Abstract
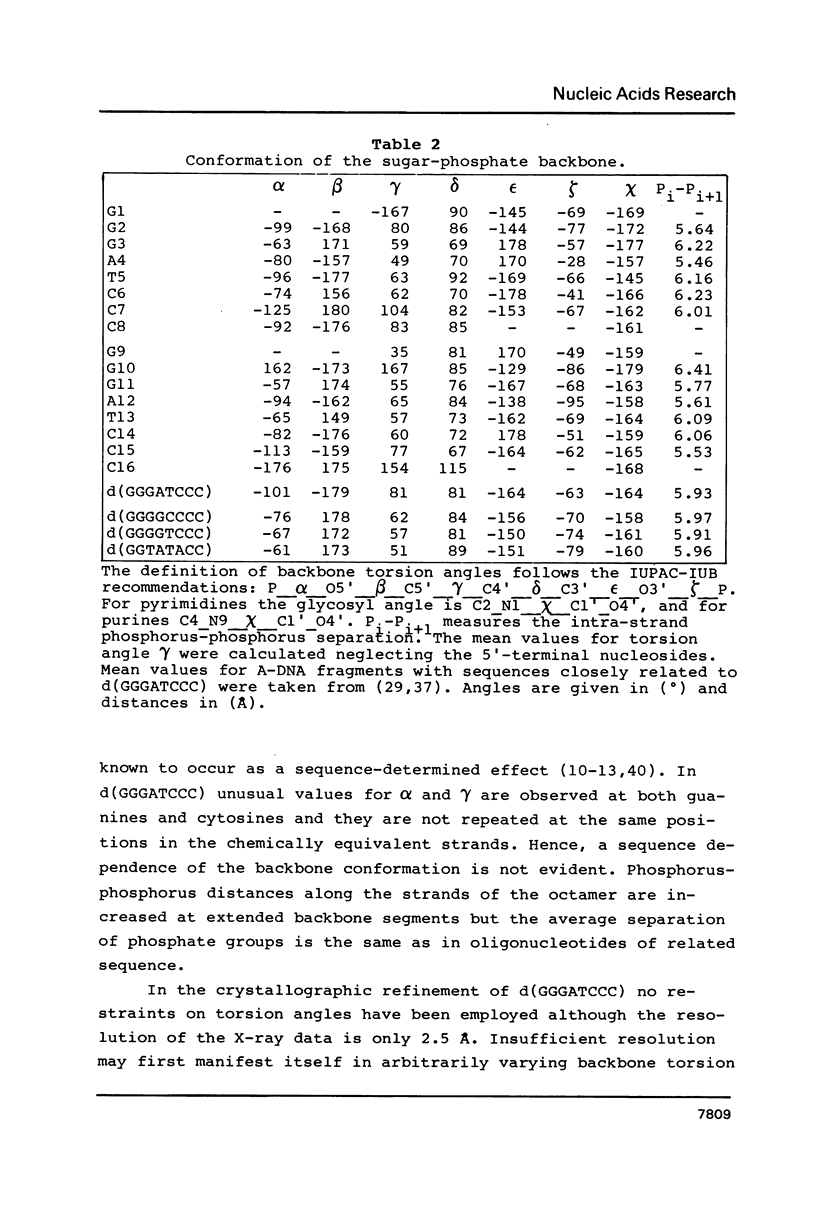
The structure of the self-complementary octamer d(GGGATCCC) has been analysed by single crystal X-ray diffraction methods at a nominal resolution of 2.5 A. With acceptable stereochemistry of the model the crystallographic R factor was 16.6% after restrained least-squares refinement. In the crystal, d(GGGATCCC) forms an A-DNA double helix with slightly varying conformation of the two strands. The average displacement of the base pairs from the helix axis is unusually large and is accompanied by pronounced sliding of the base pairs along their long axes at all dinucleotide steps except for the central AT. With 12 base pairs per complete turn the helix is considerably underwound. As observed with most oligodeoxyribonucleotides analysed by X-ray crystallography so far, the octamer displays reduced base pair tilt, increased rise per base pair and a more open major groove compared with canonical A-DNA. We propose that, based on these parameters, three A-helical sub-families may be defined; d(GGGATCCC) then is a representative of the class with intermediate tilt, rise, and major groove width.
Full text
PDF





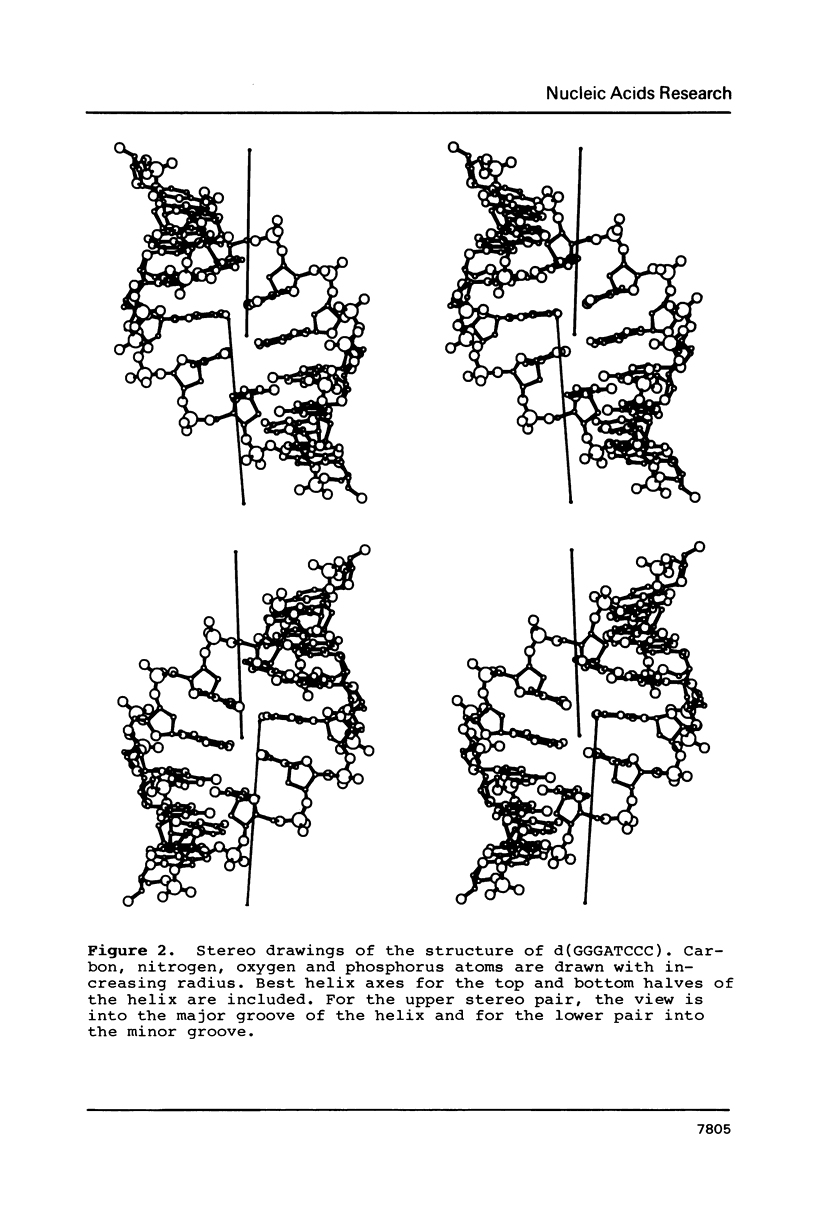
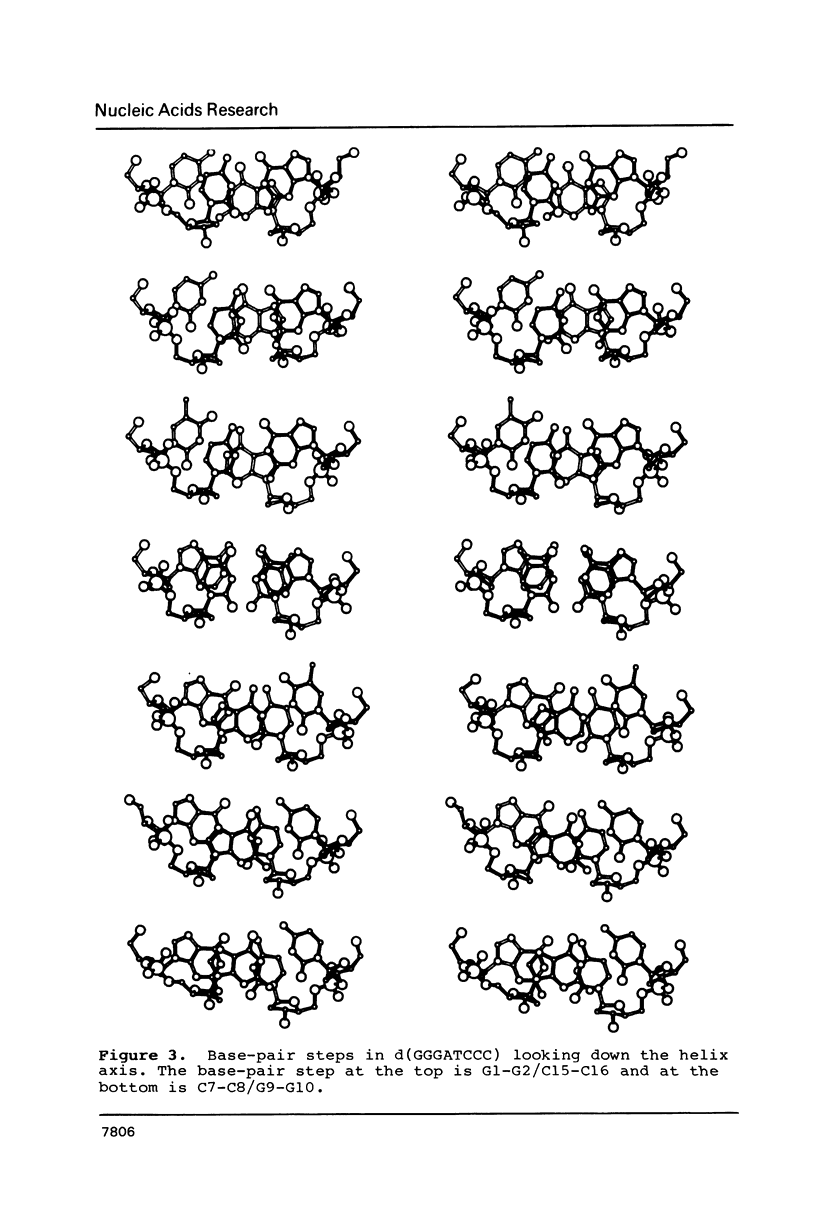
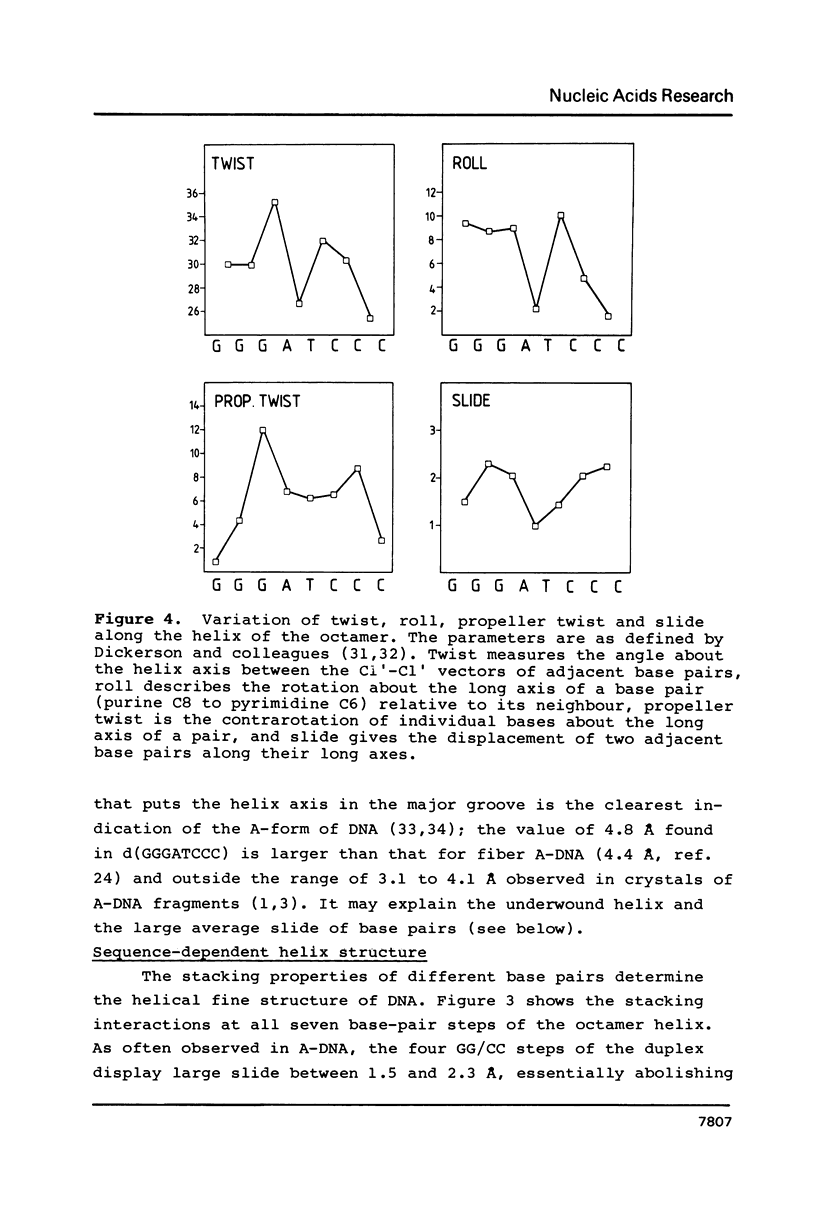








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C., Walker J. K., Wang M. DNA secondary structures: helices, wrinkles, and junctions. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):53–65. doi: 10.1101/sqb.1983.047.01.008. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Letter: The structure of polydeoxyguanylic acid with polydeoxycytidylic acid. J Mol Biol. 1974 Sep 15;88(2):551–552. doi: 10.1016/0022-2836(74)90502-6. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bram S. Polynucleotide polymorphism in solution. Nat New Biol. 1971 Oct 6;233(40):161–164. doi: 10.1038/newbio233161a0. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. A base-centred explanation of the B-to-A transition in DNA. J Mol Biol. 1984 Sep 25;178(3):773–782. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Conner B. N., Yoon C., Dickerson J. L., Dickerson R. E. Helix geometry and hydration in an A-DNA tetramer: IC-C-G-G. J Mol Biol. 1984 Apr 25;174(4):663–695. doi: 10.1016/0022-2836(84)90089-5. [DOI] [PubMed] [Google Scholar]
- Dattagupta J. K., Fujiwara T., Grishin E. V., Lindner K., Manor P. C., Pieniazek N. J., Saenger R., Suck D. Crystallization of the fungal enzyme proteinase K and amino acid composition. J Mol Biol. 1975 Sep 15;97(2):267–271. doi: 10.1016/s0022-2836(75)80039-8. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Goodsell D. S., Kopka M. L., Pjura P. E. The effect of crystal packing on oligonucleotide double helix structure. J Biomol Struct Dyn. 1987 Dec;5(3):557–579. doi: 10.1080/07391102.1987.10506413. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Haran T. E., Shakked Z., Wang A. H., Rich A. The crystal structure of d(CCCCGGGG): a new A-form variant with an extended backbone conformation. J Biomol Struct Dyn. 1987 Oct;5(2):199–217. doi: 10.1080/07391102.1987.10506390. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Lauble H., Frank R., Blöcker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987 Nov 25;15(22):9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Jain S., Zon G., Sundaralingam M. The potentially Z-DNA-forming sequence d(GTGTACAC) crystallizes as A-DNA. J Mol Biol. 1987 Sep 5;197(1):141–145. doi: 10.1016/0022-2836(87)90616-4. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Kennard O. The crystal structure of d(G-G-G-G-C-C-C-C). A model for poly(dG).poly(dC). J Mol Biol. 1985 Jun 5;183(3):385–396. doi: 10.1016/0022-2836(85)90009-9. [DOI] [PubMed] [Google Scholar]
- Mei H. Y., Barton J. K. Tris(tetramethylphenanthroline)ruthenium(II): a chiral probe that cleaves A-DNA conformations. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1339–1343. doi: 10.1073/pnas.85.5.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Ikehara M., Söll D. Recent developments in the chemical synthesis of polynucleotides. Nucleic Acids Res. 1982 Nov 11;10(21):6553–6570. doi: 10.1093/nar/10.21.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peticolas W. L., Wang Y., Thomas G. A. Some rules for predicting the base-sequence dependence of DNA conformation. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2579–2583. doi: 10.1073/pnas.85.8.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich D., Haran T., Eisenstein M., Shakked Z. Structures of the mismatched duplex d(GGGTGCCC) and one of its Watson-Crick analogues d(GGGCGCCC). J Mol Biol. 1988 Mar 5;200(1):151–161. doi: 10.1016/0022-2836(88)90340-3. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Cruse W. B., Egert E., Kennard O., Sala G., Salisbury S. A., Viswamitra M. A. Crystalline A-dna: the X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc R Soc Lond B Biol Sci. 1981 Nov 24;213(1193):479–487. doi: 10.1098/rspb.1981.0076. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Kennard O., Cruse W. B., Salisbury S. A., Viswamitra M. A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J Mol Biol. 1983 May 15;166(2):183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D. The effect of the base sequence on the fine structure of the DNA double helix. Prog Biophys Mol Biol. 1986;47(3):159–195. doi: 10.1016/0079-6107(86)90013-1. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., Rich A. Molecular structure of the octamer d(G-G-C-C-G-G-C-C): modified A-DNA. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3968–3972. doi: 10.1073/pnas.79.13.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]