Sexual reproduction poses an evolutionary paradox. An organism that reproduces asexually passes on all of its genes to each one of its individual progeny, whereas one that reproduces sexually passes on only half to each. Other things being equal, natural selection favors asexual reproduction, because given the same number of progeny, the asexual individual has double the fitness of the sexually reproducing one.
Which “things” are not equal? Two possible advantages of sex have been proposed: mixis of genes and DNA repair. The mixis argument goes as follows. Without the mixis of genes generated by sexual recombination, adaptive evolution is limited to the accumulation of favorable mutations that happen successively in each independently evolving lineage. With sexual reproduction, favorable mutations arisen in separate lineages can become combined in the same individual, providing an advantage in the adaptation to varying environments. The repair argument points out that the two haplotypes associated with diploid sex provide an error-correction mechanism for repairing genetic damage. The intact DNA of one haplotype can serve as a template for correcting the damaged DNA in the other haplotype. Moreover, deleterious mutations in one haplotype are covered up by complementary dominant mutations. Whether either one or both of these purported advantages can account for the origin and maintenance of sexual reproduction is a subject of much investigation and debate (1–3).
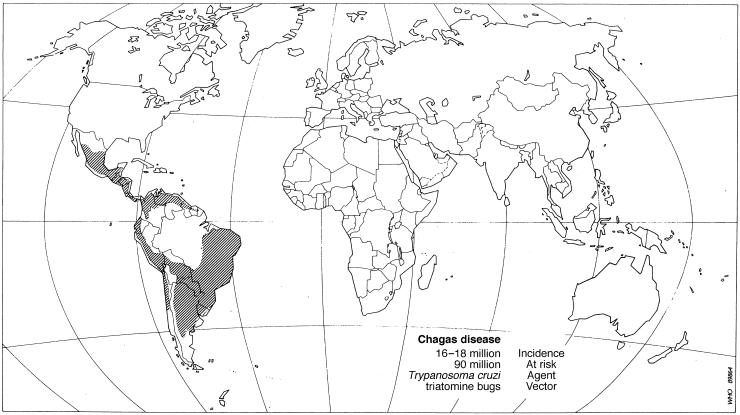
Eukaryotes, including the protozoa, are mostly diploid and have been thought to reproduce sexually. Emerging evidence from very diverse protozoan phyla brings the generality of sexual reproduction into question. (The protozoa comprise numerous phyla with older evolutionary origin and greater genetic diversity than the “higher” eukaryotic phyla, the multicellular plants, animals, and fungi.) Genetic information from increasingly detailed studies of several parasitic protozoa indicates that they have a clonal population structure; that is, that they consist of independently evolving clonal lineages without genetic recombination between them. The evidence for clonality is particularly compelling in the case of Trypanosoma cruzi (phylum Zoomastigina, class Kinetoplastida), a flagellate parasite transmitted by triatomine bugs, which is the agent of Chagas disease, an infirmity that affects some 18 million people in Latin America and is a major cause of mortality in Brazil, Bolivia, Chile, and elsewhere (Fig. 1). Oliveira et al. (4) provide new evidence that T. cruzi consists of clonally reproducing lineages. They ascertain the genetic polymorphism at eight microsatellite loci in 24 T. cruzi strains from the states of Sao Paulo and Minas Gerais, Brazil. (Some strains, isolated from a single vector or host, consist of more than one clonal lineage.) The loci consist primarily of two-bp repeats with allele length 110–319 bp. Three observations indicate that the strains represent clonal lineages: excess homozygosity, linkage disequilibrium, and strain associations coincident with those obtained with a separate locus. The eight loci are very polymorphic with a high average heterozygosity of 49.4% per locus, but well below the 78.5% heterozygosity expected with random mating. This finding indicates that allelic segregation and reassociation are not occurring according to the Mendelian laws. Second, the distribution of two-locus allele combinations that appear in the sample is different from that expected from random association between alleles at different loci, showing highly significant linkage disequilibrium for all 28 two-locus associations. Third, a strain classification based on ribosomal RNA type conforms to the classification obtained with the microsatellite loci, reflecting that strains are reproducing as clonal lineages, rather than there being independent association between loci as expected with sexual reproduction.
Figure 1.
World distribution of Chagas disease. [Reproduced with permission from ref. 42 (copyright 1991, World Health Organization).]
The clonal population structure of T. cruzi was discovered by Tibayrenc et al. (5), who analyzed 15 gene loci coding for enzymes in 121 stocks of widely dispersed geographic origins, from the United States and Mexico to Chile and Southern Brazil. One unanticipated observation was the repeated presence of some 15-locus genotypes (diplotypes) in distant localities, whereas most multilocus genotypes were absent. One 15-locus diplotype (#39) was represented by 25 stocks from remote localities throughout Brazil, Bolivia, and Chile. Two other 15-locus diplotypes (#19 and #20), different from each other by only one allele, were represented by 40 stocks from virtually the whole geographic range of the parasite. A second critical observation was the enormous genetic divergence between diplotypes: diplotype #39 had no alleles in common with diplotypes #19 and #20 at 10 of the 13 polymorphic loci (5, 6). The genetic variation was very large, yielding 7 × 1015 possible different diplotypes, so that no single diplotype should have appeared more than once in a sample of 121 stocks, let alone 25 or 40 times. The immense majority of the possible genotypes, including some expected with high frequencies, were lacking. Linkage disequilibrium was, indeed, quite large (5, 6). The great genetic distance between genotypes (as noted above between #39 and either #19 or #20) indicated that the clonal lineages were ancient. Genetic distance between #39 and either #19 or #20 was several times larger than between orangutans and humans, suggesting that the clonal lineages had been evolving independently for millions of years (5–7). Great diversity between strains occurs also within the limited geographic sample studied by Oliveira et al. (4): 17 mutational steps is the minimum distance between any two strains, indicating ancient divergence.
Explicit statistical methods have been developed (8–9) for analyzing genetic polymorphism data available in the literature for several parasitic protozoa. These methods are based on the fundamental Mendelian properties of segregation and independent assortment, which are constrained when sexual reproduction is restrained. Segregation is a property of individual diploid loci, which is shown to be curbed by: (a) fixed heterozygosity, (b) absence of segregating genotypes, or (c) deviations from Hardy–Weinberg equilibrium frequencies. When information is available for more than one locus, impeded recombination is manifested (in either haploids or diploids) by (d) over-represented or widespread identical genotypes, (e) absence of recombinant genotypes, (f) linkage disequilibrium, or (g) conformity between independent genetic markers. Oliveira et al. (4) rely on (c), (f), and (g) to conclude that T. cruzi has a clonal population structure.
Several studies have shown that one or several of (a)–(g) obtain for T. cruzi (5–11). The analysis performed by Tibayrenc et al. (8–9) has demonstrated clonal population structures for several other parasitic protozoa, on the basis of the following published evidence: Leishmania braziliensis (12), L. infantum (13), L. major (14), L. tropica (15–16), T. brucei, the agent of human sleeping sickness and of magana and other animal diseases (17–21), and T. vivax (22–23). Strong evidence of clonality has been shown (8–9) for Entamoeba histolytica (24, 25), Giardia (26–28), and T. congolese (29); and suggestive but not definitive for Naglaeria australiensis (30), N. fowleri (30), N. gruberi (30), Toxoplasma gondii (31, 32), Trichomonas foetus (33), and Trichomonas vaginalis (33), as well as for the fungi Candida albicans (34), Candida tropicalis (35), and Cryptococcus neoformans (36, 37).
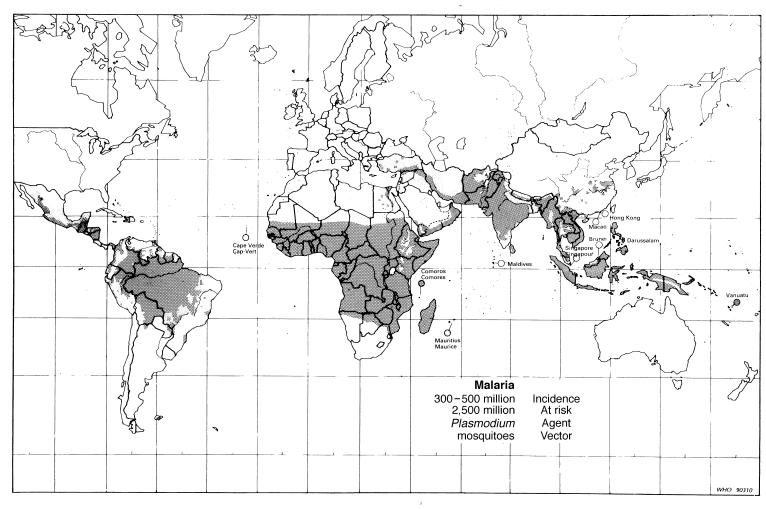
The most puzzling instance of clonal population structure comes from Plasmodium falciparum, the agent of malignant malaria. Malaria counts among mankind’s worst scourges: there are 300–500 million clinical cases in the world each year and more than 1 million child deaths in Sub-Saharan Africa alone (38) (Fig. 2). The human infective form of the parasite is haploid, but diploidy occurs in the mosquito vector, where fertilization takes place and haploid sporozoites are formed that are transmitted by the mosquito to humans. The sexual stage required for the completion of the life cycle would seem to make P. falciparum an unlikely candidate for a clonal population structure. Indeed, clonality has been excluded for P. falciparum because of evidence of genetic recombination (e.g., ref. 39). Yet a recent study of 25 strains from different regions of the world suggests that P. falciparum may have a clonal population structure (40). Detailed analysis of the gene coding for the circumsporozoite protein (Csp), a highly polymorphic antigen, contradicts the expectations ensuing from sexual (meiotic) recombination in three ways: (i) the incidence of recombination events does not increase with nucleotide distance along the DNA sequence; (ii) the strength of linkage disequilibrium between nucleotides is independent of distance; and (iii) nucleotide sequences in the two end regions of the gene are correlated with each other, but not with the sequences in the central region they span, which consists of multiple repeats of 12 bp-long motifs. These observations are contrary to what is expected from meiotic recombination, but are consistent with intragenic mitotic recombination and, hence, with a clonal population structure. It would seem that sexuality in the physiological sense does not involve sexuality in the genetic sense of mixis, because meiosis in P. falciparum either (i) does not take place, or (ii) occurs between identical haplotypes. Alternative (ii) may be the likely explanation in malaria regions of low infectivity. If the male and female gametocytes taken in the mosquito’s blood meal derive from a single haplotype (i.e., from genetically identical schizonts), recombination between them will yield once again the original haplotype. Most of the samples analyzed in ref. 40 come from South East Asia and tropical America, which are regions of low infectivity. Whether or not clonality prevails in the highly infective regions of Africa and New Guinea remains to be examined by the methods of ref. 40. It may well be that P. falciparum exhibits a clonal population structure in some but not other regions of the world. But it also may be that there is some physiological or genetic mechanism that inhibits fertilization in P. falciparum between genetically different gametes.
Figure 2.
World distribution of malaria. [Reproduced with permission from ref. 42 (copyright 1991, World Health Organization).]
Why does clonality matter? Whether the population structure of a unicellular parasite is primarily clonal or panmictic has evolutionary and public health import (4–11). First, in a sexually reproducing organism the individual genotype is ephemeral; the entity that persists and evolves is the gene pool, and a few individuals encompass most of the genetic variability of the species. In a clonally propagating organism, the entity that persists and evolves is the clonal lineage; the genetic diversity of the species can be captured only by extensive sampling of distinct lineages. Second, extensive genetic divergence among clonal lineages may imply proportionally diverse biological characteristics, including pathogenicity, host and vector propensity, vulnerability to drugs and vaccines, and other medically significant attributes. For example, Chagas disease may be chronic or acute, affect the heart or gastrointestinal organ systems, varies in virulence, and so on (4–6, 38). This is hardly surprising because extant T. cruzi lineages diverged from one another much before human origins (4, 7, 41), so that specific adaptation to human hosts will have evolved independently in separate lineages. Third, in clonally evolving organisms, epidemiological surveys and medical typing, as well as the search for specific vaccines and drugs should not proceed randomly; rather, they are likely to be more successful if they are preceded by identification and characterization of clonal lineages, targeting those that are more pathogenic or ubiquitous.
References
- 1.Williams G C. Sex and Evolution. Princeton: Princeton Univ. Press; 1975. [Google Scholar]
- 2.Maynard Smith J. The Evolution of Sex. Cambridge: Cambridge Univ. Press; 1978. [Google Scholar]
- 3.Michod R E, Levin B R, editors. The Evolution of Sex. Sunderland, MA: Sinauer; 1987. [Google Scholar]
- 4.Oliveira R P, Broude N E, Macedo A M, Cantor C R, Smith C L, Pena S D J. Proc Natl Acad Sci USA. 1998;95:3776–3780. doi: 10.1073/pnas.95.7.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibayrenc M, Ward P, Moya A, Ayala F J. Proc Natl Acad Sci USA. 1986;83:115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tibayrenc M, Ayala F J. Evolution. 1988;42:277–292. doi: 10.1111/j.1558-5646.1988.tb04132.x. [DOI] [PubMed] [Google Scholar]
- 7.Ayala F J. Biol Res. 1993;26:47–63. [PubMed] [Google Scholar]
- 8.Tibayrenc M, Kjellberg F, Ayala F J. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Brenière S F, Dardé M-L, Ayala F J. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Skarecky D, Ayala F J. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tibayrenc M. Annu Rev Microbiol. 1996;50:401–429. doi: 10.1146/annurev.micro.50.1.401. [DOI] [PubMed] [Google Scholar]
- 12.Desjeux P, Dedet J P. Trans R Soc Trop Med Hyg. 1989;83:610–612. doi: 10.1016/0035-9203(89)90373-8. [DOI] [PubMed] [Google Scholar]
- 13.Moreno G, Rioux J A, Lanotte G, Pratlong F, Serres E. Leishmania: Taxonomie et Phylogénèse: Applications Eco-Epidémiologiques, Colloques Internationaux de Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale. Paris: CNRS/INSERM; 1986. pp. 105–119. [Google Scholar]
- 14.Maazoun R, Pratlong F, Lanotte G, Rioux J A. Leishmania: Taxonomie et Phylogénèse: Applications Eco-Epidémiologiques, Colloques Internationaux de Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale. Paris: CNRS/INSERM; 1986. pp. 119–129. [Google Scholar]
- 15.Pratlong F, Lanotte G, Ashford R W, Rioux J A. Leishmania: Taxonomie et Phylogénèse: Applications Eco-Epidémiologiques, Colloques Internationaux de Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale. Paris: CNRS/INSERM; 1986. pp. 129–139. [Google Scholar]
- 16.Lanotte G, Rioux J A, Serres E. Leishmania: Taxonomie et Phylogénèse: Applications Eco-Epidémiologiques, Colloques Internationaux de Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale. Paris: CNRS/INSERM; 1986. pp. 27–40. [Google Scholar]
- 17.Gibson W C, Marshall T F deC, Godfrey D G. Adv Parasitol. 1980;18:175–246. doi: 10.1016/s0065-308x(08)60400-5. [DOI] [PubMed] [Google Scholar]
- 18.Gibson W C, Gashumba J K. Trans R Soc Trop Med Hyg. 1983;77:114–118. doi: 10.1016/0035-9203(83)90033-0. [DOI] [PubMed] [Google Scholar]
- 19.Gibson W C, Wellde B T. Trans R Soc Trop Med Hyg. 1985;79:671–676. doi: 10.1016/0035-9203(85)90187-7. [DOI] [PubMed] [Google Scholar]
- 20.Mihok S, Otieno L H, Darji N. Parasitology. 1990;100:219–233. doi: 10.1017/s0031182000061229. [DOI] [PubMed] [Google Scholar]
- 21.Mehlitz D, Zillmann U, Scott C M, Godfrey D G. Trop Med Parasitol. 1982;33:113–118. [PubMed] [Google Scholar]
- 22.Allsopp B A, Newton S D. Int J Parasitol. 1985;15:265–270. doi: 10.1016/0020-7519(85)90063-3. [DOI] [PubMed] [Google Scholar]
- 23.Kilgour V, Godfrey D G. Ann Trop Med Parasitol. 1975;69:329–335. doi: 10.1080/00034983.1975.11687016. [DOI] [PubMed] [Google Scholar]
- 24.Proctor E M, Wong Q, Yang J, Keystone J S. Am J Trop Med Hyg. 1987;37:296–301. doi: 10.4269/ajtmh.1987.37.296. [DOI] [PubMed] [Google Scholar]
- 25.Sargeaunt P G, Williams J E, Jackson T F H G, Simjee A E. Trans R Soc Trop Med. 1982;76:401–402. doi: 10.1016/0035-9203(82)90200-0. [DOI] [PubMed] [Google Scholar]
- 26.Meloni B P, Lymbery A J, Thompson R C A. Am J Trop Med Hyg. 1988;38:65–73. doi: 10.4269/ajtmh.1988.38.65. [DOI] [PubMed] [Google Scholar]
- 27.Nash T E, McCutchan T, Keister D, Dame J B, Conrad J D, Gillin F D. J Infect Dis. 1985;152:64–73. doi: 10.1093/infdis/152.1.64. [DOI] [PubMed] [Google Scholar]
- 28.Meloni B P, Lymbery A J, Thompson R C A. Am J Trop Med Hyg. 1989;40:629–637. doi: 10.4269/ajtmh.1989.40.629. [DOI] [PubMed] [Google Scholar]
- 29.Gashumba J K, Baker R D, Godfrey D G. Parasitology. 1988;96:475–486. doi: 10.1017/s0031182000080112. [DOI] [PubMed] [Google Scholar]
- 30.Cariou M L, Pernin P. Genetics. 1987;115:265–270. doi: 10.1093/genetics/115.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dardé M L, Bouteille B, Pestre-Alexandre M. Am J Trop Med Hyg. 1988;39:551–558. doi: 10.4269/ajtmh.1988.39.551. [DOI] [PubMed] [Google Scholar]
- 32.Dardé M L, Bouteille B, Pestre-Alexandre M. Bull Soc Fr Parasitol. 1990;8:238. (abstr.). [Google Scholar]
- 33.Nadler S A, Honigberg B M. J Parasitol. 1988;74:797–801. [PubMed] [Google Scholar]
- 34.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Mallié M, Bastide J-M. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann P F, Kemker B J, Hsiao C B, Dev S J. Clin Microbiol. 1989;27:2514–2521. doi: 10.1128/jcm.27.11.2514-2521.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safrin R E, Lancaster L A, Davis C E, Braude A I. Am J Clin Pathol. 1986;86:204–208. doi: 10.1093/ajcp/86.2.204. [DOI] [PubMed] [Google Scholar]
- 37.Brandt M E, Bragg S L, Pinner R W. J Clin Microbiol. 1993;31:2819–2823. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. Tropical Disease Report, Twelfth Programme Report. Geneva: W.H.O.; 1995. [Google Scholar]
- 39.Babiker H, Walliker D. Parasitol Today. 1997;13:262–267. doi: 10.1016/s0169-4758(97)01075-2. [DOI] [PubMed] [Google Scholar]
- 40.Rich S M, Hudson R R, Ayala F J. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maslov D A, Simpson L. Parasitol Today. 1995;11:30–32. [Google Scholar]
- 42.World Health Organization. Tropical Diseases, Progress in Research 1989–1990, Tenth Programme Report. Geneva: W.H.O.; 1991. [Google Scholar]