacted with the 20S proteasome which was done by measuring capacity to degrade the short peptide substrate Suc-LLVY-AMC (Fig. 4A). Addition of Pa200 to 20S proteasome produced a 4 fold increase in proteolytic activity. Next the capacity of the 20S proteasome-Pa200 complex to degrade native and oxidized hemoglobin was measured. Unlike the increased degradation of Suc-LLVY-AMC, Pa200 appeared to almost completely block the 20S proteasome from degrading both native and oxidized hemoglobin (Fig. 4B). The ability of Pa200 to enhance the capacity of 20S proteasome to degrade Suc-LLVY-AMC and to block degradation of a normal protein has been reported previously [25] but it seemed surprising that Pa200 could also essentially block degradation of oxidized Hb. To expand upon this it was investigated whether Pa200 could also inhibit degradation of other oxidized proteins and, because of the oxidative-stress-induced association of Pa200 with chromatin, native and oxidized histone proteins seemed a good target substrates for degradation by either 20S proteasome, or 20S proteasome plus Pa200. 20S proteasome degraded oxidized histones, significantly better than native histones (t-test, p≤0.05), but when Pa200 was added to the 20S proteasome it actually increased the degradation of native histones and decreased that of oxidized histones (Fig. 4C). This suggests that Pa200 actually enhances selectivity for degradation of normal proteins, while reducing selectivity for oxidized histones. Thus, while Pa200 may play a role in turnover of normal histones, it does not appear to be involved in the specific removal of oxidatively modified histones from the chromatin.
Fig. 4.
(A) Pa200 increases the capacity of 20S proteasome to degrade the peptide substrate Suc-LLVY-AMC. 20S proteasome was incubated with a 4 fold molar excess of Pa200 for 30 minutes. Suc-LLVY-AMC was then added to samples and proteolytic capacity measured over the next 4 h. Values are means ± SE where n = 3. (B) Pa200 reduces the capacity of 20S proteasome to degrade both oxidized and naive hemoglobin substrates. 20S Proteasome ± Pa200 was prepared as in panel A. Hb-AMC or HbOX-AMC was then added to samples and proteolytic capacity measured over the next 4 h. Values are means ± SE, where n = 8. (C) Pa200 enhances the capacity of proteasome to degrade normal histones but reduces proteasome’s capacity to degrade oxidized histones. 20S Proteasome ± Pa200 was prepared as in panel A and AMC labeled normal histones, or oxidized AMC labeled histones were used as substrates. Oxidized histones were prepared by treatment with either 1mM or 2mM H2O2 for 1 h. Proteolysis values are means ± SE, where n = 6. (D) Immunoprecipitation with either anti-Pa200 or anti-β1 both result in co- immunoprecipitation of 20S proteasome subunits, however only anti-β1 antibodies co-precipitate PARP. MEF cells were grown to 10% confluence, then immunoprecipitation with either anti-β1 antibody, anti-Pa200 antibody, or anti-porin antibody (as a non-specific binding control) was performed. Samples were run on Western blots and screened for co-immunoprecipitation of anti-β1 or anti-PARP.
Another nuclear regulator of proteasome is poly-ADP ribose polymerase (PARP) which has been shown to enhance the proteasome selective degradation of oxidized histones, and to play an important role in the oxidative stress induced adaptive increase in proteolytic capacity in the nucleus [10]. Because of this, it seemed possible that perhaps PARP might work in association with Pa200. To test this hypothesis an immunoprecipitation assay was performed, using pull-down with either 20S proteasome or Pa200 antibodies. The samples were then screened for the presence of either 20S proteasome or PARP (Fig. 4D). While both of the samples contained 20S proteasome subunits, only the 20S proteasome pull-down contained PARP. These results indicate that PARP only associates with proteasome that does not contain the Pa200 regulator and, therefore, acts to increase proteasome activity and selectivity for oxidized histones [10] in a mechanism independent of Pa200.
Pa28γ and 20S Proteasome
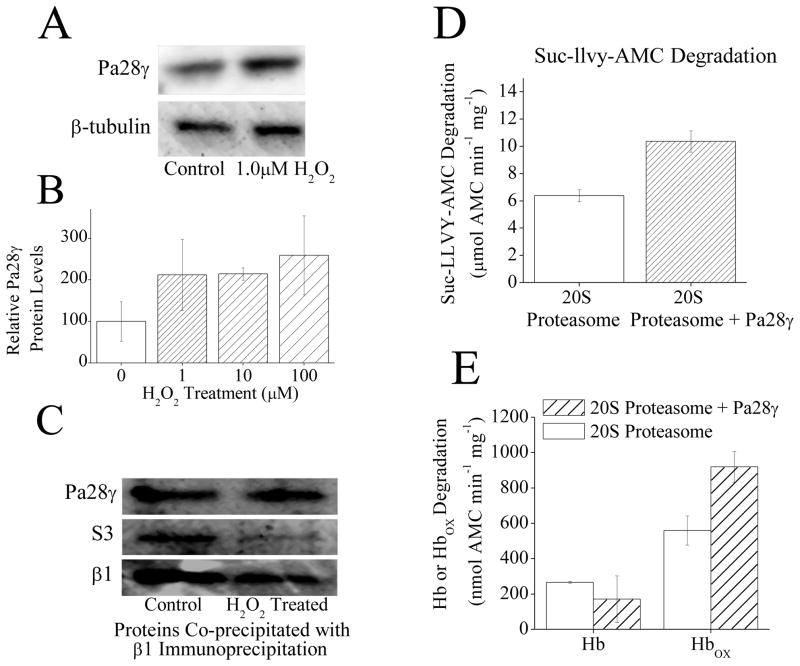
The other proteasome regulator is Pa28γ whose subunits are highly similar to both Pa28α and Pa28β, but Pa28γ is primarily a nuclear protein, whereas Pa28α and Pa28β are found in the cytoplasm. It is thought that Pa28γ forms a homoheptameric ring which can attach to either end of the 20S proteasome cylinder. The function of this, regulator, however is largely unknown [24, 31]. It appears that like Pa28αβ and Pa200, H2O2 adaptation caused an increase in expression of Pa28γ (Fig. 5A, B). To further investigate this increase the levels of Pa28γ were examined over a range of doses of H2O2. From this somewhat of a progressive rise in Pa28γ levels with H2O2 exposures was observed between 1μM and 100μM: peaking at a 2.5 fold increase in Pa200 levels with H2O2 treatment of 100μM (Fig. 5B). Interestingly however, unlike Pa28αβ (Fig. 2A), There was no observed immediate increase in Pa28γ binding to 20S proteasome with H2O2 induced dissociation of 26S proteasome (Fig. 5C). Dissociation of 20S and 19S components of the 26S proteasome was again confirmed, but this time the S3 subunit of the 19S regulator was used (for independent verification) and it was found that less of it co-precipitated with the 20S β1 subunit after H2O2 treatment (Fig. 5C). It seems that addition of Pa28γ to 20S proteasome appears to cause a weak stimulation (60% increase) of the capacity of proteasome to degrade the peptide substrate Suc-LLVY-AMC (Fig. 5D). By comparison the addition of purified Pa28αβ to 20S proteasome appears to cause a 50-fold increase in the capacity of proteasome to degrade the peptide substrate Suc-LLVY-AMC (Fig. 2B).
Fig. 5.
(A) H2O2 exposure causes an increase in Pa28γ protein levels. MEF cells were pretreated with ± 1μM H2O2. Then, 24 h later samples were harvested and run on Western blots which were then screened with antibodies against either Pa200 or the loading control β-tubulin. (B) H2O2 exposure causes an increase in Pa28γ protein levels. MEF cells were prepared as in panel A in triplicate. Sample band intensity was then measured and adjusted based on β-tubulin band intensity. Values are means ± SE, where n = 3. (C) Following H2O2 treatment there is no change in Pa28γ association with 20S proteasome. MEF cells were exposed to 1mM H2O2 for 1 h, after which immunoprecipitation of anti-α5 was performed. Samples were run on Western blots and screened for co-immunoprecipitation of anti-Pa28γ, or anti-S3 (a subunit of the 19S regulator) as a measure of 26S proteasome disassembly. anti-α5 (not shown) was used as a loading control to confirm similar levels of 20S proteasome. The β1 blot was taken from a separate set of samples run under the same conditions. (D) Pa28γ slightly increases the capacity of 20S proteasome to degrade the peptide substrate Suc-LLVY-AMC. Purified 20S proteasome was incubated with a 4 fold molar excess of Pa28γ for 30 minutes. Suc-LLVY-AMC was then added to samples and proteolytic capacity measured. Values are means ± SE, where n = 3. (E) Pa28γ increases the capacity of 20S proteasome to degrade oxidized hemoglobin but not native hemoglobin. Purified 20S proteasome was incubated with a 4 fold molar excess of Pa28γ for 30 minutes. Hb-AMC or HbOX-AMC was then added to samples and proteolytic capacity measured. Values are means ± SE, where n = 8.
As with Pa28αβ and Pa200 it was examined whether Pa28γ might stimulate the selective degradation of oxidized proteins. In this assay no change in the capacity to degrade native hemoglobin was observed with the addition of Pa28γ to purified 20S proteasome. There was however, an ≈70% rise in the capacity to degrade oxidized hemoglobin with the addition of Pa28γ to 20S proteasome (Fig. 5E). From these results it appears that Pa28γ might play a similar, if weaker role, to Pa28αβ through facilitating the selective degradation of oxidized proteins under oxidative stress. Given that Pa28αβ is primarily located in the cytoplasm and Pa28γ in the nucleus [32] it is possible that both these regulators have the function of enhancing the removal of damaged proteins, in different cellular regions.
Regulation of the Immunoproteasome by Pa28αβ, Pa200, and Pa28γ
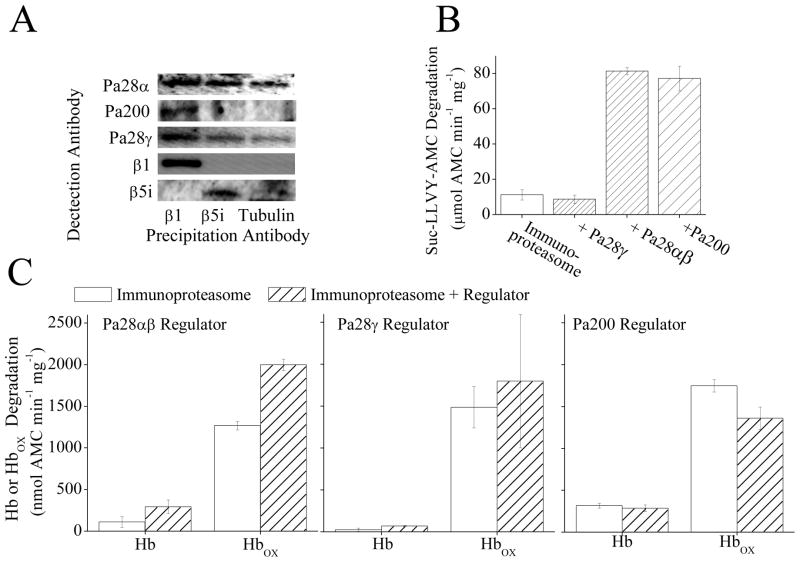
The immunoproteasome is another important protein involved in oxidative stress responses, whose expression is induced during oxidative stress adaptation [2, 23]. Immunoproteasome is at least as capable as the 20S proteasome of the selective degradation of oxidized proteins, and contributes significantly to the adaptive response [2, 23]. Both the immunoproteasome and Pa28αβ are induced by interferon-γ and both of them have been suggested to participate in similar functions in the cell [33]. Because of this co-association of Pa28αβ and immunoproteasome it has been speculated that they might act together. Immunoprecipitation of MEF cell lysate with an antibody against the 20S proteasome subunit β1 did cause co-precipitation of Pa28αβ, Pa200, and Pa28γ (showing that all three regulators do bind to 20S proteasome), however, immunoprecipitation with an antibody against the immunoproteasome subunit β5i caused rather weaker co-precipitation of Pa28αβ and Pa28γ but not of Pa200 (Fig. 6A). These results would suggest that the immunoproteasome can bind to (and be regulated by) Pa28αβ and Pa28γ, but that Pa200 does not share this capacity. To further test this conclusion purified immunoproteasome was incubated with purified samples of Pa28αβ, Pa28γ or Pa200. Addition of Pa28αβ or Pa200 to immunoproteasome enhanced the capacity to degrade the short peptide substrate Suc-LLVY-AMC by 5- to 7-fold, but Pa28γ actually decreased peptide degradation slightly (Fig. 6B). These results suggest that immunoproteasome can actually be controlled by all three regulators. Addition of purified Pa28αβ to immunoproteasome increased its capacity to degrade oxidized Hb (Fig. 6C, left panel) but, since degradation of native Hb also increased, the increase in selectivity for oxidized proteins was not as great as that observed (in Fig. 2C) with 20S proteasome. It must be said, however, that the actual specific activity of immunoproteasome plus Pa28αβ (Fig. 6C) was about twice that of 20S proteasome plus Pa28αβ (Fig. 2C). Addition of Pa28γ to immunoproteasome did not appear to cause a notable change in the capacity of immunoproteasome to degrade either oxidized or native hemoglobin (Fig. 6C, central panel). Addition of Pa200 to immunoproteasome also did not appear to have a strong effect on the capacity of immunoproteasome to degrade labeled hemoglobin proteins (Fig. 6C, right panel). This is in contrast to results with 20S proteasome (Fig. 4B,C) where Pa200 appeared to block the majority of the capacity of 20S proteasome to degrade either oxidized or native Hb. From the results of Fig. 6 it would appear clear that Pa28αβ positively regulates the selective degradation of oxidized proteins by immunoproteasome. Pa28γ appears to bind to immunoproteasome, but has little or no effect on its activity with peptide substrates or either normal or oxidized proteins. It should be noted however that there is no direct evidence that Pa200 actually binds to immunoproteasome, though it does increase peptidase activity yet slightly decrease the degradation of an oxidized Hb.
Fig. 6.
(A) Immunoprecipitation of MEF cell lysate with an antibody against the 20S proteasome subunit β1 causes co-precipitation of Pa28αβ, Pa200, and Pa28γ. However immunoprecipitation with an antibody against the immunoproteasome subunit β5i causes weak co-precipitation of Pa28αβ, and Pa28γ but not Pa200. Immunoprecipitation of MEF cells was performed using anti-β1, anti-β5i, or anti-β-tubulin (as a control for non-specific binding) antibodies. The Immunoprecipitate was analyzed by Western blotting and samples were screened for co-immunoprecipitation of anti-Pa28α, anti-Pa200, anti-Pa28γ, anti-β1, or anti-β5i. (B) Pa28αβ and Pa200 but not Pa28γ increase the capacity of immunoproteasome to degrade the peptide substrate Suc-LLVY-AMC. Immunoproteasome was incubated with a 4 fold molar excess of either Pa28γ Pa28αβ or Pa200 for 30 minutes. Suc-LLVY-AMC was then added to samples and proteolytic capacity measured. Values are means ± SE, where n = 3. (C) Binding of Pa28αβ (left panel) but not Pa28γ (center panel) or Pa200 (right panel) significantly increases the selective degradation of oxidized hemoglobin by immunoproteasome. Samples were prepared as in A, then AMC labeled hemoglobin or AMC labeled oxidized hemoglobin were added to samples as substrates. Values are means ± SE, where n = 8.
Multiple forms of Hybrid Proteasomes
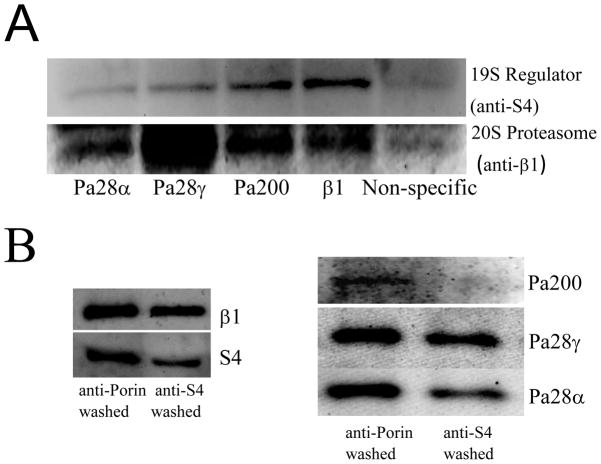
The majority of proteasome in mammalian cells (≈70–80%) is thought to be free 20S proteasome while some 15% of total proteasome is believed to be 26S proteasome [32, 34]. The 20S proteasome lacks the 19S regulatory cap that would enable it to degrade ubiquitinated proteins in an ATP-stimulated manner, but both 20S and immunoproteasome are very good at selectively degrading oxidized proteins, which 26S is not [1–5, 9–11, 35–37]. It has been shown that both of the regulators Pa28αβ, and Pa200 are capable of forming so called ‘hybrid’ proteasome [38, 39]. These hybrid proteasomes have been described as containing the 19S regulator on one end of the 20S proteasome core and a different regulator (either Pa28αβ or Pa200) on the other end (i.e. 19S-20S-regulator complexes). It is, however, unclear whether 20S proteasomes may form some sort of ‘hybrid’ proteasomes without involvement of 19S regulators. To test this co-immunoprecipitation was used to isolate either 20S proteasome or the regulators Pa28αβ, Pa28γ, or Pa200 from MEF cells (Fig. 7A). Western blotting was then used to determine what was bound to the isolated proteins (see also the results of Fig 6A). It was found that each of the regulator antibodies was capable of pulling down large quantities of 20S proteasomes (measured through anti-β1).
Fig. 7.
(A) Immunoprecipitation of MEF cell lysate with anti-Pa28α, anti-Pa28γ, anti-Pa200 or anti-β1 antibodies causes co-immunoprecipitation of both 20S and 26S proteasome subunits, however while anti-β1 and anti-Pa200 pull out a comparable amount of S4, anti-Pa28α and anti-Pa28γ antibodies are relatively poor at co- immunoprecipitation of S4. Immunoprecipitation protocols were as reported in the legends to Figs. 3 and 4. Non-specific binding was measured through immunoprecipitation of anti-porin. (B) Depletion of S4 through immunoprecipitation causes a small loss of Pa28α and Pa28γ but a very large loss of Pa200. MEF cell lysates were subjected to three cycles of immunoprecipitation with either anti-S4 or anti-porin. Non-immunoprecipitated supernatants were then taken and run on Western blots.
To look at the amount of interaction of ‘hybrid’ 26S proteasome we next screened the samples with an antibody against the S4 subunit specific to the 19S regulator of the 26S proteasome. All of the samples immunoprecipitated with Pa28αβ, Pa28γ, or Pa200 antibodies also had the 19S subunit bound, indicating that the regulators do indeed form some hybrid (19S-20S-regulator) proteasomes (Fig. 6A). The Pa200 sample had a comparable amount of S4 in it to the proteasome sample, indicating that a moderate proportion of Pa200 within the cell is hybrid. Both the Pa28α and Pa28γ samples, however, despite containing as much if not more 20S proteasome than the β1 (20S) proteasome samples, contained a very small amount of the 19S regulator subunit S4 (Fig. 7A). This suggested that a significant proportion of the proteasome to which Pa28αβ and Pa28γ are bound might actually be free 20S proteasomes. To test this hypothesis the reverse experiment was performed: Using multiple immunoprecipitation washes with the S4 antibody, ≈75% of the 19S regulator was removed from the sample (Fig. 7B, left panel). This resulted in a complete loss of Pa200 from the sample, but only a ≈50% reduction in Pa28α and Pa28γ (Fig. 7B, right panel). From the results of Figs. 6A, 6A, and 6B, it can conclude that the majority of Pa200 is bound to hybrid 26S proteasome (19S-20S-Pa200) while Pa28αβ and Pa28γ hybrid proteasomes have much greater flexibility, and may be found in 19S-20S-Pa28αβ/Pa28γ complexes, or Pa28αβ/Pa28γ-20S complexes or even, perhaps, Pa28αβ/Pa28γ-20S- Pa28αβ/Pa28γ complexes.
DISCUSION
Perhaps surprisingly, it appears that both the proteasome regulators, Pa28αβ, and Pa200 exhibit significantly increased levels in cells transiently adapted to oxidative stress (t-test, p≤0.05). With a weak increase also observed in Pa28γ under oxidative stress adaptation. In contrast, both in this report and previous reports there was no observed increase in 19S proteasome regulator levels (or ATP-stimulated proteolysis) under identical conditions [2]. Previous work [2, 23] has led to the hypothesis that the Pa28αβ and Pa28γ regulators might actually enhance the capacity of the 20S proteasome to degrade oxidized proteins. In this report it is now shown directly, that indeed, this hypothesis appears to be correct and the finding provides support to the hypothesis that Pa28αβ and Pa28γ may augment oxidative stress adaptive responses by enhancing the capacity of proteasome to selectively remove damaged proteins. Given also that these two regulator are orthologs and have very different cellular localizations [32], the Pa28αβ regulator is primarily localized in the cytoplasm while the Pa28γ regulator in the nucleus, it is likely that these two regulators might play similar roles in different cell compartments.
In further support of the potential role for Pa28αβ in response to acute oxidative stress, it appears that when cells are exposed to hydrogen peroxide there is not only an increase in expression of both Pa28αβ and Pa28γ over the subsequent 24 h but there is also an almost immediate increase in Pa28αβ binding to 20S proteasome that is evident within one hour of exposure to the oxidant. In previous work, using WI-38 cells, it was reported that there is a detachment of the 19S regulator from 26S proteasomes to form more free 20S proteasomes for a period of 1–3 hours following H2O2 exposure. This increase was argued to result in an increased capacity to degrade oxidized proteins [3]. Similar disassembly of 26S proteasomes following H2O2 treatment was also reported in yeast by Wang et al. [8] who have proposed Ecm29 as the protein that actually induces disassembly. While work from this group implicated HSP70 as the chaperone that preserves 19S regulators after dissociation, until 26S proteasomes are re-assembled within 5 hours of H2O2 exposure [3]. This paper now confirms oxidant-induced 26S disassembly in a different mammalian cell line, using two new measures: diminished (19S) subunits S3 and S4 binding to 20S proteasomes. The finding that many of these newly detached 20S complexes immediately bind to Pa28αβ regulators is exciting, given that, it also directly demonstrates the increased ability of 20S-Pa28αβ proteasome complexes to selectively degrade oxidatively damaged proteins. Interestingly, however, there was no corresponding immediate increase in the formation of 20S-Pa28γ (nuclear) complexes under oxidative stress. This may be because nuclear 20S proteasomes was bound to PARP, whose binding increases under oxidative stress[10], instead during this period, and Pa28γ may be more important later on in adaptive responses after it is synthesized de novo.
The immunoproteasome has long been believed to be linked with Pa28αβ due to induction of their co-expression by interferon-γ and their similar localization [2, 40, 41]. In this paper it is directly demonstrated, for the first time, that Pa28αβ is capable of binding to immunoproteasome and that this binding enhances the capacity of immunoproteasome to selectively degrade oxidized proteins. In contrast Pa28γ and Pa200 do not appear to modify immunoproteasome activity. Considering the extremely large induction of both immunoproteasome and Pa28αβ during transient adaptation to mild oxidative stress [2, 23], the direct demonstration that 20S proteasome-Pa28αβ complexes are much more capable than other proteasome forms of rapidly removing oxidized proteins is surely of physiological significance.
Pa200, like Pa28αβ and Pa28γ, is induced by hydrogen peroxide exposure. It does not, however, appear to possess the same ability as the Pa28αβ and Pa28γ regulators to enhance the capacity of 20S proteasome to selectively degrade oxidized proteins. In fact addition of Pa200 to 20S proteasome resulted in an almost complete loss of capacity to degrade both oxidized and native hemoglobin, although it did increase the degradation of native (but not oxidized) histones, and of the Suc-LLVY-AMC peptide. There is a moderate body of evidence that Pa200 has a role DNA repair [25, 39]. This paper confirmed previous reports that Pa200 will form punctuate foci in the nucleus under toxic H2O2 exposure, but now shows that this also occurs (albeit to a lesser extent) at low (μM) adaptive H2O2 concentrations. Pa200 punctuate nuclear foci are believed to be the product of Pa200 binding to the chromatin at sites of DNA damage [25, 39]. Interestingly Pa200 was found to increases 20S proteasome’s ability to degrade normal histones and the Suc-LLVY-AMC peptide, however it significantly decreases (t-test, p≤0.05) proteasome’s ability to degrade oxidized histones and both normal and oxidized Hb. Previous work has shown that Poly-ADP ribose polymerase (PARP) will, among its many functions, bind to 20S proteasome and activate its ability to selectively degrade oxidized histones [10]. In this paper we show that while PARP will bind to 20S proteasome it will not bind to 20S-Pa200 proteasome complexes. From these results it seems possible that both Pa200 and PARP regulate the 20S proteasome in similar but separate ways. One hypothesis to draw from these results is that PARP-proteasome complexes may induce the removal of damaged histones so that DNA may be repaired, while Pa200-proteasome complexes may catalyze the proteolytic removal of undamaged histones from damaged DNA such that the DNA may be unwound and then repaired.
Our studies reveal that the interactions between the 20S and 26S proteasome and its regulators are even more complex and intricate than was previously thought. We also observe a highly dynamic quality to regulator binding in which different regulators may be switched in and out of proteasome as a product of changing cellular conditions. We demonstrate that the three proteasome regulators: Pa28αβ, Pa28γ and Pa200 all appear to bind to 20S proteasome, with and without the addition of a 19S regulator, forming multiple types of hybrid proteasomes, although the Pa200 regulator appears to prefer 20S proteasome with a 19S regulator at the other end. We also show that immunoproteasome forms a hybrid complex with Pa28αβ. In summary we see that the proteasome regulators all appear to be important in response to oxidative stress but appear to posses very different roles. In particular, the 20Sproteasome- Pa28αβ complex appears to have much greater significance in successful adaptation to oxidative stress than was previously realized.
Highlights.
Pa28αβ & Pa28γ enhance selective degradation of oxidized proteins by 20S proteasome
Pa28αβ enhances selective degradation of oxidized proteins by the immunoproteasome
Oxidative-stress adaptation induces synthesis of Pa28αβ, Pa28γ, & Pa200 regulators
Pa200 and poly-ADP polymerase may cooperate in enabling initiation of DNA repair
Pa28αβ, Pa28γ & Pa200 regulators are all important for oxidative stress adaptation
Acknowledgments
This research was supported by grant #RO1-ES003598, and by American Recovery and Reinvestment Act (ARRA) Supplement 3RO1-ES 003598-22S2, both from the NIH/NIEHS to KJAD. Purified Pa200 [25] was the kind gift of Dr Martin Rechsteiner of the University of Utah, to whom we are also grateful for helpful suggestions.
Abbreviations
- MEF
murine embryonic fibroblasts
- H2O2
hydrogen peroxide
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- AMC
7-amino-4-methycoumarin (a fluorophore)
- Suc-LLVY-AMC
a succinylated peptide consisting of Leucine-Leucine-Valine-Tyrosine-AMC (used as a peptide substrate to measure proteolytic capacity)
- Hb
hemoglobin
- Hbox
oxidized hemoglobin
- PARP
poly-ADP ribose polymerase
- 19S
Pa28αβ (or 11S), Pa28γ and Pa200, various regulators of the 20S proteasome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davies KJA. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 2.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJA. Biochem J. 2010;432:585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grune T, Catalgol B, Licht A, Ermak G, Pickering AM, Ngo JK, Davies KJA. Free Radic Biol Med. 2011;51:1355–1364. doi: 10.1016/j.freeradbiomed.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies KJA. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 5.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJA, Grune T. Biochem J. 1998;335(Pt 3):637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Q, Reinacker K, Dimayuga E, Nukala V, Drake J, Butterfield DA, Dunn JC, Martin S, Bruce-Keller AJ, Keller JN. FEBS Lett. 2003;546:228–232. doi: 10.1016/s0014-5793(03)00582-9. [DOI] [PubMed] [Google Scholar]
- 7.Ding Q, Dimayuga E, Keller JN. Antioxid Redox Signal. 2006;8:163–172. doi: 10.1089/ars.2006.8.163. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Yen J, Kaiser P, Huang L. Sci Signal. 2010;3:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grune T, Reinheckel T, Davies KJA. J Biol Chem. 1996;271:15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- 10.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJA. Proc Natl Acad Sci U S A. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shringarpure R, Grune T, Mehlhase J, Davies KJA. J Biol Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 12.Raymond J, Segre D. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 13.Keller JN, Schmitt FA, Scheff SW, Ding Q, Chen Q, Butterfield DA, Markesbery WR. Neurology. 2005;64:1152–1156. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- 14.Chondrogianni N, Stratford FL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. J Biol Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- 15.Fucci L, Oliver CN, Coon MJ, Stadtman ER. Proc Natl Acad Sci U S A. 1983;80:1521–1525. doi: 10.1073/pnas.80.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang F, Taylor A. Biochem J. 1995;307(Pt 1):297–303. doi: 10.1042/bj3070297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whittier JE, Xiong Y, Rechsteiner MC, Squier TC. J Biol Chem. 2004;279:46135–46142. doi: 10.1074/jbc.M406048200. [DOI] [PubMed] [Google Scholar]
- 18.Ahn K, Erlander M, Leturcq D, Peterson PA, Fruh K, Yang Y. J Biol Chem. 1996;271:18237–18242. doi: 10.1074/jbc.271.30.18237. [DOI] [PubMed] [Google Scholar]
- 19.Hussong SA, Kapphahn RJ, Phillips SL, Maldonado M, Ferrington DA. J Neurochem. 2010;113:1481–1490. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotamraju S, Matalon S, Matsunaga T, Shang T, Hickman-Davis JM, Kalyanaraman B. Free Radic Biol Med. 2006;40:1034–1044. doi: 10.1016/j.freeradbiomed.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 21.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Kruger E. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 22.Wiese AG, Pacifici RE, Davies KJA. Arch Biochem Biophys. 1995;318:231–240. doi: 10.1006/abbi.1995.1225. [DOI] [PubMed] [Google Scholar]
- 23.Pickering AM, Linder RA, Zhang H, Forman HJ, Davies KJA. J Biol Chem. 2012. pp. 10021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Rechsteiner M. Biochimie. 2001;83:373–383. doi: 10.1016/s0300-9084(01)01236-6. [DOI] [PubMed] [Google Scholar]
- 25.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. Embo J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickering AM, Davies KJA. Free Radic Biol Med. 2012;52:239–246. doi: 10.1016/j.freeradbiomed.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies KJA, Goldberg AL. J Biol Chem. 1987;262:8227–8234. [PubMed] [Google Scholar]
- 28.Bota DA, Davies KJA. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 29.Ngo JK, Davies KJA. Free Radic Biol Med. 2009;46:1042–1048. doi: 10.1016/j.freeradbiomed.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo JK, Pomatto LC, Bota DA, Koop AL, Davies KJA. J Gerontol A Biol Sci Med Sci. 2011;66:1178–1185. doi: 10.1093/gerona/glr145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao I, Liu J, Li X, Luo H. Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks P, Fuertes G, Murray RZ, Bose S, Knecht E, Rechsteiner MC, Hendil KB, Tanaka K, Dyson J, Rivett J. Biochem J. 2000;346(Pt 1):155–161. [PMC free article] [PubMed] [Google Scholar]
- 33.Teoh CY, Davies KJA. Arch Biochem Biophys. 2004;423:88–96. doi: 10.1016/j.abb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Bose S, Brooks P, Mason GG, Rivett AJ. Biochem J. 2001;353:291–297. doi: 10.1042/0264-6021:3530291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies KJA. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 36.Grune T, Reinheckel T, Joshi M, Davies KJA. J Biol Chem. 1995;270:2344–2351. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- 37.Pacifici RE, Kono Y, Davies KJA. J Biol Chem. 1993;268:15405–15411. [PubMed] [Google Scholar]
- 38.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K. J Biol Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 39.Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N. Proc Natl Acad Sci U S A. 2008;105:16165–16170. doi: 10.1073/pnas.0803145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A, Ghazal P, Lee JD, Fourie AM, Wu Y, Pang J, Ngo K, Peterson PA, Fruh K, Yang Y. Science. 1999;286:2162–2165. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- 41.Rivett AJ, Bose S, Brooks P, Broadfoot KI. Biochimie. 2001;83:363–366. doi: 10.1016/s0300-9084(01)01249-4. [DOI] [PubMed] [Google Scholar]