Abstract
Protein misfolding due to missense mutations is a common pathogenic mechanism in cystathionine beta-synthase (CBS) deficiency. In our previous studies, we have successfully expressed, purified and characterized nine CBS mutant enzymes containing the following patient mutations: P49L, P78R, A114V, R125Q, E176K, R266K, P422L, I435T and S466L. These purified mutants exhibited full heme saturation, normal tetrameric assembly and high catalytic activity. In this work, we used several spectroscopic and proteolytic techniques to provide a more thorough insight into the conformation of these mutant enzymes. Far-UV circular dichroism, fluorescence and second derivative-UV spectroscopy revealed that the spatial arrangement of these CBS mutants is similar to the wild-type although the microenvironment of the chromophores may be slightly altered. Using proteolysis with thermolysin under native conditions, we found that the majority of the studied mutants is more susceptible towards cleavage suggesting their increased local flexibility or propensity to local unfolding. Interestingly, the presence of the CBS allosteric activator, S-adenosylmethionine (AdoMet), increased the cleavage rate of wild-type and the AdoMet-responsive mutants, while the proteolytic rate of the AdoMet-unresponsive mutants was not significantly changed. Pulse proteolysis analysis suggested that the protein structure of the R125Q and E176K mutants is significantly less stable than that of wild-type and the other mutants. Taken together, the proteolytic data show that the conformation of pathogenic mutants is altered despite retained catalytic activity and normal tetrameric assembly. This study demonstrates that the proteolytic techniques are a useful tool for the assessment of the biochemical penalty of missense mutations in CBS.
INTRODUCTION
Cystathionine beta-synthase (CBS) deficient homocystinuria (CBSDH; OMIM# 236200) is the most common inherited defect in sulfur amino acids metabolism characterized by severely elevated levels of plasma homocysteine, methionine and S-adenosyl-L-homocysteine (1). The worldwide frequency is estimated to be around 1:330,000 (1), thus classifying CBSDH as a rare disease. Interestingly, this number may be underestimated as several molecular epidemiological studies revealed the prevalence of homozygosity or compound heterozygosity for pathogenic mutations to be around 1:10,000 (2–5). If untreated, CBSDH manifests clinically with connective tissue symptoms such as dislocated lenses and skeletal abnormalities, mental retardation and vascular complications, particularly thromboembolic episodes (1). About half of CBS deficient patients respond to a treatment with pharmacological doses of pyridoxine (vitamin B6) with a significant lowering of plasma homocysteine levels and an alleviation of the clinical phenotype. The treatment of pyridoxine non-responsive patients involves a low methionine diet and supplementation with betaine which lowers homocysteine by promoting its remethylation to methionine via betaine:homocysteine S-methyltransferase.
Cystathionine beta-synthase (EC# 4.2.1.22) is a pyridoxal-5′-phosphate (PLP) dependent hemeprotein which catalyzes condensation of serine and homocysteine to form cystathionine as the first committed step in transsulfuration and subsequent biosynthesis of cysteine, glutathione and taurine. Human enzyme has a modular structure and a complex regulatory behavior (reviewed in (6,7)). More than 160 mutant alleles have been described in CBSDH patients so far (http://cbs.lf1.cuni.cz/cbsdata/cbsmain.htm) with missense mutations as the most common variants in the CBS gene accounting for 87% of analyzed patients alleles. Our previous study proposed that misfolding of CBS mutants may be responsible for pathogenicity in CBS deficiency as majority of tested CBS mutants formed large inactive aggregates devoid of heme (8). This proposed mechanism was later supported by several studies exploring the beneficial effect of chemical chaperones on the recovery, activity and assembly of various CBS mutant proteins (9–11). The presence of chemical chaperones such as ethanol or dimethylsulfoxide during expression in bacteria permitted the purification of several CBS mutants which exhibit normal heme saturation, native tetrameric assembly and the same or higher specific activities than the wild type CBS (12). Although the recovery of fully active CBS mutants suggests an improved folding, the final conformation of the purified mutants most likely differs from that of wild-type protein as the purified fully active CBS mutants varied in their response to exogenous addition of PLP cofactor as well as in their response to S-adenosyl-L-methionine (AdoMet) stimulation (12). In addition to studying spatial arrangement of proteins by spectroscopic monitoring of their conformation, proteolytic techniques such as proteolysis under native conditions and pulse proteolysis proved to be useful and effective to examine some aspects of protein structure (13,14); this approach uses thermolysin, an endoprotease that preferentially cleaves peptide bonds in regions containing hydrophobic amino acids. The rate of proteolysis with thermolysin under native conditions reveals the extent of unfolding of the studied proteins because this endoprotease can only cleave flexible regions and partially or globally unfolded structures (15). Further, pulse proteolysis is a technique that uses thermolysin to monitor urea-induced unfolding. After a short proteolytic pulse, the fraction of folded proteins remains intact whereas the locally and/or globally unfolded species are digested. Using this premise, the protein is analyzed in varying concentrations of urea and the cm value, a measure of protein thermodynamic stability in the urea gradient, may be determined (14). The recent study of several CBS mutants in crude extracts showed that the extent of protein unfolding inversely correlated with catalytic activity of the mutant enzymes.
In this study we applied several spectroscopic and proteolytic techniques to get an insight into the changes in CBS protein conformation induced by disease-causing missense mutations. We studied nine purified mutants P49L, P78R, A114V, R125Q, E176K, R266K, P422L, I435T and S466L. The purification and biochemical characterization of these mutants was reported elsewhere (12,17); the determination of their conformational properties adds additional new information thus permitting an understanding of pathogenicity of missense mutations in CBS.
EXPERIMENTAL PROCEDURES
Purification of CBS proteins
The wild-type and mutant CBS enzymes except for the R266K CBS were expressed as fusion proteins with the N-terminal GST in E. coli Rosetta2 (DE3) cells and purified essentially as described previously (12). The R266K mutant enzyme was expressed in E. coli Rosetta2 (DE3) cells with a C-terminal 6xHis tag and isolated following the previously reported procedure (17).
Circular dichroism (CD) and fluorescence spectroscopy
The CD spectra of CBS proteins (0.5 mg/ml; 50 mM phosphate buffer, pH 7.5) were recorded by chiroptic spectrometer Jasco J-810. The intrinsic fluorescence of CBS proteins was measured in the same buffer using Perkin Elmer LS55 fluorescence spectrometer. The excitation wavelength was 295 nm (slit width 5 nm) for tryptophans and 420 nm (slit width 10 nm) for internal aldimines with an emission signal scanned from 300 to 450 nm (slit width 5 nm) and from 430 to 700 nm (slit width 10 nm), respectively.
Second-derivative UV spectroscopy
The UV spectra were recorded between 10 and 90°C with 2.5°C increments and a 3 min equilibrium time at each temperature. Measurements were made on an Agilent diode array model 8453 UV-visible spectrophotometer equipped with a Peltier temperature controller. Two ml of protein samples (0.2 mg/ml) were prepared by diluting the stock protein with the appropriate amount of buffer (20 mM HEPES pH 7.4, 1 mM Tris-(2-carboxyethyl)-phosphine, 0.01% Tween 20) and then placed in a quartz cuvette with a 1 cm path length. A micro stir bar (100 rpm) was put into the cuvette improve the heat-exchange in the sample. The second derivative UV spectra at each temperature were used to compare the changes of tertiary structure of proteins. The peak positions for aromatic amino acids tryptophan, tyrosine and phenylalanine were plotted as a function of temperature.
Proteolysis with thermolysin under native conditions and pulse proteolysis
Proteolytic techniques were performed and evaluated according to the previously published procedures (18). To carry out the proteolytic experiments in the presence of AdoMet, the CBS proteins (0.5 mg/ml) were pre-incubated with 300 μM AdoMet at room temperature for 10 min. Each experiment was repeated at least twice.
The rate of proteolysis under native conditions was expressed as kp, the constant in a single exponential equation.
Pulse proteolysis revealed the cm value, i.e. concentration of urea at which fraction of folded proteins comprises 50% of the entire protein population. To assess accuracy of the determined cm values, we observed proteolytic cleavage of the CBS proteins in 2M urea solution. The kinetic constant for proteolysis of all proteins with determined cm was lower than 0.2 min−1 indicating negligible cleavage of the protein during the proteolytic pulse at the beginning of the transition zone and thus the absence of a systematic error (19).
Protein Structure Modeling
Model of the full-length human CBS was built by homology modeling package Modeller 9v4 (Discovery Studio v2.5, Accelrys) using the structure of Drosophila melanogaster CBS (PDB 3PC2) as a template. The input sequence alignment was constructed by Align123 package (Discovery Studio v2.5, Accelrys). Among the resulting five models, the model with the lowest probability density function (PDF) total energy was selected. Since D. melanogaster CBS is highly active and does not respond to AdoMet stimulation (31), the presented model most likely corresponds to an activated conformation of human CBS.
In-gel digestion and mass spectrometric analysis
To identify fragments observed during native proteolysis, we excised the bands of interest and performed in-gel digestion according to previously described procedure (20). Mass spectra were acquired using MALDI-TOF MS (Autoflex II, Bruker Daltonics) and processed as published previously (18).
RESULTS
Mutants analyzed in this study
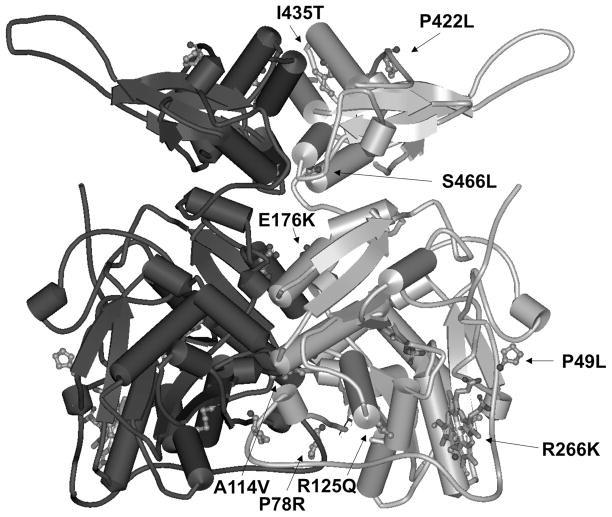
All studied mutants were purified into homogeneity. They retained catalytic activity and their saturation with heme and tetrameric assembly were similar to wild-type CBS as previously reported (12,17). All of the studied CBS mutants, except R266K and I435T, were expressed in the presence of a particular chemical chaperone included in the growth medium as described previously (12). Parallel expression of the selected mutants (R125Q, E176K, P422L and S466L) in the absence or the presence of chemical chaperone in the medium and their subsequent purification did not show significant differences between “chaperoned” CBS mutants and their “non-chaperoned” counterparts in terms of catalytic activity, AdoMet response, oligomeric status or heme saturation (data not shown). The only detectable difference was in the increased yield and recovery of the purified enzymes (data not shown). The studied mutations are located a) in the proximity of the heme-binding pocket (P49L, R125Q and R266K), b) at the dimer interface (P78R, A114V and E176K), and c) in the regulatory domain (P422L, I435T and S466L) (Fig. 1).
Figure 1.
Model of hCBS. Model of the full-length human CBS based on the crystal structure of the dimeric Drosophila melanogaster CBS (PDB: 3PC2). The CBS cofactors (heme and PLP) are displayed as sticks. The arrows are pointing to the mutated residues (displayed as scaled balls and sticks) in one of the subunit of WT CBS. The analyzed pathogenic mutations are located in the proximity of the heme-binding pocket (P49L, R125Q and R266K), at the dimer interface (P78R, A114V and E176K) and in the regulatory AdoMet-binding domain (P422L, I435T and S466L).
Far-UV CD spectroscopy
To assess the impact of mutations on the formation of the secondary structure of the protein, we analyzed CBS mutants by far UV-CD spectroscopy. Far-UV CD spectra (Fig. S1 in the Supporting Information) showed maxima at ~208 and ~222 nm indicating a large content of helical structures. No noticeable differences in the content of secondary structure were observed among the studied CBS mutants compared to wild-type enzyme with the single exception of the E176K mutant. This mutant exhibited a different shape of the CD spectrum indicating a partial decrease in proportion of helical structures together with formation of a different type of secondary structure; nevertheless, these changes are subtle and the overall helical content is retained. Far-UV CD spectra obtained in the presence of AdoMet did not show any change in the overall secondary structure compared to the CD spectra in the absence of AdoMet (data not shown).
These observations are consistent with the structural topology of the studied mutations. Only the R125Q and E176K mutations could affect hydrogen bonding with polar residues that form the adjacent helices; the available structural data show that the R125 residue forms hydrogen bond with E234 and the E176 interacts with hydroxyl moiety of T383 (21,22). Our data demonstrate that the E176K mutation, causing the significant changes in charge and sterical disproportions, leads to a partial destabilization of the helical structures in the CBS enzyme and, in contrast, the R125Q does not affect substantially the helical content of the CBS protein. Alternatively, the presence of either pathogenic mutation might lead to a local rearrangement/fluctuations resulting in the loss of stability as detected in our previous work (12). Taken together, the CD results suggest that the majority of CBS mutants possessed unaltered secondary structure compared to that of wild-type enzyme and the presence of AdoMet does not alter the secondary structure in CBS enzymes.
Fluorescence spectroscopy
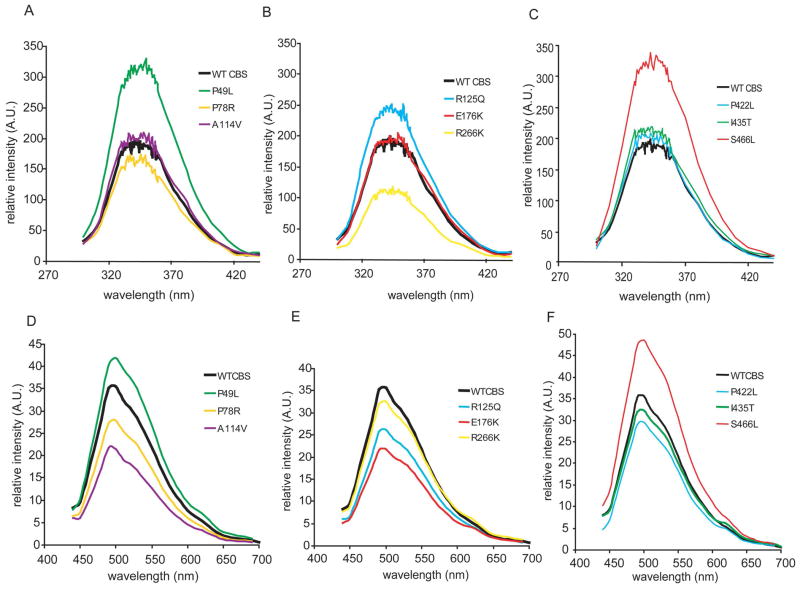
Using fluorescence spectroscopy, we studied conformation of the CBS mutants by analyzing the microenvironment of the tryptophans. Tryptrophan fluorescence spectra for all CBS enzymes exhibited broad maxima resulting from the fluorescence of eight tryptophan residues in each CBS subunit (Fig. 2). These spectra did not show any major differences in the position of emission maxima indicating an unaltered polarity in the tryptophan environment. However, significant changes were observed for relative intensities of the emission, namely, increased intensity for P49L, R125Q and S466L and, on the contrary, decreased intensity for R266K in comparison to the wild-type. The altered intensity of emission may be caused by a different microenvironment of tryptophans. The altered relative orientation of one or more tryptophans towards possible quenching groups, such as polar amino acid residues, and/or mutual position of tryptophan and tyrosine residues, affecting resonance energy transfer from hydroxyphenyl to indolyl groups, could have caused the intensity changes observed (20).
Figure 2.
Fluorescence spectroscopy of CBS proteins. A–C Emission spectra of tryptophan residues (excitation at λ = 295 nm); D–F Emission spectra of internal aldimines (excitation at λ = 420 nm).
Fluorescence spectroscopy was further used for analyses of PLP that is covalently bound in the active site of CBS in the form of aldimine. These measurements revealed that the majority of the mutants exhibited decreased intensity of emission (Fig. 2). These changes may be caused by increased extent of quenching by surrounding amino acid residues or by the heme moiety (21). Alternatively, lower intensity of fluorescence may be attributed to a decreased saturation of CBS proteins with PLP that was previously observed for R125Q, E176K and partially also for the C-terminal mutants P422L, I435T and S466L (12). In summary, both fluorescence-based measurements showed that the majority of the studied mutants are similar to wild-type CBS with minor differences in the microenvironment of the fluorophores.
Second-derivative UV spectroscopy
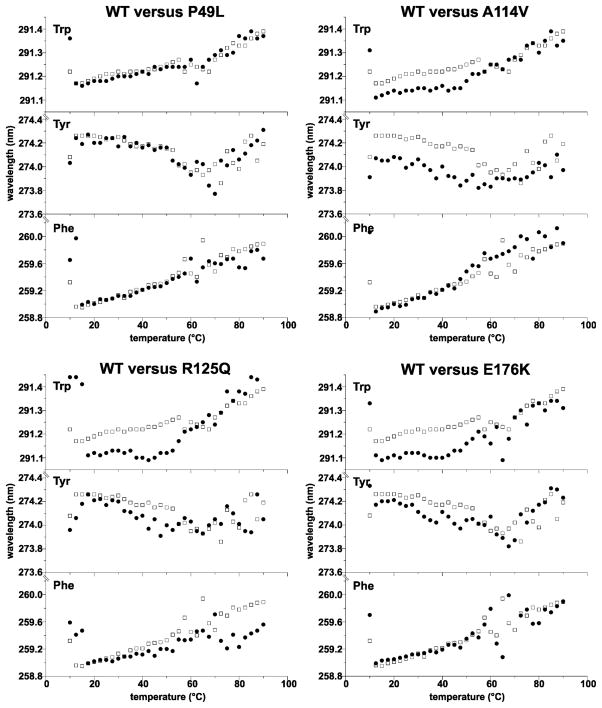
To determine possible changes in the microenvironment of chromophores, we also performed a second-derivative UV spectroscopy that may reveal information about the microenvironments of tryptophan, tyrosine and phenylalanine residues (22). Positions of the maxima were unaltered for the majority of the studied mutants (see P49L as an example for the unaltered mutant CBS spectrum in Fig. 3). A decrease in the wavelength of the maximum for tryptophan and tyrosine were recorded for A114V, R125Q and E176K (Fig. 3; spectral traces are shown at Fig. S2 in the Supporting Information) suggesting possible differences in the microenvironment of these chromophores. It should be noted that these changes were subtle, typically around 0.1 nm. Nevertheless, the altered positions of maxima were consistently observed for all temperatures below the melting point, i.e. from 10 °C to 55 °C (Fig. 3). These data further suggest that some of the studied mutants may differ from the wild-type in the local spatial arrangement of the CBS protein, such as A114V, R125Q or E176K, and/or in the mutual position of the catalytic core and the C-terminal AdoMet-binding regulatory domain, such as P422L or S466L.
Figure 3.
Second-derivative UV spectroscopy: peak position for aromatic amino acids (Phe, Tyr and Trp) as a function of temperature. The exact peaks for a particular aromatic amino acid were determined by using 2nd derivative UV spectra recorded every 2.5°C for temperatures from 10°C to 90°C (see Supplementary Information for raw spectra) and subsequently plotted as a function of temperature. The wild type enzyme (WT, open squares) is separately compared with P49L, A114V, R125Q and E176K CBS mutants (filled circles). The P49L represent the CBS mutants that were similar to wild-type enzyme. On the contrary, A114V, R125Q and E176K exhibited blue shifts for tyrosines and tryptophans.
Proteolysis with thermolysin under native conditions
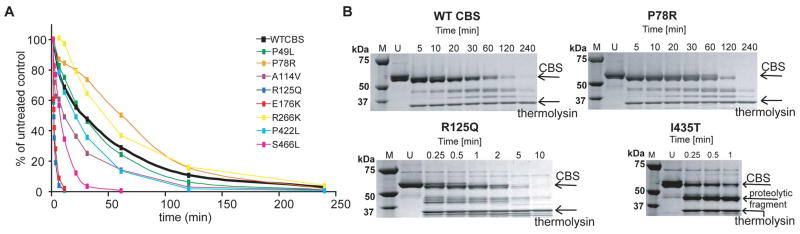
To determine the structural flexibility and tendency towards unfolding of the CBS mutants, we assessed their proteolytic susceptibility towards thermolysin under native conditions.
Three mutants, namely P49L, P78R and R266K, were resistant towards proteolysis to an extent similar or even higher than the wild-type. The remaining six CBS mutants showed higher susceptibility to proteolysis and extremely rapid cleavage was observed for R125Q and E176K (Table 1, Fig. 4 and Fig. S3 in the Supporting Information). In case of the mutant I435T we were not able to determine proteolytic kinetics since this mutant was almost instantly, within several seconds, cleaved to a fragment having molecular weight of ≈ 40 kDa (Fig. 4B). This proteolytic fragment corresponds to the catalytic core of CBS as was determined by in-gel digestion followed by MALDI-TOF mass spectrometry (Tab. S1 and Fig. S4 in the Supporting Information). These data show that the I435T mutant is cleaved predominantly in the connecting loop between the catalytic core and the C-terminal regulatory domain. Thus one of the requirements for this technique, i.e. the cleavage without formation of major fragments, has not been met.
Table 1.
CBS specific activities in the absence and presence of AdoMet and rate constants (kp) of proteolytic kinetics under native conditions.
| Protein | CBS specific activity (U/mg of protein)*
|
kp (min−1) | kp in presence of AdoMet (min−1) | Adomet +/AdoMet − ratio of kp | |
|---|---|---|---|---|---|
| − AdoMet | + AdoMet | ||||
|
| |||||
| wtCBS | 148 ± 21 | 530 ± 45 | 0.026 ± 0.005 | 0.056 ± 0.005 | 2.15 |
| P49L | 122 ± 12 | 388 ± 39 | 0.025 ± 0.001 | 0.057 ± 0.005 | 2.28 |
| P78R | 110 ± 15 | 501 ± 27 | 0.0097 ± 0.001 | 0.031 ± 0.002 | 3.20 |
| A114V | 100 ± 11 | 401 ± 31 | 0.054 ± 0.011 | 0.5 ± 0.08 | 9.26 |
| R125Q | 139 ± 18 | 106 ± 9 | 0.87 ± 0.18 | 1.105 ± 0.008 | 1.27 |
| E176K | 138 ± 14 | 111 ± 8 | 0.97 ± 0.18 | 1.19 ± 0.18 | 1.22 |
| R266K | 191 ± 17 | 386 ± 26 | 0.014 ± 0.001 | 0.041 ± 0.02 | 2.93 |
| P422L | 226 ± 28 | 238 ± 41 | 0.036 ± 0.003 | 0.041 ± 0.001 | 1.14 |
| I435T | 564 ± 44 | 582 ± 60 | N.D. | N.D. | N.D. |
| S466L | 626 ± 48 | 639 ± 43 | 0.109 ± 0.008 | 0.063 ± 0.007 | 0.58 |
Figure 4.
Proteolysis of CBS mutants with thermolysin under native conditions. A. Comparison of wild- type CBS with mutant proteins. Each point represents a mean from at least two independent experiments. B. Representative gels depicting proteolytic cleavage of selected mutants. P78R represents the proteolytically resistant mutants, while R125Q belongs to the more rapidly cleaved proteins. The mutant I435T is rapidly cleaved with the formation of the major fragment of molecular weight ≈ 40 kDa. “M“ refers to molecular weight marker, “U“ refers to untreated control without added thermolysin.
We also studied the effect of AdoMet on the proteolytic kinetics (Tab. 1). We have demonstrated previously that allosteric activation of CBS is associated with opening of the protein conformation and that the wild-type CBS is cleaved in the presence of AdoMet more rapidly with doubling of the kinetic constant (18). The similar increase in cleavage rate was also observed in this study for all AdoMet-responsive CBS mutants, namely the P49L, P78R, A114V and R266K. In case of the A114V mutant, the presence of the AdoMet in the proteolytic assay promoted an extensive opening of conformation accompanied with a dramatic 9-fold increase in the rate of cleavage compared to the rate in the absence of the ligand. In contrast, virtually no change in the cleavage rate in the presence of AdoMet was observed for the mutants with the highest cleavage rates (R125Q and E176K) and for the C-terminal mutants (P422L and S466L). Interestingly, the same mutants did not show any response to AdoMet allosteric stimulation of catalytic activity in our previous study (12).
Data from proteolysis with thermolysin under native conditions suggest that the majority of the studied CBS mutants has higher structural flexibility and is more susceptible to proteolytic cleavage than the wild-type enzyme despite only subtle changes in protein conformation observed by spectroscopic techniques. Additionally, increased proteolytic rates of the wild-type and AdoMet-responsive mutant enzymes in the presence of AdoMet are indicative of conformational rearrangement upon AdoMet binding. On the contrary, the cleavage rates of AdoMet-unresponsive mutants were not altered suggesting their inability to change conformation upon AdoMet binding and thus explaining their inability to be allosterically stimulated by AdoMet.
Pulse proteolysis
Using pulse proteolysis, we have complemented the proteolytic data under native conditions by determining the resistance of wild-type and mutant CBS proteins against urea-induced denaturation. Interestingly, the majority of the mutants were found to be more resistant towards urea-induced denaturation than the wild-type (Tab. 2; a representative gel with corresponding curve of the P49L mutant is shown at Fig. 3 in the Supporting Information). On the other hand, mutants R125Q and E176K - sensitive to rapid proteolytic cleavage - exhibited abnormal behavior in pulse proteolysis which resulted in the non-sigmoidal curves of the urea-induced unfolding (Fig. S4 in the Supplementary Information); this finding indicates very low cooperativity of the proteins tertiary structure. Furthermore, parallel pulse proteolysis of R125Q and E176K mutant proteins expressed in the absence or the presence of chemical chaperone showed essentially the same profiles. This result further supports our previous notion that a chemical chaperone present during mutant CBS expression does not alleviate the impact of the pathogenic mutation on the CBS protein, but rather results in enrichment of the mutant protein in the crude extract thus increasing the total yield and total recovery during the purification.
The presence of AdoMet increased the stability of wild-type CBS as reported previously (18,23) whereas the cm values for the majority of mutants were not changed (Tab. 2). Taken together, the stability of the CBS mutants in the absence of AdoMet did not change after addition of AdoMet and was comparable to the stability of wild-type CBS incubated with AdoMet. Different behavior was observed only for the R266K mutant which, unlike the remaining wild-type and mutant CBS enzymes, exhibited a significantly lowered protein stability upon AdoMet binding with concomitantly decreased cm value.
DISCUSSION
The cause of pathogenicity of the eight disease-causing CBS mutant enzymes that have normal catalytic activity was not apparent from our initial study (12). Purification and initial characterization of the CBS mutants expressed in the presence of various chemical chaperones showed that the inclusion of a chemical chaperone induced a specific increase of the DnaJ molecular chaperone (12). The likely involvement of the DnaJ/DnaK/GrpE machinery resulted in an enhanced folding efficiency of the mutants thus shifting the equilibrium between the native active and misfolded inactive forms in favor of the folded fraction. However, a parallel expression of the selected mutants in the absence and presence of a chemical chaperone resulted in lower yields and lower total recovery of CBS from “non-chaperoned” samples compared to “chaperoned” ones, supporting the notion that the presence of a chemical chaperone during the mutant CBS expression mainly affects the equilibrium between folded and misfolded forms (data not shown). Additionally, our previous results suggest that the properly folded active conformations of these CBS mutants were enriched in the crude extracts (12). Furthermore, our analyses of several CBS mutants in bacterial lysates (P49L, P78R, A114V and R125Q) revealed that their expression in the presence of various chemical chaperones did not significantly alter their proteolytic susceptibility (data not shown).
In this study, we compared conformational properties of the nine purified mutant CBS enzymes with the wild-type enzyme using far-UV CD, fluorescence and second-derivative UV spectroscopy. In the next step, we determined their structural flexibility by proteolysis with thermolysin under native conditions as well as their sensitivity towards urea-induced denaturation by pulse proteolysis. Only subtle conformational differences between the wild-type and the mutants studied were detected by the spectroscopic methods used. These findings are consistent with our previous work showing that the studied mutants did not exhibit dramatic abnormalities in the specific activities, heme saturation or native tetramer formation (12).
The major differences in the properties of CBS mutants were observed when tested for their proteolytic susceptibility under native conditions. These data suggest that the mutant proteins adopted conformation, which differs from the wild-type CBS, being more flexible and exposing more hydrophobic residues for the thermolysin to attack. Moreover, the less compact structure of the mutants A114V, R125Q and E176K can also be assumed from their spectra of the second-derivative UV spectroscopy which revealed subtle decrease in the wavelength for the maxima assigned for tryptophan and tyrosine residues; similar blue shift was observed in unfolding of several model proteins (24).
Interestingly, the increased structural mobility of the mutants is accompanied by impaired protein stability in urea solution only for the extensively flexible CBS mutants, R125Q and E176K, while the other CBS mutants exhibit unaffected global protein stability as was demonstrated by pulse proteolysis. The same or even higher resistance against urea-induced unfolding compared to the wild-type is also consistent with the thermostability of these mutants previously determined by absorption spectrophotometry (12). Analogous increased proteolytic susceptibility associated with subtle conformational changes and with unaltered thermodynamic stability was reported for the yeast phosphoglycerate kinase compared to its ortholog from E. coli (15). It was proposed that the discrepancies in the phosphoglycerate kinase orthologs were caused by a divergent interdomain cooperativity and consequently different mechanism of unfolding in these modular proteins. It is tempting to speculate that the increased structural mobility of majority of the CBS mutants is not caused by their low thermodynamic stability but more likely by the lower kinetic barrier of the protein unfolding; the altered interdomain communication, despite only minor conformational changes in each particular domain, may be responsible for the increased unfolding rates of these mutants.
The findings of increased proteolytic susceptibility of CBS mutants towards thermolysin are consistent with our previous study conducted directly in bacterial lysates for a different set of CBS mutants (16). In the previous study (16), it was proposed that higher proteolytic susceptibility of the misfolded CBS mutants in vitro may mirror their accelerated turnover in vivo indicating possibly an important role of proteolysis in the pathophysiology of CBS deficiency (10,28). However, higher proteolytic susceptibility of all studied mutants in the previous study was associated with their increased sensitivity toward urea-induced denaturation. This discrepancy may be due to a different panel of studied CBS mutations and different degree of purity of CBS proteins in each study. In the present study, we analyzed catalytically active CBS mutants that were successfully purified to homogeneity after expression in E. coli whereas the previous work (16) was carried out mainly on mutants exhibiting decreased levels of catalytic activity that were not amenable to purification due to an excessive aggregation. It indicates that decreased global protein stability may be observed only for the severely affected CBS mutants but not for the mutants exhibiting subtle conformational changes. This notion is also supported by the present study showing that only the R125Q and E176K CBS mutants exhibited impaired protein stability in a urea gradient. Our present study indicates that even subtle changes in protein conformation of the catalytically active CBS mutants with normal structural stability in vitro may lead to more rapid degradation of these variants in vivo.
Since the altered response to AdoMet was proposed as one of the possible pathogenic mechanisms in CBS deficiency, particularly for the C-terminal missense CBS mutations, we compared the kinetics of proteolytic cleavage of wild-type and mutant CBS enzymes in the presence of this allosteric activator. The most rapidly cleaved CBS mutants, namely R125Q and E176K, exhibited unaltered kp in the presence of AdoMet compared to that obtained in the absence of AdoMet. This indicates that these mutants cannot bind AdoMet and/or are locked in a specific conformation that prevents allosteric change upon AdoMet binding. This suggestion is also supported by the previously reported lack of stimulation of the catalytic activity by AdoMet and by heating of mutants (12). More surprisingly, extremely increased proteolytic susceptibility of the A114V mutant in the presence of AdoMet suggests that its allosteric activation is likely associated with an extensive opening of the folded structure exposing naturally buried hydrophobic residues on the protein surface. Interestingly, the effect of AdoMet on native proteolysis of the R266K mutant was similar to wild type CBS but different behavior of this mutant was observed using pulse proteolysis. The presence of AdoMet led to a lower cm value indicating that this CBS ligand decreases thermodynamic stability of the R266K mutant. This result correlates well with the previously observed decreased thermal stability and AdoMet activation of this mutant compared to wild-type CBS (17). The impaired response to AdoMet activation was also observed for the C-terminal mutants. The proteolytic cleavage of the P422L mutant was not significantly increased and, moreover, the S466L mutant was cleaved even less rapidly in the presence of AdoMet. These findings further support the previous notion that the C-terminal mutants are locked in a specific conformation, which results in a permanently activated mutant CBS enzymes lacking proper response to AdoMet stimulation (25,26). Interestingly, our results indicate that these mutant proteins may be more flexible in the absence of AdoMet than the wild-type. Even though the S466L mutant does not respond to AdoMet, it is still capable of binding it as reported previously (25). As CBS domains in the C-terminal region very likely fold independently of the catalytic core (27), these mutants may still bind AdoMet, but are apparently unable to rearrange their conformation. It is tempting to speculate that this locked conformation is in vivo recognized as a misfolded structure by the cellular control machinery and are consequently targeted for degradation (28).
It should be noted that the mutants P49L and P78R did not exhibit structural abnormalities by the approaches used in this study, which correlates well with their biochemical properties very similar to that of wild type enzyme (12). The P49L exhibited high catalytic activity when expressed in the pKK expression vector without any additional tags or fusion partners (29). On the contrary, the P78R possessed a decreased enzyme activity in the same study using the pKK construct which is consistent with a study analyzing the purified mutant reported by the Banerjee group (30). Data on P78R indicate that pathogenicity of these mutants may be revealed by employing different expression system or using specific conditions. Nevertheless, the P49L mutation is often associated with very mild clinical manifestions (CBS Mutation Database; http://cbs.lf1.cuni.cz/cbsdata/cbsmain.htm and Sally Stabler, oral communication at the 8th Conference on Homocysteine Metabolism, Lisboa 2011).
It should be noted that the application of a bacterial expression systems that produce the target protein with an affinity tag may have an artificial effect on quality of the purified proteins. For instance, the GST-tag that was used for the majority of the mutants may increase protein stability during the expression and subsequent purification (31). The dramatic effect of the type and position of the employed purification affinity tag (i.e. bulky fusion partner, such as GST versus short flexible tag, such as 6xHis) and its position on the proper folding was recently demonstrated in the expression studies of the R266K mutant (17). Nevertheless, using of affinity tags is necessary for production of target proteins in sufficient yields for their conformational analysis.
In this study, we demonstrated that protein structures of the studied CBS mutants are more locally flexible than that of the wild-type despite their normal catalytic activity and unaffected sensitivity towards urea-induced denaturation. In conclusion, the conformational analysis of the mutants using spectroscopic and proteolytic approach proved to be a useful tool for the assessment of the biochemical penalty of the CBS mutations.
Supplementary Material
Table 2.
The midpoint values of urea-induced unfolding (cm) for CBS mutants in the absence and in the presence of AdoMet.
| Protein | cm in the absence of AdoMet (M) | cm in the presence of AdoMet (M) |
|---|---|---|
|
| ||
| wtCBS | 2.7 ± 0.08 | 3.26 ± 0.08 |
| P49L | 3.31 ± 0.13 | 3.40 ± 0.15 |
| P78R | 3.39 ± 0.06 | 3.42 ± 0.05 |
| A114V | 2.94 ± 0.20 | 3.46 ± 0.13 |
| R125Q | N.D. | N.D. |
| E176K | N.D. | N.D. |
| R266K | 3.08 ± 0.10 | 2.62 ± 0.11 |
| P422L | 3.85 ± 0.12 | 4.00 ± 0.09 |
| I435T | N.A. | N.A. |
| S466L | 3.39 ± 0.18 | 3.18 ± 0.12 |
The values were determined using pulse proteolysis followed by nonlinear data fitting into a sigmoidal equation. N.D. - not determined due to non-sigmoidal behavior of the mutant proteins (see Fig. S3 in the Supplementary Information). N.A. - not applicable; pulse proteolysis could not have been used due to a rapid cleavage in the absence of urea
Acknowledgments
FUNDING SOURCE STATEMENT: This work was supported by the research programs of the Charles University in Prague PRVOUK-P24/LF1/3, UNCE 204011, SVV 264502 and by the grant No. 7310 from the Grant Agency of the Charles University in Prague (to AH and VK), by Postdoctoral Fellowship 0920079G from the American Heart Association (to TM), by National Institutes of Health Grant HL065217, by American Heart Association Grant In-Aid 09GRNT2110159 and by a grant from the Jerome Lejeune Foundation (to JPK), by Ramon y Cajal research contract RYC-2009-04147 from the Spanish Ministry of Science and Innovation (to ALP) and grant P11-CTS-7187 from the Junta de Andalucia (to ALP).
We thank to Ms. Kateřina Raková and to Jitka Honzíková, MSc. for technical assistance and to Dr. Jose M. Sanchez-Ruiz for useful comments on the manuscript.
ABBREVIATIONS
- AdoMet
S-adenosylmethionine
- CBS
cystathionine beta-synthase
- CBSDH
cystathionine beta-synthase deficient homocystinuria
- CD
circular dichroism
- cm
midpoint of urea-induced unfolding determined by pulse proteolysis
- kp
rate constant of proteolysis under native conditions
- PLP
pyridoxal-5′-phosphate
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional data can be found in the Supporting Information including detailed results from CD and second derivative UV spectroscopy, and pulse proteolysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mudd SH, Levy HL, Kraus JP. Disorders of transsulfuration. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler K, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. McGraw-Hill; New York: 2001. [Google Scholar]
- 2.Gaustadnes M, Ingerslev J, Rutiger N. Prevalence of congenital homocystinuria in Denmark. N Engl J Med. 1999;340:1513. doi: 10.1056/NEJM199905133401915. [DOI] [PubMed] [Google Scholar]
- 3.Refsum H, Fredriksen A, Meyer K, Ueland PM, Kase BF. Birth prevalence of homocystinuria. J Pediatr. 2004;144:830–832. doi: 10.1016/j.jpeds.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Janosik M, Sokolova J, Janosikova B, Krijt J, Klatovska V, Kozich V. Birth prevalence of homocystinuria in Central Europe: frequency and pathogenicity of mutation c. 1105C>T (p R369C) in the cystathionine beta-synthase gene. J Pediatr. 2009;154:431–437. doi: 10.1016/j.jpeds.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linnebank M, Homberger A, Kraus JP, Harms E, Kozich V, Koch HG. Haplotyping of wild type and I278T alleles of the human cystathionine beta-synthase gene based on a cluster of novel SNPs in IVS12. Hum Mutat. 2001;17:350–351. doi: 10.1002/humu.36. [DOI] [PubMed] [Google Scholar]
- 6.Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004;279:29871–29874. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- 7.Singh S, Madzelan P, Banerjee R. Properties of an unusual heme cofactor in PLP-dependent cystathionine beta-synthase. Nat Prod Rep. 2007;24:631–639. doi: 10.1039/b604182p. [DOI] [PubMed] [Google Scholar]
- 8.Janosik M, Oliveriusova J, Janosikova B, Sokolova J, Kraus E, Kraus JP, Kozich V. Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet. 2001;68:1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh LR, Chen X, Kozich V, Kruger WD. Chemical chaperone rescue of mutant human cystathionine beta-synthase. Mol Genet Metab. 2007;91:335–342. doi: 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh LR, Kruger WD. Functional rescue of mutant human cystathionine beta-synthase by manipulation of Hsp26 and Hsp70 levels in Saccharomyces cerevisiae. J Biol Chem. 2009;284:4238–4245. doi: 10.1074/jbc.M806387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopecka J, Krijt J, Rakova K, Kozich V. Restoring assembly and activity of cystathionine beta-synthase mutants by ligands and chemical chaperones. J Inherit Metab Dis. 2011;34:39–48. doi: 10.1007/s10545-010-9087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majtan T, Liu L, Carpenter JF, Kraus JP. Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: purification and characterization of eight CBS mutant enzymes. J Biol Chem. 2010;285:15866–15873. doi: 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park C, Marqusee S. Probing the high energy states in proteins by proteolysis. J Mol Biol. 2004;343:1467–1476. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- 14.Park C, Marqusee S. Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 15.Young TA, Skordalakes E, Marqusee S. Comparison of proteolytic susceptibility in phosphoglycerate kinases from yeast and E. coli: modulation of conformational ensembles without altering structure or stability. J Mol Biol. 2007;368:1438–1447. doi: 10.1016/j.jmb.2007.02.077. [DOI] [PubMed] [Google Scholar]
- 16.Hnizda A, Jurga V, Rakova K, Kozich V. Cystathionine beta-synthase mutants exhibit changes in protein unfolding: conformational analysis of misfolded variants in crude cell extracts. J Inherit Metab Dis. 2012;35:469–477. doi: 10.1007/s10545-011-9407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majtan T, Kraus JP. Folding and activity of mutant cystathionine beta-synthase depends on the position and nature of the purification tag: Characterization of the R266K CBS mutant. Protein Expr Purif. 2012;82:317–324. doi: 10.1016/j.pep.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hnizda A, Spiwok V, Jurga V, Kozich V, Kodicek M, Kraus JP. Cross-talk between the catalytic core and the regulatory domain in cystathionine beta-synthase: study by differential covalent labeling and computational modeling. Biochemistry. 2010;49:10526–10534. doi: 10.1021/bi101384m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park C, Marqusee S. Quantitative determination of protein stability and ligand binding by pulse proteolysis. In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT, Ploegh HL, editors. Current protocols in protein science. Wiley; Hoboken: 2006. pp. 20.11.1–20.11.14. [DOI] [PubMed] [Google Scholar]
- 20.Hnizda A, Santrucek J, Sanda M, Strohalm M, Kodicek M. Reactivity of histidine and lysine side-chains with diethylpyrocarbonate -- a method to identify surface exposed residues in proteins. J Biochem Biophys Methods. 2008;70:1091–1097. doi: 10.1016/j.jbbm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. Embo J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003;1647:206–213. doi: 10.1016/s1570-9639(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 23.Willaert K, Engelborghs KW. The quenching of tryptophan fluorescence by protonated and unprotonated imidazole. Eur Biophys J. 1991;20:177–182. [Google Scholar]
- 24.Kery V, Poneleit L, Meyer JD, Manning MC, Kraus JP. Binding of pyridoxal 5′-phosphate to the heme protein human cystathionine beta-synthase. Biochemistry. 1999;38:2716–2724. doi: 10.1021/bi981808n. [DOI] [PubMed] [Google Scholar]
- 25.Mach H, Middaugh CR. Simultaneous monitoring of the environment of tryptophan, tyrosine, and phenylalanine residues in proteins by near-ultraviolet second-derivative spectroscopy. Anal Biochem. 1994;222:323–331. doi: 10.1006/abio.1994.1499. [DOI] [PubMed] [Google Scholar]
- 26.Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci U S A. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wetlaufer DB. Ultraviolet spectra of proteins and amino acids. Adv Protein Chem. 1962;17:303–390. doi: 10.1016/s0065-3233(08)60022-4. [DOI] [PubMed] [Google Scholar]
- 28.Singh LR, Gupta S, Honig NH, Kraus JP, Kruger WD. Activation of mutant enzyme function in vivo by proteasome inhibitors and treatments that induce Hsp70. PLoS Genet. 2010;6:e1000807. doi: 10.1371/journal.pgen.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 30.Sen S, Yu J, Yamanishi M, Schellhorn D, Banerjee R. Mapping peptides correlated with transmission of intrasteric inhibition and allosteric activation in human cystathionine beta-synthase. Biochemistry. 2005;44:14210–14216. doi: 10.1021/bi051046d. [DOI] [PubMed] [Google Scholar]
- 31.Koutmos M, Kabil O, Smith JL, Banerjee R. Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine {beta}-synthase. Proc Natl Acad Sci U S A. 2010;107:20958–20963. doi: 10.1073/pnas.1011448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta S, Wang L, Hua X, Krijt J, Kozich V, Kruger WD. Cystathionine beta-synthase p. S466L mutation causes hyperhomocysteinemia in mice. Hum Mutat. 2008;29:1048–1054. doi: 10.1002/humu.20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozich V, Sokolova J, Klatovska V, Krijt J, Janosik M, Jelinek K, Kraus JP. Cystathionine beta-synthase mutations: effect of mutation topology on folding and activity. Hum Mutat. 2010;31:809–819. doi: 10.1002/humu.21273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen S, Banerjee R. A pathogenic linked mutation in the catalytic core of human cystathionine beta-synthase disrupts allosteric regulation and allows kinetic characterization of a full-length dimer. Biochemistry. 2007;46:4110–4116. doi: 10.1021/bi602617f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.