Abstract
The risk of exposure to Listeria monocytogenes (L. monocytogenes) when consuming Ready-to-Eat (RTE) seafood was assessed in the Veneto Region (Italy). Thirty-eight samples were analyzed, each sample consisted of three subunits belonging to the same batches. The first of the three units was examined immediately, the second was stored at +4°C (for all of its shelf-life) and the third at +10°C (for the latter third of its shelf-life) before the analysis. Chemical-physical and microbiological parameters were tested simultaneously. Culture results showed the presence of viable L. monocytogenes in 9 (23,68%) of the 38 samples analysed, 3 (33,33%) of which with a concentration >100 cfu/g. PCR tests yielded 12 L. monocytogenes positive samples. Semipreserves with aw (water activity) and pH values that favour L. monocytogenes growth were the only ones to result positive to microbiological and PCR tests. Temperature proved to be an important factor as it limits the growth of L. monocytogenes, including products with potentially high competitive microbial charges. Four different serotypes were recovered and ribotyping has helped to highlight the genomic variability of L. monocytogenes strains in food. This supports the hypothesis that L. monocytogenes continues to evolve genetically to the detriment of phenotypic conservation.
1. Introduction
L. monocytogenes is an intracellular pathogen which, especially if foodborne, may induce what is known as listeriosis. Once ingested, L. monocytogenes can penetrate the intestinal endothelial barrier, the placental or the hematoencephalic barrier [1, 2]. The groups at higher risk of contracting the disease are young people, the old, (over 65), pregnant women, and immune compromised people, the YOPI, an acronym coined by De Cesare et al. [3, 4]. In healthy subjects, L. monocytogenes can lead to episodes of gastroenteritis and fever [5, 6].
Listeriosis is considered a rare disease, its incidence in humans ranges between 0.1 and 11.3 cases/million [7], with a high mortality rate, up to 30% in the categories most at risk (YOPI) [8]. Based on an EFSA 2010 report, the incidence in Europe was of 3 cases/million of inhabitants [9]. Because the incubation period can span from 3 up to 60 days, this disease is often difficult to trace because it is not easy to isolate the food that is responsible for the infection. Europe has some well-documented episodes particularly in France [10], Finland [11], Switzerland [12], the UK [11], Belgium [13], and Ireland [14]. Distinct psychrotolerant characteristics allow L. monocytogenes to adapt to acidic conditions and to low water activity environments, making it an insidious threat to some kinds of food as ready-to-eat food (RTE) that is characterized by mild treatments and a medium-to-long shelf-life—a highly sought-after quality by today's consumer [15–22]. The fish products that pose potential risks include mainly cold smoked fish, raw carpaccio, and marinated fish. Smoked fish products in particular were reported to cause human infections [23–25]. In Europe, smoked salmon, more than all the other products, was reported to have surpassed the maximum threshold limits allowed for L. monocytogenes contamination [9].
The risk of consuming RTE seafood does not so much entail the contamination of the raw product, which will often have L. monocytogenes, but at low concentrations, as much as the product's characteristics which in time encourage its growth [26–28].
Inadequate consumer knowledge on how to store RTE food at home, at the right refrigerated temperature, has led to higher risks of L. monocytogenes growth [29, 30].
The goal of this study is to determine the distribution of L. monocytogenes in RTE fish semipreserves, as set out by EC Regulations 2073/2005 related to this category which distinguishes the foods that favour L. monocytogenes growth from those that do not. Different types of repfed products were examined, these include marinated seafood salads (with cephalopods, surimi, crustaceans, bivalves), marinated shrimps, cephalopods and salmon carpaccio, marinated mackerel, smoked herrings, and cold smoked salmon.
Packaged products marketed in the Veneto Region (Italy) were sampled to determine the levels of L. monocytogenes. The products inspected included ones with intrinsically favourable characteristics, thus ideal for L. monocytogenes growth, and those with unfavourable characteristics (pH ≤ 4.4, a w ≤ 0.92; pH ≤ 5.0 and a w ≤ 0.94). Tests were carried on both these types of products stored, at 4°C and at 10°C, the latter being a more realistic simulation of household conditions which may experience thermal abuse. Each sample unit, aside from examining L. monocytogenes (qualitative and quantitative tests) also analysed the following: total aerobic mesophilic count, total psychrophiles count, total psychrophiles H2S producers, moulds, yeasts, and lactic bacteria to establish whether correlation exists in relation to the presence of L. monocytogenes Furthermore, enrichment broths were also tested for L. monocytogenes using PCR and serotyping and ribotyping of isolated strains and culture tests.
2. Materials and Methods
The samples, each consisting of 3 sample units coming from a production batch, upon reaching the laboratory, were stored in a refrigerator in their original package at a temperature of 4°C and 10°C until their shelf-life expiry (storage at 10°C in the last third of their shelf-life expiry). Product shelf-life ranged from 8 days (raw salmon carpaccio) to 70 days (seafood salads). During the collection phase at the various traders, only the samples which had not already surpassed half of their expiry period were considered. The products selected were both national and international products taken from 9 different stores. Upon reaching the laboratory, one sample unit was examined immediately, the other two were on the last day of expiry. In total 38 samples were analysed, amounting to 114 sample units.
2.1. Microbial Analysis
Total aerobic mesophilic count on agar plates with incubation in aerobiosis at 30°C for 72 hours (ISO 4833:2003).
Total psychrotolerant count on iron agar plates (Lyngby) with incubation in aerobiosis at 15°C for 7 days.
Total psychrophiles producing hydrogen sulphide count on iron plates (Lyngby) with incubation in aerobiosis at 15°C for 7 days.
Count of the moulds and yeasts on Rose Bengal Chloramphenicol Agar plates with incubation in aerobiosis at 25°C for 5 days.
Lactic bacteria count on MRS agar plates (final pH 6.4) with incubation in aerobiosis at 30°C for 72 hours.
L. monocytogenes count on ALOA agar at 37°C for 48 hours (ISO 11290-2:1998/Amd 1 2004).
Detection of L. monocytogenes on ALOA agar and PALCAM agar plates at 37°C for 48 hours following selective enrichment in Half Fraser and Fraser Broth (ISO 11290-1:1996/Amd 1 2004).
The quantitative and qualitative methods were carried out at the same time, the same day.
2.2. Chemical and Physical Analysis
pH measure using the Mettler Toledo MP 220 instrument, with temperature autocompensation.
Water activity (a w) using Rotronic 29539 instrument (ISO 21807:2004).
2.3. Genomic DNA Extraction
Detection of L. monocytogenes consisted in taking 1 ml of enrichment broth (Half Fraser), after 24 hours of incubation, to extract DNA and carry out subsequent PCR tests for Listeriaspp and L. monocytogenes. DNA extraction was carried out on pellet, obtained after centrifugation of the enrichment broth (2,000 g for 2 minutes, followed by 12,000 g for 5 minutes on the surnatant). Once the surnatant was removed, PBS was added and 16,000 g underwent centrifugation for another 2 minutes. GenElute Bacterial Genomic DNA Mini Kit (Sigma) was used, following the manufacturer's instructions, protocol for gram positive bacteria.
2.4. Listerial Genus Detection by PCR
Nested PCR was the method used to target the codifying gene for 16S rRNA in which the amplified product of the first reaction becomes the template for subsequent nested reactions. The primers used in the first reaction were the forward primer LI1 5′-CTC CAT AAA GGT GAC CCT-3′ and the reverse primer U1 5′-CAG CMG CCG CGG TAA TWC-3′ [31]. The reaction took place in a final volume of 25 μL with concentrations of 1X GeneAmp PCR Buffer II (Applied Biosystems), 1.5 mM of MgCl2, 0.2 mM of each dNTP, 0.2 μM of both primers, 1.25 U of Ampli Taq DNA polymerase (Applied Biosystems), and 5 μL of extracted DNA. For amplification, the thermal cyler GeneAmp PCR System 9700 (Applied Biosystem) was used with temperatures set at initial denaturation at 95°C for 3 min, followed by 25 cycles, each with a denaturation phase at 95°C for 90 sec, and an annealing phase at 50°C for 90 sec and an extension phase at 72°C for 2 min, followed by a final extension phase at 72°C for 10 min. The primers used for the nested reactions were forward primer LS1 5′-ACG ACC GCA ADG TTG AAA CT-3′ and reverse primer LS2 5′-GAC GTC ATC CCC ACC TTC CT-3′ manufactured at the Nucleic Acids Technology laboratory applied to foods at the Istituto Zooprofilattico Sperimentale of Brescia. The reaction was prepared in a final volume of 25 μL with concentrations of 1X GeneAmp PCR Buffer II (Applied Biosystems), 1.5 mM of MgCl2, 0.2 mM of each dNTP, 0.2 μM of both primer, with 0.75 U of Ampli Taq DNA polymerase (Applied Biosystems) and 2.5 μL of extracted DNA. For amplification the thermal cycler GeneAmp PCR System 9700 (Applied Biosystem) was used with temperatures for initial denaturation set at 95°C for 3 min, followed by 35 cycles, each comprising a denaturation phase at 95°C for 30 sec, an annealing phase at 59°C for 30 sec, an extension phase at 72°C for 30 sec, followed by a final extension phase at 72°C for 5 min. Analysis of foreseen amplification, of 301 bp, was carried out after electrophoresis in agarose gel at 2.5% stained with ethidium bromide (final concentration on gel: 0.5 μg/mL), by means of exposure to UV radiation.
2.5. L. monocytogenes Detection by PCR
A method to detect the HLY gene target (haemolytic secreted pathogenic factor or hemolysin) was used with forward primer LIS1 5′-CGG AGG TTC CGC AAA AGA TG-3′ and reverse primer LIS2 5′-CCT CCA GAG TGA TCG ATG TT-3′ [32]. Reaction was prepared using an end volume of 25 μL with concentrations of 1X GeneAmp PCR Buffer II (Applied Biosystems), 1.5 mM of MgCl2, 0.2 mM for each dNTP, 0.2 μM of both primers, 0.75 U of Ampli Taq DNA polymerase (Applied Biosystems) and 5 μL of extracted DNA. Amplification was carried out in the thermal cycler GeneAmp PCR System 9700 (Applied Biosystem) with a temperature profile for initial denaturation set at 95°C for 3 min, followed by 35 cycles, each comprising a denaturation phase at 95°C for 30 sec, an annealing phase at 58°C for 30 sec and an extension phase at 72°C for 30 sec, followed by a final extension phase at 72°C for 5 min. Analysis of foreseen amplification, of 234 bp, was then conducted after electrophoresis in agarose gel at 2.5% stained with ethidium bromide (final concentration on gel: 0,5 μg/mL), by means of exposure to UV radiation.
2.6. L. monocytogenes Serotyping by Multiplex PCR (M-PCR)
Serotyping by M-PCR was performed using primers as described by Doumith et al.[33]. Primers enable the identification of Listeria and the subdivision of strains belonging only to the L. monocytogenes species into four distinct serogroups.
Serogroup 1 comprises serotype 1/2a and 3a; serogroup 2 of serotypes 1/2c and 3c; serogroup 3 serotypes 1/2b and 3b; and serogroup 4, serotypes 4b, 4d and 4e.
The PCR mix included PCR Master Mix 1X (Qiagen, Milan, Italy), mix of primers (lmo0737, ORF2819, ORF2110, lmo1118 and prs), sterile distilled penta-H2O and the DNA extracted. PCR reaction conditions included an initial step of denaturation at 94°C for 3 minutes, 35 cycles at 94°C for 0.40 minutes, 53°C for 1.15 minutes, 72°C for 1.15 minutes and a final step at 72°C for 7 minutes.
PCR products underwent 2% agarose gel electrophoretic separation at 90 V for 90 minutes. Then, after being stained with ethidium bromide (10 ng/ml), they were visualised on UV transilluminator.
2.7. L. monocytogenes Serotyping with Seroagglutination
Serotyping with antisera yielded 12 different serotypes of L. monocytogenes in terms of cellular surface, and the somatic “O” and flagellar “H” antigens.
Seroagglutination was carried out to confirm the serotypes and serogroups obtained using the molecular method. The “SEIKEN” Listeria Antisera Kit (Denka Seiken co. LTD, Tokyo, Japan) was used according to a modified method outlined by Seeliger and Hohne [34].
2.8. L. monocytogenes Ribotyping
Positive L. monocytogenes colonies were detected and characterized using the Du Pont Qualicon Ltd. RiboPrinter Q system [3, 4]. The colonies were taken from agar plates using a plastic stick. They were suspended in a sample buffer. The solution was lysed at 85°C for 20 minutes, and two specific lysing agents were added. Subsequently, a batch containing eight containers to hold eight samples was inserted in the automated ribotyping instrument. In brief, RiboPrinter generates an enzymatic digestion of the solutions using the EcoRI restriction enzyme, and electrophoresis of DNA fragments was transferred onto a membrane. The membrane was hybridized using a specific chemiluminescent probe. Finally, the instrument detects the signal emitted by a CCD camera and software converts the images in ribotyping patterns. Ribotyping (of the species) is an automated process if more than 85% of the sample patterns resemble the reference patterns of the instrument's database. The latter were obtained from various international collections (e.g., ATCC, DSMZ, or JMC) and identified with a DUP-ID code (Dupont Identification). Genotypic characterization of bacterial strains consisted of comparing the strains of a batch, assigning to them what is called a ribogroup code (RG). Each ribotyped strain was compared with all the patterns of the profiles contained in the database. In general, if there was a similarity ≥93% between the profile of the strain investigated and that of the databank, the strain was assigned to the corresponding ribogroup of that profile. Ribogroup attribution is only possible if strains undergo simultaneous analysis or if analysis is conducted a few days later since the database has to be updated on an ongoing basis, accessing international databanks available on the internet.
To obtain phylogenetic trees the results were extracted from a Pearson correlation (UPGMA method) with BioNumerics software version 6.1.
3. Results
The analyses were carried out on a range of 38 different commercial products 28 (73,68%) of which had a w and pH values favourable for growth of L. monocytogenes, while 10 (26,32%) had unfavourable values instead (Table 1).
Table 1.
Types of ready-to-eat seafood, country of origin.
| Type of product | No. of samples with characteristics favourable to L. monocytogenes | No. of samples with characteristics unfavourable to L. monocytogenes | Country of origin |
|---|---|---|---|
| Cold smoked salmon | 12 | 2 | Other EU country |
| Marinated seafood salad | 5 | 3 | Italy |
| Shrimps in brine | 3 | 3 | Other EU country |
| Salmon Carpaccio | 4 | — | Italy |
| Octopus Carpaccio | 2 | — | Italy |
| Marinated cuttlefish | — | 1 | Italy |
| Cooked marinated mackerel | 2 | — | Italy |
| Cold smoked herrings | — | 1 | Other EU country |
|
| |||
| Total | 28 | 10 | — |
Of the 28 samples with favourable characteristics for growth of the pathogen conditions, nine were positive for L. monocytogenes (32,14%), according to the standard culture methods (ISO 11290-1:1996/Amd 1 2004); of these, 3 (10,71%) had values exceeding the limit (100 cfu/g) established by EC Reg. no. 2073/2005. No sample with unfavourable characteristics was found positive for viable L. monocytogenes in microbiological testing.
Table 2 illustrates the corresponding values of L. monocytogenes qualitative test, quantitative test, and PCR results. Listerial genus was present in all 38 samples analysed according to the amplification results of the genus specific PCR reaction. Nonetheless, the species specific PCR demonstrated the presence of L. monocytogenes after enrichment in 12 samples only, of which 9 were also found to contain the pathogen by classical culture tests. The microbiological parameters are reported in Table 3. In general they rose when thermal abuse at 10°C occurred. In the 3 samples with a charge of >100 cfu/g, L. monocytogenes grew despite the presence of competitive flora, both lactic and alternating. Despite the increase of microbial values at the end of shelf-life, there were no significant changes in the product pH and a w values. In evidence that of three samples with a higher than 100 cfu/g value (sample no. 2, 8 and 10), two were analysed after thermal abuse at 10°C (last third of its shelf-life), and one proved unfit at the time it was being taken. It is worth noting that the container of sample no. 2, at the end of its shelf-life, had bulging but no unpleasant odours. Other samples, when opened, had no significant organoleptic alterations.
Table 2.
Products resulting positive to L. monocytogenes.
| Sample no. | Type of product | Temperature of storage | L. monocytogenesculture qualitative test | L. monocytogenesculture quantitative test (cfu/g) | L.monocytogenes PCR |
|---|---|---|---|---|---|
| 1 | Seafood salad | T0 | Positive | <5 | Positive |
| T0 | Positive | <5 | Positive | ||
| 2 | Salmon carpaccio | 4°C | Positive | <5 | Positive |
| 10°C | Positive | 330 | Positive | ||
| 3 | Marinated seafood Cocktail | 4°C | Positive | <5 | Positive |
| 10°C | Neg. | <5 | Positive | ||
| 4 | Octopus Carpaccio | 4°C | Neg. | <5 | Positive |
| 5 | Cooked and marinated mackerel fillets | T0 | Positive | <5 | Positive |
| 10°C | Neg. | <5 | Positive | ||
| 6 | smoked salmon | 4°C | Neg. | <5 | Positive |
| 10°C | Neg. | <5 | Positive | ||
| T0 | Positive | <5 | Positive | ||
| 7 | Fresh salmon in protective atmosphere | 4°C | Positive | <5 | Positive |
| 10°C | Positive | <5 | Positive | ||
| T0 | Neg. | <5 | Positive | ||
| 8 | Smoked salmon | 4°C | Positive | <5 | Positive |
| 10°C | Positive | 140,000 | Positive | ||
| 9 | Fresh salmon in protective atmosphere | 4°C | Positive | <5 | Positive |
| 10°C | Positive | <5 | Positive | ||
| 10 | Smoked salmon | T0 | Positive | 6,600 | Positive |
| 11 | Salmon Carpaccio | 10°C | Neg. | <5 | Positive |
| 12 | Cooked and marinated mackerel fillets | T0 | Positive | <5 | Positive |
Table 3.
Detailed description of microbial charges, aw and pH values of each L. monocytogenes-positive sample (microbiological and PCR tests). Values expressed in CFU/g.
| Sample n. | Types of product | Temperature of storage | Mesophiles count | Psicrophiles count | Psicrophiles H2S count | Moulds count | Yeasts count | Lactic count | aw | pH |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Seafood salad | T0(∗) | 9 × 108 | 6.6 × 105 | <1 | <1 × 102 | <1 × 102 | 2.4 × 108 | 0.0680556 | 5:01 |
| T0 | 2.8 × 104 | 1.4 × 104 | 6 | 4 × 102 | 9 × 102 | 9.1 × 102 | 0.0666667 | 6:01 | ||
| 2 | Salmon carpaccio | 4°C(∗∗) | 6.5 × 105 | 3.8 × 105 | 3.5 × 102 | 2 × 102 | 2.4 × 103 | 6.4 × 105 | 0.0673611 | 6:01 |
| 10 °C(∗∗∗)(∗∗∗∗) | 1.9 × 10 8 | 1.3 × 10 8 | 4.7 × 10 4 | 9 × 10 2 | 1.7 × 10 4 | 1.5 × 10 8 | 0.0673611 | 6:01 | ||
| 3 | Marinated seafood cocktail | 4 °C | 7.5 × 104 | 4.5 × 104 | <1 | < 1 × 102 | <1 × 102 | 3.7 × 104 | 0.0673611 | 5:01 |
| 4 | Octopus carpaccio | 10°C | 5.5 × 107 | 3 × 106 | <1 | <1 × 102 | <1 × 102 | 4.6 × 107 | 0.0680556 | 4:09 |
| 5 | Cooked and marinated Mackerel Fillets(∗∗∗∗∗) | 4°C | 4.2 × 106 | 2.4 × 104 | <1 | < 1 × 102 | <1 × 102 | 7.3 × 106 | 0.0680556 | 5:03 |
| 6 | Smoked salmon | T0 | 7.8 × 104 | 7.4 × 103 | <1 | <1 × 102 | 7 × 102 | 3.6 × 102 | 0.0638889 | 6:01 |
| 10°C | 7.1 × 107 | 6.1 × 107 | 1.1 × 107 | <1 × 102 | 1.5 × 105 | 4.4 × 102 | 0.0652778 | 6:03 | ||
| 4°C | 1.6 × 105 | 9.6 × 104 | 6.9 × 103 | 4.9 × 103 | <1 × 102 | 2 × 103 | 0.0673611 | 6:01 | ||
| 7 | Fresh salmon in protective atmosphere | 10°C | 1.2 × 106 | 2.2 × 106 | 8 × 104 | 6 × 102 | 5.9 × 103 | 6.1 × 103 | 0.0680556 | 6:00 |
| T0 | 2.7 × 106 | 2.2 × 106 | 2 × 105 | <1 × 102 | 3.6 × 103 | 9.8 × 103 | 0.0673611 | 6:00 | ||
| 8 | Smoked salmon | T0 | 7.6 × 107 | 4 × 107 | 1.8 × 105 | <1 × 102 | 3 × 102 | 2.8 × 107 | 0.0673611 | 5:08 |
| 4°C | 4.2 × 10 8 | 1.5 × 10 8 | 1.2 × 10 7 | <1 × 10 2 | 4.5 × 10 4 | 7.3 × 10 7 | 0.0673611 | 6:02 | ||
| 9 | Fresh salmon in protective atmosphere | 10°C | 3.6 × 106 | 2.9 × 106 | 5.2 × 103 | <1 × 102 | 3.9 × 103 | 5.1 × 103 | 0.0673611 | 6:02 |
| 4°C | 6.7 × 106 | 1.7 × 105 | 1.4 × 103 | <1 × 102 | 3 × 102 | 4.1 × 106 | 0.0673611 | 6:02 | ||
| 10 | Smoked salmon | 10°C | 1.4 × 10 7 | 1.4 × 10 7 | <1 | <1 × 10 2 | <1 × 10 2 | 4.3 × 10 6 | 0.0666667 | 6:02 |
| 11 | Salmon carpaccio | T0 | 7.5 × 106 | 3.2 × 106 | 1.3 × 105 | 1 × 103 | 3.5 × 103 | 1.7 × 105 | 0.0673611 | 6:00 |
| 12 | Cooked and marinated mackerel fillets (∗∗∗∗∗) | 10°C | 4.1 × 107 | 1.2 × 105 | <1 | <1 × 102 | <1 × 102 | 2.7 × 108 | 0.0680556 | 5:03 |
Note. The lines with bold lettering refer to the 3 samples with a L. monocytogenes concentration of >100 cfu/g.
(∗): zero time, analysis of one of the three sample units on the day they reached the laboratory.
(∗∗): 4°C, analysis at the end of shelf-life at 4°C storage conditions.
(∗∗∗): 10 °C, analysis at the end of shelf-life at 10°C storage conditions in the last third before final expiration.
(∗∗∗∗): packaging with bulging at the end of shelf- life with no unpleasant odour.
(∗∗∗∗∗): samples from the same company but from different batches.
Serotyping of 15 isolated L. monocytogenes strains coming from RTE fish semipreserves was carried out using Multiplex PCR to separate the 4 main serotypes (1/2a, 1/2b, 1/2c and 4b) into 4 distinct groups. Confirmation of the serotype of single serogroups was achieved with the seroagglutination method (Table 4 and Figure 1).
Table 4.
Serotyping and ribotyping results on isolated L. monocytogenes strains.
| Sample no. | Types of product | Temperature of storage | Serotype | Ribotype | Ribogroup |
|---|---|---|---|---|---|
| 1 | Seafood salad | T0 | 4b | DUP 1042 | 1369 |
| T0 | 1/2a | DUP 1043 | 568 | ||
| 2 | Salmon Carpaccio | 4°C | 1/2a | DUP 20243 | 568 |
| 10°C | 1/2a | DUP 20243 | 1340 | ||
| 3 | Marinated seafood cocktail | 4°C | 1/2b | DUP 1052 | 564 |
| 10°C | |||||
| 4 | Octopus carpaccio | 4 °C | |||
| 5 | Cooked and marinated mackerel fillets | T0 | 4b | DUP 1042 | 1296 |
| 10°C | |||||
| 6 | Smoked salmon | 4°C | |||
| 10°C | |||||
| T0 | 1/2a | DUP 20243 | 568 | ||
| 7 | Fresh salmon in protective atmosphere | 4 °C | 1/2a | DUP 1042 | 568 |
| 10 °C | 1/2a | DUP 1042 | 568 | ||
| T0 | |||||
| 8 | Smoked salmon | 4°C | 1/2a | DUP 1042 | 568 |
| 10°C | 1/2a | DUP 1043 | 568 | ||
| 9 | Fresh salmon in protective atmosphere | 4°C | 1/2a | DUP 1042 | 568 |
| 10°C | 1/2a | DUP 1042 | 568 | ||
| 10 | Smoked salmon | T0 | 4d | DUP 1042 | 1533 |
| 11 | Salmon carpaccio | 10°C | |||
| 12 | Cooked and marinated mackerel fillets | T0 | 1/2a | DUP 1042 | 1296 |
Figure 1.
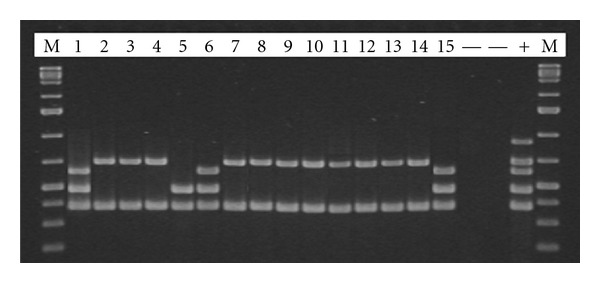
Multiplex PCR on isolated L. monocytogenes strains. M: marker; line 1: sample no. 1; lines 2-3-4: sample no. 7; line 5: sample no. 3; line 6: sample no. 5; lines 7-8-9: sample no. 2; lines 10-11: sample no. 8; lines 12-13: sample no. 9; line 14: sample no. 12; line 15: sample no. 10.
The 15 strains of L. monocytogenes analysed came from 4 different serotypes: the highest percentage was related to serotype 1/2a (73,33%), followed by serotype 4b (13,33%), 1/2b (6,67%) and 4d (6,67%).
Ribotyping of L.monocytogenes colonies in microbiological tests (see Table 4) has yielded the broad variation of isolated strains, identifying 15 particular strains on the basis of species. The most diffuse type was DUP-1042, found in 9 samples (60%). DUP-20243, DUP-1043, and -DUP1052 were characterised in the strains examined, in 3 (20%), 2 (13.33%) and 1 (6.66%) sample, respectively.
In terms of ribogroup distribution, RG-568 was the most diffuse, found in 9 of the 15 samples (60%), and the other ribogroups were each found in only one sample, except RG-1296 which was found in 2 samples. Data from phylogenetic analysis on the restriction pattern (Figure 2) yielded a classification of three main strain groups identified with ribogroup 568, being highly phylogenetically related, in turn distant, though still phylogenetically related to the 4 strains grouped in the upper part of the dendrogram. The third and last group comprises of only one sample, no. 3. It was analysed after a shelf-life at a temperature of 4°C being very different from rest of the L. monocytogenes that were characterized.
Figure 2.
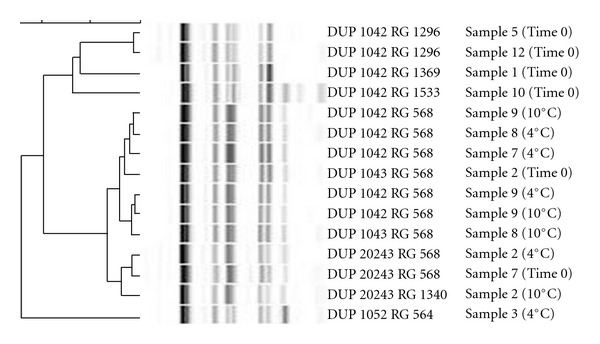
Phylogenetic distances between the various isolated L. monocytogenes strains.
4. Discussion
The data demonstrate that there is a rather low probability of L.monocytogenes exceeding the 100 cfu/g limit in the RTE seafood distributed in the Veneto Region, which would at any rate be associated with improper storage of the product or with thermal abuse. In fact, of the 3 positive samples out of the 38, 2 had been stored at 10°C in the last third period of their lifecycle and only one was defective at the time it was collected.
Analysis conducted at the end of shelf-life provides additional confirmation of the above-mentioned data.
The total charge of psychrophiles and of lactic bacteria found in the three positive samples was high, differently from Jameson's theory which affirms that the lactic charge should develop a sort of microbial competition against L. monocytogenes [35].
It is worth noting that 9 semi-preserves resulted positive at the qualitative microbiological test for L. monocytogenes (found in 25 g).
If at the end of shelf-life the above mentioned 9 samples had a <5 cfu level, the producer would still have to demonstrate, with appropriate studies and challenge tests, that L. monocytogenes will not grow and multiply.
Our report reiterates that smoked and fresh salmon are particularly hazardous products.
In case of marinated products, just three samples had viable L. monocytogenes. At the end of shelf-life however no L. monocytogenes growth was detected. Differently from salmon, these products have much lower a w and pH values, a factor that no doubt affects the growth of L. monocytogenes [36]. Moreover, smoked salmon production will have L. monocytogenes in the raw ingredient which cannot be eliminated in the phases leading to final packaging but can only be contained [37]. Seafood salads instead, since they are made with precooked raw ingredients, do not have L. monocytogenes. If there is any L. monocytogenes present in the end product, it implies that contamination occurred after the processing phases, after marinating.
The higher number of positive samples in the molecular biology tests and the inability to isolate culture underscore the presence of viable not culturable (VNC) organisms or no longer viable cells [38].
The PCR test compared to the culture is greatly targeted and more sensitive. Used on first enrichment broth it may prove useful in routine laboratory work because at least in PCR-negative cases it does not require microbiological tests.
The samples examined, all being positive to PCR tests for Listeria, confirm that these kinds of products are often contaminated with Listeria, but that such condition rarely evolves into hazardous L. monocytogenes concentrations.
In all the samples with unfavourable a w and pH values for L. monocytogenes growth, the PCR test for L. monocytogenes was negative, confirming that even if the product was contaminated in the production phase, L. monocytogenes would be overwhelmed by the product's unfavourable environment.
The M-PCR method adopted was rapid, reproducible, and cost-effective. It can therefore be used in any equipped laboratory to perform molecular analysis. Its use however is not suggested to identify rare serotypes. For a serotype-specific analysis of isolated strains the traditional sero-agglutination method is recommended. The latter is no doubt less reproducible, more expensive, and requires specially trained staff, but it enables the detection of rare serotypes which in the years to come may be more frequently associated to cases of listeriosis.
It is striking that 10 of the 11 L. monocytogenes strains isolated from salmon belonged to serotype 1/2a, as already confirmed by other scientific sources [39–41].
The agglutination method to confirm the serotypes of the single serogroups detected with M-PCR has highlighted a rare strain among the isolates belonging to the 4d serotype.
Rare serotypes have been linked to an epidemic outbreak, as confirmed in a Finnish study by Maijala et al. in 2000 [11]. An epidemic case of listeriosis caused by the strain L. monocytogenes serotype 3a, isolated in packaged butter, was described therein. The epidemic outbreak affected 25 people, causing 6 deaths.
Figure 2 shows the variability of strains found in the environment underscoring the need to develop methods that can assess the characterisation of pathogenicity, since same strains may correspond to different levels of pathogenicity.
Literature, in fact, documents L. monocytogenes and considers it a ubiquitous microorganism capable of adapting to different environmental conditions [42]. However, despite the various reports on genotyping and characterization [43, 44], there is still very little empirical evidence on the correlation between its presence in food and the ensuing pathologies its consumption generates in humans. This ought to encourage a more ample use of molecular characterization to provide information, even at the epidemiological level, on the distribution of L. monocytogenes in the environment, which though it maintains phenotypic homogeneity, undergoes ongoing phenomena of clonal evolution with small, yet significant, genome variations.
In addition, it is interesting to note that the 4 strains grouped in the upper part of the dendrogram identified as RG 1296, RG 1369, and RG 1533 were all isolated in samples analysed at time 0. While ribogroups 564, 568, and 1340 were instead isolated in samples analysed both at the beginning as well as at the end of the shelf-life period, this suggests that the latter may be more resistant in time to different storage conditions.
Because of the relatively small number of tested samples, this work should be considered as a preliminary study. Results should be confirmed throught further studies on different kind of products and increasing the number of samples.
5. Conclusions
Temperature plays a key role in preventing the growth of L. monocytogenes. In fact, the refrigeration chain is not always respected especially in case of products with a few weeks of shelf-life, as the products in this report, which may be subjected to temperature changes which do not bring about any evident organoleptic alterations. Production companies must therefore bear this in mind when certifying their products, using challenge tests to establish how safe a product is in terms of L. monocytogenes. They must also foresee temperature tests, in particular for products such as smoked salmon, traditionally from Northern European countries having lower average temperatures than countries like Italy also having less difficulty in managing the cold chain.
Products with a w and pH characteristics that favour L. monocytogenes growth were the only ones to result positive to microbiological and PCR tests in this work, in support of the potential hazard of these products.
The highest percentage related to serotype 1/2a confirm similar investigations in seafood products and the genetic characterization using ribotyping demonstrates the genomic variability of L. monocytogenes strains also in this kind of food. This distinctive features could be interesting related to L. monocytogenes survival capacity under different storage conditions.
Nearly 30 years have gone by since Canadians Schelch [45] demonstrated that L. monocytogenes can infect human via contaminated food. The significant number of studies conducted since then has provided much data on this pathogen. Currently a lot is known about pathogenesis in humans and about its ability to resist and grow in different types of food.
Rightfully defined as an “evolving pathogen” by Bryan [46], it changes its phenotypic and genotypic characteristics under outside-induced stress conditions, with acidic substances, new additives, and with the innovative food technologies applied by food industries. The latter, when producing all the latest food products, have to constantly monitor them for this versatile pathogen. In tandem education campaigns geared at the consumers who may be exposed to L. monocytogenes must provide general rules and guidelines for proper food storage and for food preparation in the domestic environment.
References
- 1.Goldfine H, Shen H. Listeria monocytogenes, Pathogenesis and Host Response. New York, NY, USA: Springer; 2007. [Google Scholar]
- 2.Ramaswamy V, Cresence VM, Rejitha JS, et al. Listeria—review of epidemiology and pathogenesis. Journal of Microbiology, Immunology and Infection. 2007;40(1):4–13. [PubMed] [Google Scholar]
- 3.De Cesare A, Manfreda G. Use of automated ribotyping for epidemiological investigations. Annals of Microbiology. 2002;52:181–190. [Google Scholar]
- 4.De Cesare A, Manfreda G, Macrì M, Cantoni C. Application of automated ribotyping to support the evaluation of the Listeria monocytogenes source in a Taleggio producing plant and to predict the risk human health linked to the accidental sale of contaminated cheese. Archivio Veterinario Italiano. 2006;57:231–240. [Google Scholar]
- 5.Franciosa G, Tartaro S, Wedell-Neergaard C, Aureli P. Characterization of Listeria monocytogenes strains involved in invasive and noninvasive listeriosis outbreaks by PCR-based fingerprinting techniques. Applied and Environmental Microbiology. 2001;67(4):1793–1799. doi: 10.1128/AEM.67.4.1793-1799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piana A, Are R, Orrù M, et al. Listeria monocytogenes meningoencephalitis: molecular methods for diagnosis and for monitoring the response to chemotherapy. Italian Journal of Public Health. 2005;2:29–34. [Google Scholar]
- 7.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes and Infection. 2007;9(10):1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Duffes F, Corre C, Leroi F, Dousset X, Boyaval P. Inhibition of Listeria monocytogenes by in situ produced and semipurified bacteriocins of Carnobacterium spp. on vacuum-packed, refrigerated cold-smoked salmon stored at 4 °C and 8 °C. Journal of Food Protection. 1999;62(12):1394–1403. doi: 10.4315/0362-028x-62.12.1394. [DOI] [PubMed] [Google Scholar]
- 9.European Food Safety Authority (EFSA) The community summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European union in 2008. European Food Safety Authority. 2010;8(1, article 1496) doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Valk H, Vaillant V, Jacquet C, et al. Two consecutive nationwide outbreaks of listeriosis in France, October 1999—February 2000. American Journal of Epidemiology. 2001;154(10):944–950. doi: 10.1093/aje/154.10.944. [DOI] [PubMed] [Google Scholar]
- 11.Maijala R, Lyytikäinen O, Johansson T, et al. Exposure of Listeria monocytogenes within an epidemic caused by butter in Finland. International Journal of Food Microbiology. 2001;70(1-2):97–109. doi: 10.1016/s0168-1605(01)00532-3. [DOI] [PubMed] [Google Scholar]
- 12.Bille J, Blanc DS, Schmid H, et al. Outbreak of human listeriosis associated with tomme cheese in northwest Switzerland, 2005. Euro Surveillance. 2006;11(6):91–93. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=633/ [PubMed] [Google Scholar]
- 13.Yde M, Botteldoorn N, Bertrand S, Collard JM, Dierick K. Microbiological and molecular investigation of an increase of human listeriosis in Belgium, 2006-2007. Eurosurveillance. 2010;15(6):p. 5. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19482/ [PubMed] [Google Scholar]
- 14.Jensen AK, Ethelberg S, Smith B, et al. Substantial increase in listeriosis, Denmark 2009. Eurosurveillance. 2010;15(12) http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19522/ [PubMed] [Google Scholar]
- 15.Eklund MW, Poysky FT, Paranjpye RN, Lashbrook LC, Peterson ME, Pelroy GA. Incidence and sources of Listeria monocytogenes in cold-smoked fishery products and processing plants. Journal of Food Protection. 1995;58(5):502–508. doi: 10.4315/0362-028X-58.5.502. [DOI] [PubMed] [Google Scholar]
- 16.Gandhi M, Chikindas ML. Listeria: a foodborne pathogen that knows how to survive. International Journal of Food Microbiology. 2007;113(1):1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Graves LM, Swaminathan B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. International Journal of Food Microbiology. 2001;65(1-2):55–62. doi: 10.1016/s0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 18.Huss HH, Jørgensen LV, Vogel BF. Control options for Listeria monocytogenes in seafoods. International Journal of Food Microbiology. 2000;62(3):267–274. doi: 10.1016/s0168-1605(00)00347-0. [DOI] [PubMed] [Google Scholar]
- 19.Rørvik LM, Caugant DA, Yndestad M. Contamination pattern of Listeria monocytogenes and other Listeria spp. in a salmon slaughterhouse and smoked salmon processing plant. International Journal of Food Microbiology. 1995;25(1):19–27. doi: 10.1016/0168-1605(94)00080-p. [DOI] [PubMed] [Google Scholar]
- 20.Ross T, Dalgaard P, Tienungoon S. Predictive modelling of the growth and survival of Listeria in fishery products. International Journal of Food Microbiology. 2000;62(3):231–245. doi: 10.1016/s0168-1605(00)00340-8. [DOI] [PubMed] [Google Scholar]
- 21.Ryser ET, Marth EH. Listeria, Listeriosis, and Food Safety. New York, NY, USA: Marcel Dekker; 2007. [Google Scholar]
- 22.Vescovo M, Scolari G, Zacconi C. Inhibition of Listeria innocua growth by antimicrobial-producing lactic acid cultures in vacuum-packed cold-smoked salmon. Food Microbiology. 2006;23(7):689–693. doi: 10.1016/j.fm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Brett MSY, Short P, McLauchlin J. A small outbreak of listeriosis associated with smoked mussels. International Journal of Food Microbiology. 1998;43(3):223–229. doi: 10.1016/s0168-1605(98)00116-0. [DOI] [PubMed] [Google Scholar]
- 24.Ericsson H, Eklow A, Danielsson-Tham ML, et al. An outbreak of listeriosis suspected to have been caused by rainbow trout. Journal of Clinical Microbiology. 1997;35(11):2904–2907. doi: 10.1128/jcm.35.11.2904-2907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miettinen MK, Siitonen A, Heiskanen P, Haajanen H, Bjorkroth KJ, Korkeala HJ. Molecular epidemiology of an outbreak of febrile gastroenteritis caused by Listeria monocytogenes in cold-smoked rainbow trout. Journal of Clinical Microbiology. 1999;37(7):2358–2360. doi: 10.1128/jcm.37.7.2358-2360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornu M, Beaufort A, Rudelle S, et al. Effect of temperature, water-phase salt and phenolic contents on Listeria monocytogenes growth rates on cold-smoked salmon and evaluation of secondary models. International Journal of Food Microbiology. 2006;106(2):159–168. doi: 10.1016/j.ijfoodmicro.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Gombas DE, Chen Y, Clavero RS, Scott VN. Survey of Listeria monocytogenes in ready-to-eat foods. Journal of Food Protection. 2003;66(4):559–569. doi: 10.4315/0362-028x-66.4.559. [DOI] [PubMed] [Google Scholar]
- 28.Jørgensen LV, Huss HH. Prevalence and growth of Listeria monocytogenes in naturally contaminated seafood. International Journal of Food Microbiology. 1998;42(1-2):127–131. doi: 10.1016/s0168-1605(98)00071-3. [DOI] [PubMed] [Google Scholar]
- 29.Garrido V. Quantitative microbiological risk assessment: listeriosis and ready to eat products approach [Ph.D. thesis] Pamplona, Spain: University of Navarra; 2008. [Google Scholar]
- 30.Marklinder IM, Lindblad M, Eriksson LM, Finnson AM, Lindqvist R. Home storage temperatures and consumer handling of refrigerated foods in Sweden. Journal of Food Protection. 2004;67(11):2570–2577. doi: 10.4315/0362-028x-67.11.2570. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence LM, Gilmour A. Incidence of Listeria spp. and Listeria monocytogenes in a poultry processing environment and in poultry products and their rapid confirmation by multiplex PCR. Applied and Environmental Microbiology. 1994;60(12):4600–4604. doi: 10.1128/aem.60.12.4600-4604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. Journal of Applied Bacteriology. 1991;70(5):372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 33.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. Journal of Clinical Microbiology. 2004;42(8):3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeliger HPR, Hohne K. Chapter II serotyping of Listeria monocytogenes and related species. Methods in Microbiology. 1979;13:31–49. [Google Scholar]
- 35.Françoise L. Occurrence and role of lactic acid bacteria in seafood products. Food Microbiology. 2010;27(6):698–709. doi: 10.1016/j.fm.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Andrighetto C, Lombardi A, Ferrati M, Guidi A, Corrain C, Arcangeli G. Lactic acid bacteria biodiversity in Italian marinated seafood salad and their interactions on the growth of Listeria monocytogenes . Food Control. 2009;20(5):462–468. [Google Scholar]
- 37.Porsby CH, Vogel BF, Mohr M, Gram L. Influence of processing steps in cold-smoked salmon production on survival and growth of persistent and presumed non-persistent Listeria monocytogenes . International Journal of Food Microbiology. 2008;122(3):287–295. doi: 10.1016/j.ijfoodmicro.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Dreux N, Albagnac C, Federighi M, Carlin F, Morris CE, Nguyen-The C. Viable but non-culturable Listeria monocytogenes on parsley leaves and absence of recovery to a culturable state. Journal of Applied Microbiology. 2007;103(4):1272–1281. doi: 10.1111/j.1365-2672.2007.03351.x. [DOI] [PubMed] [Google Scholar]
- 39.Corcoran D, Clancy D, O’Mahony M, et al. Comparison of Listeria monocytogenes strain types in Irish smoked salmon and other foods. International Journal of Hygiene and Environmental Health. 2006;209(6):527–534. doi: 10.1016/j.ijheh.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Dauphin G, Ragimbeau C, Malle P. Use of PFGE typing for tracing contamination with Listeria monocytogenes in three cold-smoked salmon processing plants. International Journal of Food Microbiology. 2001;64(1-2):51–61. doi: 10.1016/s0168-1605(00)00442-6. [DOI] [PubMed] [Google Scholar]
- 41.Gudmundsdóttir S, Gudbjörnsdóttir B, Lauzon HL, Einarsson H, Kristinsson KG, Kristjánsson M. Tracing Listeria monocytogenes isolates from cold-smoked salmon and its processing environment in Iceland using pulsed-field gel electrophoresis. International Journal of Food Microbiology. 2005;101(1):41–51. doi: 10.1016/j.ijfoodmicro.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Acciari VA, Torresi M, Migliorati G, Di Giannatale E, Semprini P, Prencipe V. Characterisation of Listeria monocytogenes strains isolated from soft and semi-soft cheeses sampled in a region of Italy. Veterinaria Italiana. 2011;47(1):15–23. [PubMed] [Google Scholar]
- 43.Garrido V, Torroba L, Garcia-Jalón I, Vitas AI. Surveillance of listeriosis in Navarre, Spain, 1995–2005—epidemiological patterns and characterisation of clinical and food isolates. Euro Surveillance. 2008;13(49) [PubMed] [Google Scholar]
- 44.Kérouanton A, Marault M, Petit L, Grout J, Dao TT, Brisabois A. Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. Journal of Microbiological Methods. 2010;80(2):134–137. doi: 10.1016/j.mimet.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Schlech WF, Lavigne PM, Bortolussi RA. Epidemic listeriosis—evidence for transmission by food. The New England Journal of Medicine. 1983;308(4):203–206. doi: 10.1056/NEJM198301273080407. [DOI] [PubMed] [Google Scholar]
- 46.Bryan FL. Reflections on a career in public health: evolving foodborne pathogens, environmental health, and food safety programs. Journal of Environmental Health. 2002;65(5):14–24. [PubMed] [Google Scholar]


