Abstract
Dendritic cells (DCs) are phagocytic professional antigen-presenting cells that can prime naive T cells and initiate anti-bacterial immunity. However, several pathogenic bacteria have developed virulence mechanisms to impair DC function. For instance, Salmonella enterica serovar Typhimurium can prevent DCs from activating antigen-specific T cells. In addition, it has been described that the Salmonella Pathogenicity Island 1 (SPI-1), which promotes phagocytosis of bacteria in non-phagocytic cells, can suppress this process in DCs in a phosphatidylinositol 3-kinase (PI3K) -dependent manner. Both mechanisms allow Salmonella to evade host adaptive immunity. Recent studies have shown that IgG-opsonization of Salmonella can restore the capacity of DCs to present antigenic peptide–MHC complexes and prime T cells. Interestingly, T-cell activation requires Fcγ receptor III (FcγRIII) expression over the DC surface, suggesting that this receptor could counteract both antigen presentation and phagocytosis evasion by bacteria. We show that, despite IgG-coated Salmonella retaining its capacity to secrete anti-capture proteins, DCs are efficiently capable of engulfing a large number of IgG-coated bacteria. These results suggest that DCs employ another mechanism to engulf IgG-coated Salmonella, different from that used for free bacteria. In this context, we noted that DCs do not employ PI3K, actin cytoskeleton or dynamin to capture IgG-coated bacteria. Likewise, we observed that the capture is an FcγR-independent mechanism. Interestingly, these internalized bacteria were rapidly targeted for degradation within lysosomal compartments. Hence, our results suggest a novel mechanism in DCs that does not employ PI3K/actin cytoskeleton/dynamin/FcγRs to engulf IgG-coated Salmonella, is not affected by anti-capture SPI-1-derived effectors and enhances DC immunogenicity, bacterial degradation and antigen presentation.
Keywords: dendritic cell, Fcγ receptors, IgG-opsonization, Pathogenicity Island 1, phagocytosis, Salmonella typhimurium
Introduction
The immune system recognizes pathogenic bacteria and protects the host from tissue damage induced by their mechanisms of virulence.1–6 However, several bacteria have developed various strategies to evade clearance by the immune system.7–10 An example of a highly virulent bacterium capable of subverting host immunity is Salmonella enterica serovar Typhimurium, a Gram-negative bacterium that causes typhoid-like disease in mice and gastroenteritis in humans.11–14Salmonella Typhimurium can disseminate systemically in immune competent mice by suppressing the establishment of protective anti-bacterial immunity. It is thought that the capacity of S. Typhimurium to impair dendritic cell (DC) function contributes to preventing the onset of a protective adaptive immune response against this pathogen.1,15–20 Previous studies have shown that S. Typhimurium suppresses DC activity by inhibiting both phagocytosis of bacteria and the priming of naive T cells.11,18,19,21–24 Whereas phagocytosis seems to be targeted in a phosphatidylinositol 3-kinase (PI3K) -dependent manner by effectors encoded within the Salmonella Pathogenicity Island 1 (SPI-1), inhibition of T-cell priming is thought to be mediated by SPI-1-derived and SPI-2-derived proteins.18,22–25
On the other hand, opsonization of bacteria by S. Typhimurium-specific IgG restores the capacity of DCs to process and present antigenic peptide–MHC complexes on their surface, which prime bacteria-specific T cells after challenge with virulent S. Typhimurium.23,24 Interestingly, surface expression of Fcγ receptor III (FcγRIII; CD16) in DCs is required in this process.24 However, the question of how IgG-bacterial opsonization enhances the immunogenicity of S. Typhimurium-challenged DCs remains obscure. Although it is well established that IgG promotes the phagocytosis of foreign bodies into different cell types,2,3,26,27 whether IgG can counteract the secretion of Salmonella modulatory effectors or interfere with its capacity to evade capture in DCs remains to be evaluated.
To better understand how IgG opsonization contributes to restoring the immunogenicity of DCs challenged with virulent Salmonella, we evaluated direct and indirect consequences of IgG-opsonization on the virulence of this pathogenic bacterium. We observed that IgG-opsonization enhanced DC capture of Salmonella, despite IgG-coated S. Typhimurium (IgG-ST) retaining their capacity to secrete SPI-1-derived effectors. Accordingly, IgG-ST were observed in large numbers within these cells, being rapidly routed for lysosomal degradation. In agreement with this observation, enhanced bacterial capture mediated by IgG promoted the presentation of antigens expressed by Salmonella to antigen-specific T cells, both in vivo and in vitro. Moreover, we observed that Salmonella capture promoted by IgG is an actin/PI3K/dynamin-independent process, which is in contrast to the requirement of these elements for the entry of free Salmonella into DCs. These observations suggest that (i) IgG-ST are internalized in an FcγRIII-independent manner and (ii) engagement of this receptor contributes mainly to promotion of degradation rather than to capture of the pathogen by DCs. These data provide new insights into the mechanism by which IgG-opsonization restores DC capacity to prime T cells upon challenge with virulent Salmonella.
Material and methods
Reagents and antibodies
Reagents used in this study were saponin (Sigma-Aldrich, St Louis, MO), Triton-X100 (Sigma-Aldrich), bacterial culture media LB broth (MO BIO Laboratories, Carlsbad, CA), sucrose (Merck, Darmstadt, Germany), agar (Merck), wortmannin (Wm; Sigma-Aldrich), cytochalasin D (CytD; Sigma-Aldrich), 3-methyladenine (Tocris Bioscience, Minneapolis, MN), Pi 103 hydrochloride (Tocris Bioscience), MitMab (Tocris Bioscience), OctMab (Tocris Bioscience), carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) and granulocyte–macrophage colony-stimulating factor (GM-CSF; Peprotech, Rocky Hill, NJ). Antibodies used in this study were the following: hamster anti-CD11c-allophycocyanin (APC) (clone HL3; BD Pharmingen, San Diego, CA), rat anti-CD16/CD32-phycoerythrin (PE) (clone 2.4G2; BD Pharmingen), rat anti-CD11b-PE (clone M1/70; BD Pharmingen), rat anti-class I MHC (H-2Kb) (clone AF6-88.5; BD Pharmingen), rat anti-class II MHC (IA/IE) (clone M5/114.15.2; BD Pharmingen), polyclonal goat anti-mouse IgG-APC (Invitrogen, Carlsbad, CA), polyclonal goat anti-rabbit IgG-APC (Invitrogen), polyclonal rabbit anti-Lamp1 (Abcam, Cambridge, MA), polyclonal goat anti-rabbit-AlexaFluor 555 (Invitrogen), monoclonal anti-S. Typhimurium lipopolysaccharide (IgG1 Clone 1E6; Advanced Immuno-Chemical Inc., Long Beach, CA), serum polyclonal anti-Salmonella (Antisera group O4, Ref 294401; Denka Seiken, Tokyo, Japan), mIgG1 anti-N protein of respiratory syncytial virus (mIgG1, Clone 8E4/A7 generated in BALB/c mice), Salmonella O Antiserum Factor 4 (Ref 226591; BD Pharmingen) and blocking rat anti-CD16/CD32 (clone 2.4G2; BD Pharmingen).
Bacterial strains and growth conditions
Virulent S. Typhimurium (ST, 14028s) was obtained from the American Type Culture Collection (Manassas, VA) and the SPI-1 mutant strain [ST(ΔInvC)], the green fluorescent protein (GFP) -expressing bacteria [ST(GFP) and ST(ΔInvC:GFP)], the ovalbumin (OVA) -expressing bacteria ST(OVA) and the ST(ΔInvC:OVA) were generated as described previously.23 Bacteria were grown overnight at 37° in LB media, with antibiotics when required (100 μg/ml ampicillin for GFP-expressing and OVA-expressing bacteria) and constant agitation (180 rpm) on a bacteria shaker (Labtech, Namyangju, Korea). Then, bacteria were sub-cultured at 1/1000 dilution in LB broth and incubated with constant agitation (180 rpm) at 37°. Salmonella was grown until exponential phase was reached (optical density at 600 nm 0·4–0·6), pelleted (5900 g × 6 min at 4°) and resuspended in cold PBS. Before infecting DCs, bacteria were incubated for 10 min at 37° to improve their virulence, as previously described18.
IgG-opsonization of S. Typhimurium
Wild-type and GFP-expressing S. Typhimurium strains were grown as described above and re-suspended in cold PBS, adding 2 μl of the monoclonal antibody anti-lipopolysaccharide-S. Typhimurium (IgG1 Clone 1E6; Advanced Immuno-Chemical Inc.) or 15 μl serum polyclonal anti-Salmonella (Antisera group O4, Ref 294401; Denka Seiken). As controls, the S. Typhimurium was incubated either with mIgG1 anti-N protein of respiratory syncytial virus (mIgG1, Clone 8E4/A7 generated in BALB/c mice) or Salmonella O Antiserum Factor 4 (Ref 226591; BD Pharmingen). Samples were vigorously vortexed and incubated for 1–2 hr at 4°, evaluating every 20 min for the formation of immune complexes using a Neubauer chamber (see Supplementary material, Fig. S2). Before the infection of DCs, bacteria were incubated for 10 min at 37° to improve their virulence.
DC differentiation and culture
Dendritic cells were obtained from bone marrow progenitors of wild-type (WT), FcγRIIb−/− (CD32−/−), FcγRIII−/− (CD16−/−) or γ−/−/RIIb−/− (FcγRI−/− FcγRIIb−/− FcγRIII−/−) C57BL/6 mice. Mice were obtained from The Jackson Laboratories (Bar Harbor, ME) and maintained and manipulated in specific pathogen-free conditions. Triple FcγRs knockout mice (γ−/−/RIIb−/−) were obtained from Dr Jeffrey V. Ravetch’s laboratory. All animal work was performed according to institutional guidelines at the facility of the Pontificia Universidad Católica de Chile (Santiago, Chile). Bone marrow progenitors (1 × 106 to 1·5 × 106 cells) were seeded in 24-well plates and cultured in RPMI-1640, pH 7·2, containing 10% fetal bovine serum, 1 mm pyruvate, 2 mm glutamine, 1 mm non-essential amino acids and 10 ng/ml recombinant murine GM-CSF (Peprotech). The medium was also supplemented with gentamicin, fungizone, penicillin and streptomycin. Cells were incubated and differentiated for 5 days, replacing medium every 2 days. Differentiation of DCs was determined at day 5 by flow cytometry, where the expression of CD11c, CD11b, class I MHC, class II MHC and low-affinity FcγRs was evaluated (see Supplementary material, Fig. S1). The percentage of CD11c+ was consistently > 80%.
Gentamicin protection assays
The DCs were left untreated or were pre-treated with CytD (10 μm for 15 min) or Wm (100 nm for 1 hr) at 37° to inhibit the activity of the actin cytoskeleton and PI3K, respectively. The DCs were then pulsed with either S. Typhimurium or ST-IgG at a multiplicity of infection (MOI) equal to 25 in antibiotic-free RPMI-1640 medium, centrifuged at 400 g for 3 min and incubated for 1 hr at 37° (pulse). Then, cells were washed three times with cold PBS and treated with gentamicin (100 μg/ml) for 1 hr at 37° (chase). Once chase was completed, DCs were collected and their viability was determined by trypan blue exclusion in a Neubauer chamber. A volume containing 20 000 cells was pelleted and re-suspended in 0·1% Triton X-100-PBS. Cells were incubated for 15 min at room temperature and a volume equivalent to 2000 cells was seeded on LB plates. Colony-forming units (CFUs) were quantified and analysed after 12–16 hr incubation at 37°. The toxic effect that remaining antibiotics might have in cells was ruled out by replacing the antibiotic-containing medium with new antibiotic-free RPMI-1640 (see Supplementary material, Fig. S8 and Data S1).
Confocal microscopy assays
Bone marrow precursors from WT, FcγRIIb−/− (CD32−/−), FcγRIII−/− (CD16−/−) or γ−/−/RIIb−/− (FcγRI−/− FcγRIIb−/− FcγRIII−/−) mice were cultured over 12-mm diameter coverslips in 24-well plates for 5 days and differentiated into DCs with GM-CSF. When indicated, DCs were left untreated or treated either with CytD (10 μm for 15 min) or Wm (100 nm for 1 hr) to inhibit the activity of the actin cytoskeleton and PI3K, respectively. The DCs were pulsed either with ST(GFP) or ST(GFP)-IgG at an MOI equal to 25 bacteria per cell in antibiotic-free RPMI-1640 medium, centrifuged at 400 g for 3 min and incubated for 1 hr at 37° (pulse). Cells were then extensively washed with cold PBS and incubated for 1 hr with 100 μg/ml gentamicin (chase). Once the chase was completed, DCs were washed with PBS and fixed with 2%P-formaldehyde for 10 min at 4°. Then, coverslips were washed once with PBS and mounted with DABCO (1,4-diazabicyclo[2.2.2]octane; Sigma-Aldrich, St Louis, MO) for Confocal Microscopy analyses. Extracellular bacteria were quantified by Z-stack analyses [up to two bacteria per microscopy field (60 ×) were observed, data not shown].
Lysosomal attached membrane protein-1 (Lamp-1) detection in Salmonella-challenged DCs
Bone marrow precursors were cultured on 12-mm diameter coverslips in 24-well plates for 5 days and differentiated into DCs with GM-CSF. The DCs were pulsed either with ST(GFP) or ST(GFP)-IgG at an MOI equal to 25 for 1 hr in antibiotic-free RPMI-1640 medium (pulse), washed three times with cold PBS and treated with gentamicin (100 μg/ml) for 1 hr at 37° (chase). Then, DCs were washed once with cold PBS and fixed with 2% P-formaldehyde–5% sucrose for 10 min at 4°, washed three times with cold PBS, and permeabilized with 0·05% saponin in PBS at 4° for 10 min. Coverslips bearing DCs were placed inside a moist chamber with a hydrophobic surface (parafilm-coated) and stained with 50 μl rabbit anti-Lamp1 (1/100 in saponin 0·05%). Cells were incubated overnight at 4°, washed four times with 200 μl cold PBS and stained with 50 μl goat anti-rabbit-Alexa Fluor 555 (1/100 in saponin 0·05%) for 3 hr in the dark. Cells were then extensively washed with cold PBS and mounted for confocal microscopy analyses.
Flow cytometry assays
Wild-type, FcγRIIb−/− (CD32−/−), FcγRIII−/− (CD16−/−) or γ−/−/RIIb−/− (FcγRI−/− FcγRIIb−/− FcγRIII−/−) DCs were prepared as described above. When indicated, cells were left untreated or were treated with CytD (10 μm for 15 min) or Wm (100 nm for 1 hr). Then, DCs were pulsed either with ST(GFP) or ST(GFP)-IgG at an MOI equal to 25 for 1 hr in antibiotic-free RPMI-1640 medium (pulse), washed three times with cold PBS and treated with gentamicin (100 μg/ml) for 1 hr at 37° (chase). Dendritic cells were extensively washed with PBS, collected (5 × 105) in 300 μl cold PBS and stained with an anti-CD11c-APC antibody for 1 hr at 4°. Next, DCs were washed with PBS, pelleted at 300 g for 6 min at 4° and resuspended in P-formaldehyde 2%–sucrose 5% in PBS for flow cytometry analyses.
Haemolysis assays with sheep red blood cells
Salmonella Typhimurium, ST(ΔInvC) and ST-mIgG1 were prepared as described above. Fifty microlitres of sheep red blood cells (SRBC) were seeded in 96-well plates in PBS and 1 × 108 bacteria (ST, ST(ΔInvC) or ST-mIgG1) were added to each respective well. As a positive control (100% Lysis), 100 μl of 0·5% desoxycholate was used. Equal volumes of monoclonal antibody clone 1E6 (as compared with ST-mIgG1) and sterile PBS were added as haemolysis controls. Plates were spun at 2050 g at 20° for 10 min to promote contact between bacteria and SRBC. After centrifugation, plates were incubated for 5 hr at 37° with 5% CO2 and filled to a final volume equal to 250 μl. Then, plates were spun at 2050 g at 20° for 10 min to pellet both SRBC and bacteria. Supernatants were recovered and transferred to an ELISA plate. Levels of released haemoglobin were measured at 405 nm.
Flow cytometry analyses for determining bacteria–phagosome co-localization
Wild-type bone marrow precursors (5 × 106 cells per well) were differentiated for 5 days in six-well plates into DCs as described above and pulsed in antibiotic-free RPMI-1640 medium, either with ST(GFP) or ST(GFP)-mIgG1 at an MOI equal to 50. Then, cells were spun at 400 g for 6 min and incubated for 20 min at 37°. Once infection time was completed, DCs were extensively washed three times with cold PBS and incubated for 20 or 60 min with gentamicin-containing RPMI-1640 medium (100 μg/ml). The DCs were extensively washed with cold PBS and resuspended in 300 μl homogenization buffer (20 mm HEPES, 1 mm EDTA, 0·25 m sucrose, pH 7·2) and passed 10 times through a 25G sterile syringe to lyse the cells. Cell lysates were spun at 300 g for 6 min to pellet nuclei and non-lysed cells. Supernatants were carefully recollected, centrifuged at 13 400 g for 6 min and pellets were resuspended in 300 μl permeabilization buffer (RPMI-1640–saponin 0·2%), incubated for 5 min at room temperature and stained with anti-Lamp1-PE at 4° for 1 hr. Next, intracellular compartments were washed with the same permeabilization buffer and centrifuged at 13 400 rpm for 6 min. Pellets were resuspended in cold PBS and analysed by flow cytometry.
Antigen presentation assays
Bone-marrow-derived DCs were pulsed either with free (WT or ΔInvC mutant) or mIgG1-coated (WT or ΔInvC mutant) bacteria expressing OVA at MOI equal to 25 during 2 hr in antibiotic-free RPMI-1640 medium. The OVA-expressing strains were generated as previously described.18,23 Briefly, both Salmonella WT and ΔInvC mutant were transformed with pOVA, that is a pUC-derivate plasmid encoding the full-length sequence for chicken egg OVA under the lac promoter.23 Expression of OVA was evaluated by Western blotting (data not shown). Then, cells were extensively washed with PBS and incubated for 12 hr in 50 μg/ml gentamicin-containing RPMI-1640 medium. Viability of DCs was evaluated by trypan blue exclusion and cells were co-cultured for 20 hr with 1 × 105 OT-II cells, as previously described.24 Then, supernatants were collected and analysed for interleukin-2 and interferon-γ release by ELISA.18,22–24 In addition, OT-II activation was analysed by FACS.
In vivo experiment
T-cell receptor-α−/− C57BL/6 mice were adoptively transferred with 1·5 × 106 OT-II CD4+ T cells (intravenously) as previously described.28 OT-II CD4+ cells were purified by magnetic antibody cell sorting (> 90% purity) and stained with CFSE. After 24 hr, mice were intravenously injected with 1 × 106 CFU of either ST(OVA) or ST(OVA)-mIgG1 in 200 μl PBS. Other mice were given 20 μg OVA in 300 μl alum intraperitoneally and a control group was injected intravenously with PBS. Three days later, mice were killed and spleens were analysed for total cell count, FACS and bacterial load. To determine bacterial loads, spleen cells were treated and lysed with Triton-X100 0·1%-PBS for 15 min and then seeded over ampicillin-containing LB agar. For FACS, total splenocytes were stained with an anti-CD4 antibody (rat anti-mouse CD4-APC, clone RM4-5, cat. 553051; BD Pharmingen) and T-cell proliferation was followed by CFSE dilution in the CD4+ population. In addition, total splenocytes were stained with an anti-CD11c antibody to analyse total number of DCs.
Results
Salmonella opsonization with IgG increases bacterial entry to DCs
To approach the question as to how IgG restores the capacity of DCs to degrade and present Salmonella antigens to T cells, first we evaluated whether IgG-opsonization can counteract the capacity of this pathogen to reduce phagocytosis by DCs. For this, Salmonella was IgG-opsonized as described in the Material and methods (see Supplementary material, Fig. S2) and then DCs were pulsed for 1 hr either with free ST(GFP) or IgG-opsonized ST(GFP) and, after 60 min of incubation with gentamicin, infected cells were detected by flow cytometry. As shown in Fig. 1(a,b), the percentage of infected DCs was significantly higher for cells pulsed with either mIgG1-opsonized or pIgG-opsonized bacteria, compared with DCs challenged with free bacteria. Controls for mIgG1 and pIgG serum did not show relevant increases in infected cells. Similar results were obtained by confocal microscopy, which further showed that DCs pulsed with either mIgG1 or pIgG serum-opsonized Salmonella contained several intracellular bacteria per cell, whereas DCs pulsed with free Salmonella contained only a few (Fig. 1d–h). Cells containing bacteria were quantified and plotted as percentage of infected cells per field, showing significant differences between either mIgG1 or pIgG Serum-ST-challenged and free ST-challenged DCs (Fig. 1c). Once again, no significant differences against free bacteria treatment were observed for cells pulsed with ST(GFP) that were previously incubated with control IgGs. Furthermore, because intracellular bacterial loads fluctuated between treated DCs we determined the distribution of bacteria inside these cells. As shown in Fig. 1(i), both ST-mIgG1-infected and ST-pIgG-infected DCs showed a shift of the Gaussian curve towards higher amounts of intracellular bacteria values, suggesting that each DC captures higher numbers of IgG-opsonized ST than free bacteria. In addition, to corroborate this last observation of increased bacterial load by IgG-opsonization, we performed gentamicin protection assays to quantify intracellular bacteria. Once again, significantly higher amounts of bacteria were recovered from DCs pulsed with ST-IgG (mIgG1-opsonized and pIgG serum-opsonized) (Fig. 1j), than from DCs pulsed with free Salmonella. As shown between Fig. 1 and Fig. S2, the simple presence of IgGs without the capacity to coat Salmonella is not enough to enhance uptake of bacteria by DCs, supporting the notion that IgG-opsonization is required to trigger enhanced internalization.
Figure 1.
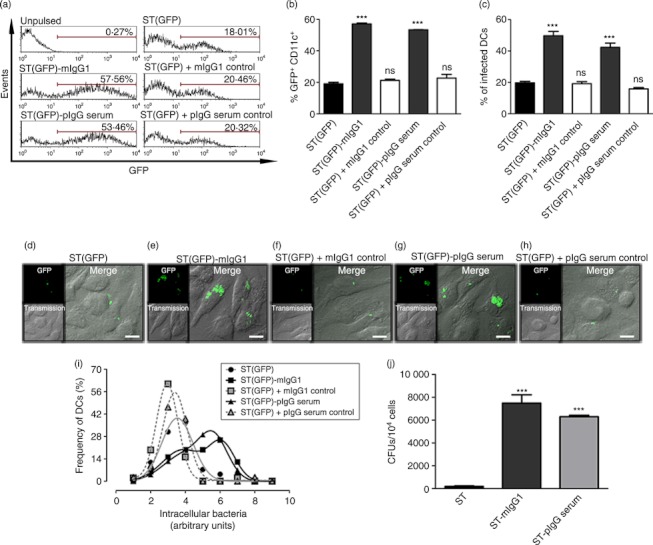
IgG-opsonization increases Salmonella entry to dendritic cells (DCs). (a) DCs were pulsed for 60 min either with free Salmonella enterica serovar Typhimurium [ST(GFP)], Salmonella coated with mIgG1 [ST(GFP)-mIgG1] or Salmonella coated with polyclonal serum [ST(GFP)-pIgG Serum]. In addition, DCs were pulsed with Salmonella incubated with control mIgG1 and control pIgG Serum [‘ST(GFP) + mIgG1 Control’ and ‘ST(GFP) + pIgG Serum Control’, respectively]. Then, cells were treated with gentamicin for 1 hr, washed and stained with anti-CD11c antibody to be analysed by flow cytometry. Cells within the marker in each histogram represent GFP+ CD11c+ population. (b) Bar graphs representing the quantification of flow cytometry data. (c) Quantification of infected cells using confocal microscopy. (d–h) Confocal microscopy images of DCs treated as described in (a). (i) Gaussian distributions for the frequencies of DCs (%) and intracellular bacterial load (Relative Units) (for details see Data S1). The correlation index (R2) obtained for ST(GFP)-, ST(GFP)-mIgG1-, ST(GFP) + mIgG1 Control-, ST(GFP)-pIgG Serum and ST(GFP) + pIgG Serum Control-infected DCs were 0·9596, 0·9969, 0·9986, 0·9994 and 0·9881, respectively. (j) Gentamicin protection assays. Bar graphs showing that IgG-opsonization increases the amount of Salmonella intracellular colony-forming units (CFUs) within DCs. DCs were incubated either with free ST, ST-mIgG1 or ST-pIgG Serum at multiplicity of infection (MOI) 25 for 1 hr, extensively washed with PBS, and treated for an additional hour with 100 μg/ml gentamicin to kill extracellular bacteria. Then, 20 000 cells were lysed and a fraction was plated on agar plates. Data were analysed by one-way analysis of variance. Data shown are means ± SEM. Each bar represents the average of at least three independent experiments. ***P < 0·001; ns, non-significant. Scale bars = 10 μm.
Taken together, IgG-opsonization of Salmonella suppressed the capacity of this pathogen to evade phagocytosis by DCs18 and a higher number of DCs internalized bacteria. Furthermore, each individual DC internalized larger bacterial loads.
IgG-opsonized Salmonella remains capable of secreting anti-capture SPI-1 effectors
The observation that IgG opsonization enhances the capacity of DCs to capture S. Typhimurium could be the result of a suppression of the release of SPI-1-derived effectors that interfere with DC phagocytosis.18 To evaluate whether IgG opsonization impairs the capacity of S. Typhimurium to secrete SPI-1-derived effectors, SRBC haemolysis was measured as previously described.29 As a positive control SRBC were challenged with free S. Typhimurium that harbour an intact SPI-1. As a negative control, SRBCs were challenged with ST(ΔInvC), a mutant strain of Salmonella that fails to produce haemolysis because it is unable to translocate SPI-1 effectors.18 The ST-IgG induced SRBC haemolysis at equivalent levels to those of free wild-type S. Typhimurium (Fig. 2a). As expected, no SRBC haemolysis was observed for the ST(ΔInvC) strain (Fig. 2a). Also, no significant SRBC haemolysis was induced by the IgG alone (Fig. 2a). These data suggest that IgG-opsonization does not prevent Salmonella from secreting SPI-1 effectors.
Figure 2.
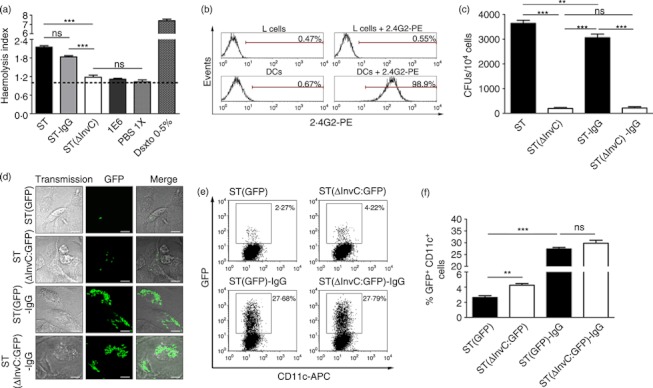
IgG-opsonized Salmonella retains the capacity to secrete SPI-1 effectors. (a) Haemolysis of sheep red blood cells (SRBCs) challenged with Salmonella enterica serovar Typhimurium (ST), ST(ΔInvC) or ST-mIgG1 was monitored by measuring haemoglobin at 405 nm. The haemolytic index was calculated as the quotient between the released haemoglobin in each treatment over the spontaneous release of unpulsed SRBC. (b) L cells do not express low-affinity Fcγ receptors (FcγRs). Surface expression of low-affinity FcγRs (FcγRIIb and FcγRIII) was measured on L cells by staining with 2.4G2-phycoerythrin (PE). Dendritic cells (DCs) were included as a positive control for 2.4G2 staining. (c) Opsonized Salmonella remain capable of infecting non-phagocytic cells. L cells were challenged either with ST, ST(ΔInvC), ST-mIgG1 or ST(ΔInvC)-mIgG1 and then intracellular conlony-forming units (CFUs) were quantified in gentamicin protection assays. (d) IgG-opsonization increases Salmonella internalization by phagocytic cells. DCs were pulsed with ST, ST(ΔInvC), ST-mIgG1 or ST(ΔInvC)-mIgG1 and mounted for confocal microscopy. (e) SPI-1-derived effectors do not impair capture of IgG-opsonized Salmonella in DCs. Dot plots show amount of GFP+ CD11c+ cells. (f) Bar graphs resuming data of dot plots shown in (e). Data were analysed by analysis of variance. Data shown are means ± SEM of three independent experiments.*P < 0·05; ***P < 0·001; ns, non-significant.
Alternatively, we evaluated whether SPI-1 effectors released from IgG-coated bacteria could still promote the entry of Salmonella into non-phagocytic cells, such as L cells.18 First, we ruled out the expression of low-affinity FcγRs on the surface of L cells by staining with an anti-CD16/CD32 antibody, which recognizes both FcγRIIb and FcγRIII (Fig. 2b). Dendritic cells were used as a positive control in these experiments. Then, L cells were infected with ST, ST-IgG, ST(ΔInvC) and ST(ΔInvC)-IgG for 60 min. As expected, large amounts of free S. Typhimurium were detected on L cells, but no significant entry could be detected for ST(ΔInvC) and ST(ΔInvC)-IgG (Fig. 2c). Interestingly, ST-IgG remained capable of invading epithelial cells, because significant amounts of bacteria could be recovered from infected L cells (Fig. 2c). These results support the notion that SPI-1-secreted effectors are fully functional on IgG-opsonized S. Typhimurium. As previously described,18 when DCs were challenged with ST(ΔInvC) strong bacterial internalization was observed because of deficienct SPI-1 effector translocation (Fig. 2d–f). In addition, both ST-IgG and ST(ΔInvC)-IgG were internalized in larger amounts by DCs but, interestingly, we recovered similar amounts of bacteria for both strains when coated with IgG (Fig. 2d–f). Because opsonized Salmonella retained the capacity to secrete SPI-1 effectors, these results suggest that DCs engulf ST-IgG using a molecular mechanism that is not inhibited by Salmonella SPI-1-derived effectors.
Actin cytoskeleton, PI3K or dynamin are not required for internalizing IgG-opsonized Salmonella by DCs
SPI-1-derived effectors modulate both the actin cytoskeleton and PI3K activity to induce Salmonella entry into non-phagocytic cells.18,30–32,34 On the other hand, these effectors negatively modulate PI3K to avoid bacterial entrance into DCs.18 To assess whether IgG-coated Salmonella was being internalized by a molecular mechanism that is not inhibited by SPI-1-associated proteins, we evaluated if the entrance of opsonized bacteria into DCs was both PI3K-independent and actin cytoskeleton-independent. In agreement with a previous report,18 DCs pre-treated either with CytD, to prevent actin polymerization,34 or Wm, to irreversibly inhibit PI3K,35 showed significant inhibition of DC infection by ST(GFP) (Fig. 3a,b). Importantly, free bacteria uptake was dependent on phagocytic class I PI3K activity (see Supplementary material, Fig. S3). However, the entry of IgG-coated bacteria could not be inhibited by either CytD or Wm (Fig. 3a,b). Similar results were observed for Salmonella coated with a polyclonal serum (see Supplementary material, Fig. S4). Contrary to the observations with free bacteria, and corroborating Wm data, we confirmed that class I PI3K was not involved in the uptake of IgG-coated bacteria (Fig. S3). Equivalent results were observed when intracellular bacterial loads were measured by gentamicin protection assays (Fig. 3c). The effect of CytD and Wm on intracellular bacterial loads was further evaluated by confocal microscopy analyses. In agreement with the data described above, we observed that inhibition of actin cytoskeleton by CytD (Fig. 3d, upper middle panels) and inhibition of PI3K by Wm (Fig. 3d, upper right panel) prevented the phagocytosis of free ST-GFP by DCs. On the contrary, no inhibition could be observed for either CytD-treated or Wm-treated DCs challenged with IgG-opsonized ST(GFP) (Fig. 3d, lower middle and right panels).
Figure 3.
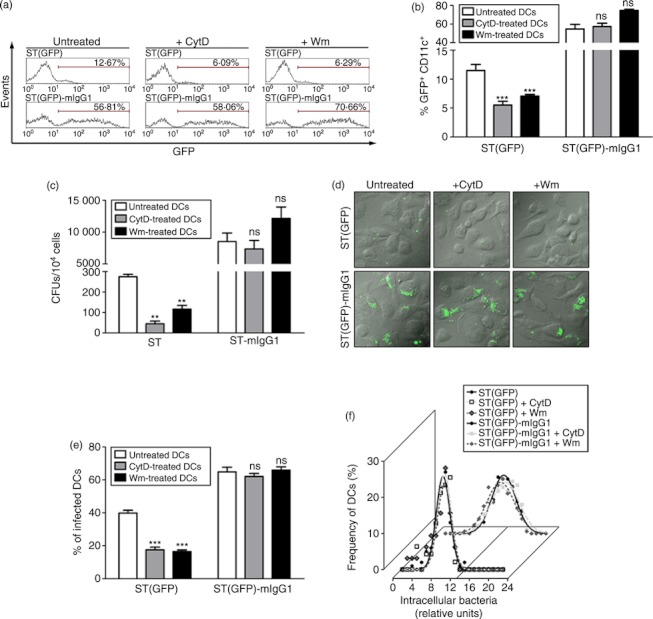
Dendritic cells (DCs) use actin cytoskeleton and phosphatidyl inositol 3-kinase (PI3K) to internalize free but not IgG-opsonized Salmonella enterica serovar Typhimurium (ST). (a) Cytochalasin D (CytD) and wortmannin (Wm) do not prevent the uptake of IgG-opsonized Salmonella. Representative histograms from three independent experiments showing GFP-derived fluorescence of CD11c+ cells challenged either with ST(GFP) or ST(GFP)-mIgG1. Left, middle and right panels show control, CytD- and Wm-treated DCs, respectively. (b) Quantification of FACS data shown in (a). Each bar is the average of three independent experiments (white: control DCs; grey: CytD-treated DCs; black: Wm-treated DCs). (c) Control, CytD-treated or Wm-treated DCs were pulsed either with ST or ST-mIgG1 and then intracellular colony-forming units (CFUs) were quantified in gentamicin protection assays as described in the Materials and methods. (d) Confocal microscopy images of control, CytD-treated or Wm-treated DCs challenged with either ST(GFP) or ST(GFP)-mIgG1. Upper and lower panels show control, CytD-treated or Wm-treated DCs challenged with ST(GFP) or ST(GFP)-mIgG1, respectively. (e) Quantification of confocal microscopy data shown in (d). Bar graph shows the percentage of infected DCs per field. Each cell that contained at least a single bacterium was considered an infected DC. (f) Gaussian distribution showing the frequency of infected DCs (%) and intracellular bacterial loads. Bacterial areas were normalized by applying a base 10 logarithm. For control DCs infected with ST(GFP) correlation index (R), mean parameter and standard deviation (SD) were 0·9862, 9·501 and 1·437, respectively (lower black curve). For CytD-treated DCs infected with ST(GFP) correlation index (R), mean parameter and SD were 0·9461, 9·818 and 1·408, respectively (lower silver dotted curve). For Wm-treated DCs infected with ST(GFP) correlation index (R), mean parameter and SD were 0·9319, 9·790 and 1·464, respectively (lower grey dotted curve). For control DCs challenged with ST(GFP)-mIgG1 correlation index (R), mean parameter and SD were 0·9898, 13·20 and 2·508, respectively (upper black curve). For CytD-treated DCs challenged with ST(GFP)-mIgG1 correlation index (R), mean parameter and SD were 0·9898, 13·52 and 2·910, respectively (upper silver dotted curve). For Wm-treated DCs challenged with ST(GFP)-IgG correlation index (R), mean parameter and SD were 0·9658, 12·53 and 2·901, respectively (upper grey dotted curve). Data were analysed by Student’s t-test between either CytD-treated or Wm-treated DCs against control cells. Data shown are means ± SEM of three independent experiments. **P < 0·01, ***P < 0·001; ns: non-significant. Scale bars = 10μm.
In agreement with the FACS data, treatment with CytD and Wm of ST(GFP)-infected DCs affects the amount of infected cells (Fig. 3e). Interestingly, the intracellular bacterial load in infected DCs remained unaltered despite treatment with CytD and Wm (Fig. 3f). This observation can be explained by the fact that although most control DCs infected with ST(GFP) showed just one bacterium inside (∼ 80% of cells, data not shown), drug treatment only reduced the fraction of DCs capturing single bacilli and not the amount of intracellular bacteria in ST(GFP)-challenged cells. Figure 3(f) shows that the Gaussian distribution obtained for DCs infected with ST(GFP)-IgG shifted to higher intervals than did the distribution for DCs infected with free ST(GFP). However, the statistical distributions of intracellular IgG-ST(GFP) inside control and drugs-treated DCs were equivalent.
Additionally, we tested whether the entry of ST-IgG was dependent on either class I or class II dynamin. We observed that only free bacteria uptake is drastically dependent on class I and II dynamin activity, compared with IgG-coated Salmonella (see Supplementary material, Fig. S5). We observed that only the amount of DCs harbouring small immune complexes were affected by class I and II dynamin inhibition, suggesting that bigger complexes (80% of total population of IgG-opsonized ST, Fig. S2) are internalized independently of dynamin. These data suggest that actin cytoskeleton, class I PI3K and class I and II dynamin are differentially involved in the phagocytosis of free versus IgG-opsonized Salmonella. This is in agreement with our previous results suggesting that ST-IgG internalization is mediated by a molecular mechanism different from that used for the engulfment of free S. Typhimurium (Fig. 2).
FcγRs are not involved in the internalization of IgG-coated Salmonella
As a consequence of the observation that internalization of ST-IgG seemed to be independent of actin, PI3K and dynamin, we tested whether the uptake of opsonized bacteria could be dependent on FcγRs.36 To evaluate whether FcγRs were involved in the phagocytosis of ST-IgG, we generated DCs from WT, FcγRIIb knockout (FcγRIIb−/−), FcγRIII knockout (FcγRIII−/−) or triple FcγRs knockout (FcγRI−/−, FcγRIIb−/− and FcγRIII−/−) mice. These cells were pulsed either with free ST(GFP) or ST(GFP)-mIgG1 and we evaluated the amount of infected DCs. First, we noted that the blockade of low-affinity FcγRs with 2.4G2 did not reduce the uptake of opsonized bacteria (Fig. 4a–c). Confocal microscopy (Fig. 4d,e) and flow cytometry analyses (Fig. 4f) showed that the amount of DCs infected either with free or mIgG1-coated ST remained unaltered, regardless of the presence or absence of either FcγRI or FcγRIIb or FcγRIII (Fig. 4d–f). Similar results were observed for serum-opsonized Salmonella (see Supplementary material, Fig. S6). Confocal microscopy and flow cytometry analyses also indicated that the intracellular bacterial loads did not significantly vary between WT, FcγRIIb−/−, FcγRIII−/− and triple FcγRs knockout cells challenged with mIgG1-coated Salmonella. These data strongly suggest that FcγRs are not required for internalizing IgG-coated ST, which is consistent with the resistance to the pharmacological inhibition of actin, PI3K and dynamin.36,37
Figure 4.
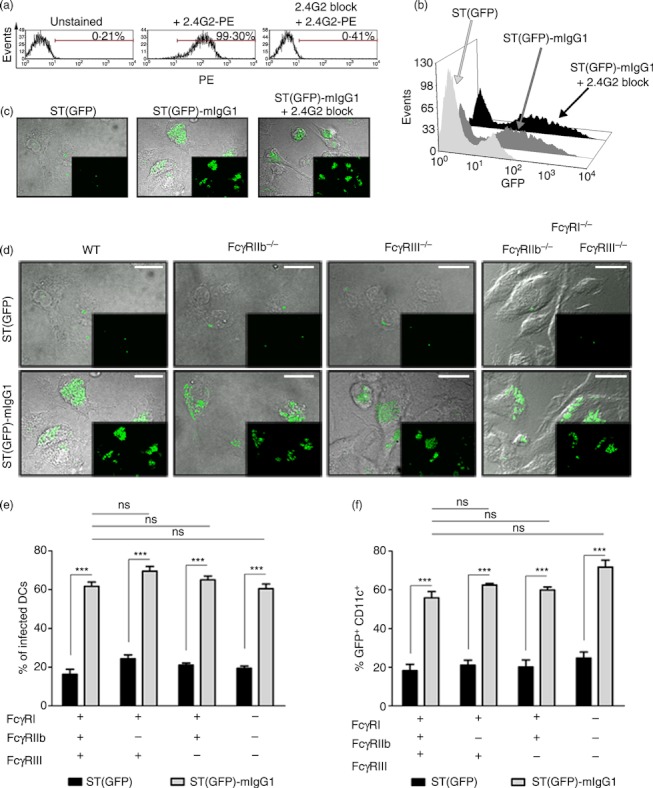
Fcγ receptors (FcγRs) are not involved in the internalization of IgG-coated Salmonella enterica serovar Typhimurium (ST). (a) Blockade of low affinity FcγRs by 2.4G2 is highly effective. Cells were left untreated or incubated with 2.4G2 blocking antibody. Unblocked low-affinity FcγRs were immune-stained with 2.4G2-phycoerythrin (PE) and analysed by FACS. (b) Blockade of low-affinity FcγRs does not reduce the percentage of ST(GFP)-mIgG1-infected dendritic cells (DCs). (c) Blockade of low-affinity FcγRs does not decrease intracellular bacterial loads. (d) Wild-type (WT), FcγRIIb−/−, FcγRIII−/− or FcγRI−/− FcγRIIb−/− FcγRIII−/− DCs were pulsed either with ST(GFP) or ST(GFP)-mIgG1. Bacterial loads were detected by confocal microscopy. No significant differences in intracellular bacterial load were observed for DCs challenged with ST(GFP)-mIgG1, independently of the availability of FcγRs. (e) Quantification of data shown in (d). Data are percentages of either ST(GFP) or ST(GFP)-mIgG1-infected DCs per field. (f) FACS analyses for DCs challenged with either ST(GFP) or ST(GFP)-mIgG1. No significant differences were observed between WT and all FcγRs-deficient DCs. Two-way analysis of variance was employed to analyse different infections between cells. One-way analysis of variance was employed to compare ST(GFP)-mIgG1-infected cells with WT and FcγR-deficient cells. Data shown are means ± SEM of three independent experiments. ***P < 0·001; ns: non-significant. Scale bars = 20 μm.
IgG-opsonized Salmonella is efficiently targeted to Lamp-1+ degradative compartments in DCs
As shown above, IgG-opsonized Salmonella was internalized more efficiently by DCs than free bacteria (Figs 1 and 2). However, increased intracellular bacterial loads would enhance antigen presentation to T cells only if bacteria are targeted to degradative compartments, such as lysosomes.38–40 Therefore, DCs pulsed either with ST(GFP) or ST(GFP)-IgG were analysed for bacteria co-localization with intracellular compartments containing Lamp-1,23,38 using both confocal microscopy and flow cytometry.
As shown in Fig. 5(a), Lamp-1+ compartments did not co-localize with single WT bacteria inside DCs (Fig. 5A.1–A.3 and lower and left histograms). Moreover, these degradative compartments were homogeneous along the selected region of interest. On the contrary, DCs infected with ST(GFP)-mIgG1 showed larger amounts of intracellular bacteria, which were either surrounded/encapsulated or co-localized with Lamp-1+ compartments (Fig. 5B.2,B.3). Figure 5(B.1) shows co-localization (yellow colour) in side-view confocal planes and superposition of histograms. Quantification of co-localization between bacteria and degradative Lamp-1+ compartments indicated that although an average of 15 intracellular ST(GFP)-IgG per cell were either surrounded or co-localized by Lamp-1+ compartments, only one single intracellular ST(GFP) bacterium was either surrounded or co-localized with this lysosomal marker (Fig. 5c).
Figure 5.
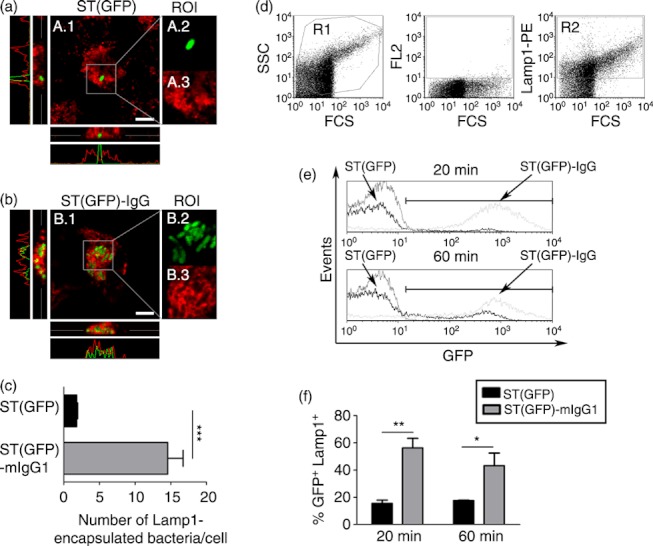
Salmonella enterica serovar Typhimurium (ST) (GFP)-IgG are rapidly targeted to Lamp1+ compartments. Dendritic cells (DCs) were challenged either with free or mIgG1-coated ST(GFP). (a) A.1 shows an overlay between ST(GFP) and Lamp1 Alexa-Fluor 555 channels. A.2 and A.3 are amplifications of region of interest (ROI). A.2 shows the ST(GFP) channel. A.3 shows the Lamp1 AF-555 channel. Lower and left microscopy merge panel show the side-view of the analysed cell (Z-stack). Grey line represents the selected focal plane shown in A.1. Histograms show the absence of overlapping between green (GFP) and red (Lamp1) emissions in the selected focal panel. (b) B.1 shows an overlay between ST(GFP)-IgG and Lamp1 Alexa-Fluor 555 (AF-555) channels. B.2 and B.3 are an amplification of the arbitrary grey squared ROI. B.2 shows the ST(GFP)-IgG channel. B.3 shows the Lamp1 AF-555 channel. Lower and left microscopy merge panel show the side-view of the analysed cell (Z-stack). Co-localizations are shown as yellow. Grey line represents the selected focal plane shown in B.1. Histograms show the presence of overlapping between green (GFP) and red (Lamp1) emissions in the selected focal panel. (c) Quantification of either ST(GFP) or ST(GFP)-IgG Lamp1-encapsulated/co-localized on each Z-stack analysed. Extracellular bacteria were discriminated by Z-stack analyses. Black and grey bars are the number of Lamp1-encapsulated/co-localized ST(GFP) or ST(GFP)-IgG, respectively. More than 100 cells were analysed for each treatment. (d) To evaluate targeting of intracellular bacteria to degradative compartments, bacteria GFP expression was assessed in Lamp1+ vesicles and analysed by flow cytometry for DCs challenged either with ST(GFP) or ST(GFP)-IgG. Left panel shows forward–side scatter dot plot of recovered intracellular compartments. Middle panel shows a dot plot for the red channel (FL2) auto-fluorescence. Right panel shows a dot plot for the positive subset of Lamp1+ compartments (both gated on R1 for each treatment). Intracellular recovered compartments showed no significant differences between free and IgG-coated ST-infected cells (data not shown). (e) Histograms showing the percentage of Lamp1+ compartments containing either ST(GFP) or ST(GFP)-IgG at 20 and 60 min. Lamp1+ compartments from Salmonella-pulsed cells were used as auto-fluorescence. Events analysed were gated from R2. (f) Graphs represent the quantification of the data from histograms shown in (e). Each bar represents the percentage of Lamp1+ compartments containing GFP-expressing bacteria. Data shown are means ± SEM of three independent experiments. Data were analysed by Student’s t-test. ***P < 0·001;**P < 0·01; *P < 0·05. Scale bars = 10 μm.
Next, we quantified by flow cytometry the frequency of Lamp-1+ compartments that contained GFP-expressing Salmonella in DCs. Hence, Lamp-1+ vesicles containing bacteria were extracted and analysed by FACS from DCs, as described in the Materials and methods. We observed that after 20 and 60 min of chase, only 20% of Lamp-1+ compartments contained ST(GFP) bacteria in DCs (Fig. 5e,f). However, the fraction of Lamp-1+ compartments containing bacteria increased significantly (up to ∼50%) when DCs were pulsed with ST(GFP)-IgG (Fig. 5e,f). These data are in agreement with the confocal microscopy analyses described above, which showed larger amounts of ST(GFP)-IgG surrounded/co-localized by Lamp-1+ compartments. These findings suggest that immediately after phagocytosis, a large amount of intracellular IgG-coated ST is directed to degradative compartments containing Lamp-1. In contrast, free virulent Salmonella can evade Lamp-1+ compartments. Such a mechanism is likely to contribute to bacterial survival and to prevent presentation of Salmonella-derived antigens to T cells.21,22
IgG-opsonization restores presentation of Salmonella-derived antigen to T cells
As shown in Fig. 6, IgG-opsonization restores the capacity of DCs to prime naive T cells. The activation of OT-II cells was measured as the capacity to secrete both interleukin-2 and interferon-γ and to up-regulate early activation markers such as CD69 in response to antigenic stimulation provided by DCs infected with OVA-expressing-ST (ST OVA:WT). In agreement with previous observations,23,24 DCs pulsed with free ST(OVA:WT) were unable to present bacteria-derived antigens to T cells (Fig. 6a,b,g). In contrast, DCs pulsed with IgG-coated ST(OVA:WT) were able to efficiently prime T cells (Fig. 6a,b,g). These results support the notion that the distribution of IgG-coated bacteria to degradative compartments inside DCs (Fig. 5) can enhance the presentation of bacteria-expressed antigens to T cells. Additionally, we observed that OT-II cells co-cultured with ST-IgG-pulsed DCs were able to strongly up-regulate markers for clonal expansion, such as interleukin-2 receptor (CD25 in Fig. 6e) and transferrin receptor (CD71 in Fig. 6f). Interestingly, ST(OVA:ΔInvC)-pulsed DCs were unable to prime T cells, despite elevated numbers of bacteria in these cells (Figs 6c,d and 2d–f). Such a large entry of bacteria was also dependent on both PI3K and actin cytoskeleton activity (data not shown). Again, antigen presentation was restored when ST(OVA:ΔInvC) was opsonized with IgG (Fig. 6c,d). These data suggest that even though ST(ΔInvC) enters DCs in greater numbers through the natural PI3K/actin cytoskeleton mechanism, the elevated bacterial load fails to ensure effective antigen presentation. Hence, it seems that opsonization allows DCs to recover antigen-specific T-cell priming by a PI3K/actin cytoskeleton/dynamin/FcγR-independent internalization mechanism to engulf IgG-coated Salmonella which, in this case, restores the presentation of Salmonella-derived antigens (Figs 3 and 6 and Figs. S4–S6).
Figure 6.
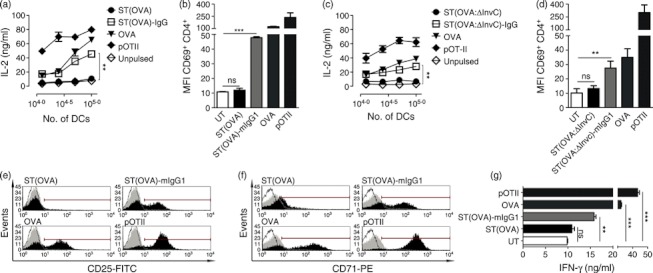
IgG-opsonization restores antigen presentation of Salmonella enterica serovar Typhimurium (ST) -derived peptides. Dendritic cells (DCs) were pulsed either with ST(WT) (a, b, e–g) or ST(ΔInvC) (c, d) bacteria both expressing ovalbumin (OVA) as free or IgG-coated. Infection was carried out by 2 hr and cells were treated with gentamicin at 50 μg/ml for 12 hr. Then, different numbers of cells were co-cultured with 1 × 105 OT-II cells (T CD4+). As a control, during co-culture, cells were pulsed with OVA protein or OT-II peptide (pOT-II). After 20 hr, supernatants were collected and analysed for interleukin-2 (IL-2) presence by ELISA. (a) IgG-opsonization of Salmonella-(OVA) restores the capacity of DCs to stimulate IL-2 secretion from OT-II cells. (b) IgG-opsonization of Salmonella-(OVA) restores activation/up-regulation of CD69 on OT II cells. (c) IgG-opsonization of Salmonella-(OVA;ΔInvC) restores the capacity of DCs to stimulate IL-2 secretion from OT-II cells. (d) IgG-opsonization of Salmonella-(OVA;ΔInvC) restores activation/up-regulation of CD69 on OT II cells. (e, f) IgG-opsonization of Salmonella-(OVA) trigger up-regulation on OT II cells of proliferation markers such as IL-2R (CD25) and Transferrin receptor (CD71), respectively. (g) IgG-opsonization of Salmonella-(OVA) restores the capacity of DCs to prime OT-II cells for secretion of interferon-γ (IFN-γ). Data shown are means ± SEM of three independent experiments. Data were analysed by analysis of variance. **P < 0·01 ***P < 0·001; ns: non-significant.
Then, we evaluated whether restoration of T-cell priming by opsonization of S. Typhimurium also occurs in infected mice. To evaluate this, groups of C57BL/6 mice were adoptively transferred with CFSE-labelled OT-II T CD4+ cells and then intravenously infected with either ST(OVA) or ST(OVA)-mIgG1. After 3 days of infection, mice displayed acute splenomegaly (see Supplementary material, Fig. S7A) and elevated loads of ST(OVA) CFUs in the spleen (Fig. S7E), which was not correlated with numbers of CD11c+ splenocytes (Fig. S7D) and suggests that infection was successful. When CD4+ CFSE+ T cells were evaluated by flow cytometry, we observed that CFSE dilution only occurred in mice infected with IgG-coated bacteria (Fig. S7B,C). These results suggest that restoration of Salmonella antigen presentation to T cells by opsonization also occurs in the spleens of infected mice.
Discussion
Virulent Salmonella strains have the capacity to subvert the function of host cells by promoting bacterial survival and dissemination.11,12,21 While SPI-1-encoded effectors promote the invasion of non-phagocytic cells,18 it has been previously shown that they impair both the phagocytic capacity and intracellular bacterial loads in DCs.17,18,41,42 Interestingly, avoidance of capture by DCs was suggested as a consequence of PI3K-impairment.18 This leads to reduced availability of the intracellular bacterial-derived antigens necessary to produce T-cell-activating antigen-loaded MHCs.43,44 IgG-opsonization of the pathogen before infection restores the capacity of DCs to prime antigen-specific naive T cells.23,24,45 However, little is known about how IgG-opsonization could enhance bacterial degradation/antigen presentation. In a previous report, we have shown that for restoration of antigen presentation by DCs in the context of an infection with ST-IgG, the superficial expression of FcγRIII (CD16)24 is required so it is likely that this receptor could also be counteracting the evasion of phagocytosis displayed by S. Typhimurium in these cells.18 To asses this hypothesis, we first evaluated whether IgG-opsonization enhances bacterial capture. We observed about a threefold increase in the percentage of DCs capturing S. Typhimurium either for mIgG1 or pIgG serum-coated bacteria. In addition, more intracellular bacteria were observed within each single DC. This shows that IgG-opsonization strongly restores the capacity of DCs to capture S. Typhimurium. Interestingly, we noted that the presence of soluble, non-specific IgG was not enough to enhance bacterial uptake, suggesting that the process of opsonization is of vital importance to counteract evasion of phagocytosis.
As IgG-opsonization abolishes the escape displayed by S. Typhimurium in DCs, we hypothesized a possible impairment in the secretion of anti-capture SPI-1-derived effectors. We observed that immune complexes (ICs) still remain able to secrete active SPI-1 effectors. Hence, there is a high probability that PI3K could still be antagonized by these virulent determinants in DCs. In this context, an alternative hypothesis suggests that DCs engulf ST-IgG by an alternative molecular mechanism, which could be PI3K-independent. We demonstrated that neither PI3K nor actin cytoskeleton (classic molecules associated with phagocytosis46–49) were involved in the internalization of Salmonella ICs. Interestingly, DCs capturing large ICs (∼80% of total ICs; see Fig. S2 for more details) did not require either class I or class II dynamin to engulf these complexes. This result supports the theory that opsonized Salmonella enters DCs via an alternative molecular pathway. This alternative pathway, which does not employ molecules that can be impaired by SPI-1, is clearly observed when compared with the uptake of a WT strain of S. Typhimurium against a ΔInvC mutant (unable to translocate SPI-1 determinants). In the non-coated state, large amounts of the ΔInvC mutant strain are captured by DCs compared with amounts of WT bacteria. However, when both strains are IgG-opsonized, no significant differences were observed, supporting the theory that SPI-1-derived effectors do not impair the entrance of ICs.
Recently, it has been described how IgG-coated bodies require FcγRs in the surface of cells to be engulfed.49–52 Likewise, both PI3K and actin cytoskeleton regulate this process.49,51 Nevertheless, IgG-coated latex beads can be engulfed independent of PI3K.53 Along these lines, it has been observed FcγR internalization by aggregated IgGs is independent of actin activity.54 Despite these findings, we did not observe crucial participation of all three classes of FcγR in the internalization of IgG-coated S. Typhimurium. Contrary to previous reports,49–51,55 we propose that IgG-opsonized Salmonella enters DCs via a new molecular mechanism that remains to be discovered. It is likely that other molecules in the pathogen’s surface can be recognized by DCs, acting as the main modulators for bacterial capture. These molecules could be, for example, flagellin, which has been characterized as an important pathogen-associated molecular pattern (PAMP) in the interaction with DCs.16,56–59 Other molecules, such as mannose, could remain uncovered by IgG, being easily recognized by mannose receptor in the surface of DCs.60 However, mannose receptor employs PI3K to engulf its ligand,61 which disagrees with our observations (Fig. 3, and Figs. S3 and S4). Alternatively, DC-SIGN (Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin), which has the capacity to recognize mannose-like patterns, could be a candidate receptor.60,62 It has recently been shown that its endocytic route requires dynamin, which could explain the capture of small ICs (Figs S2 and S5).62 Clathrin is another intra-DC molecule that can regulate IC engulfment.54,63,64 It has been recently described how internalization by way of the endocytic route using clathrin leads nascent endosomes to rapidly fuse with lysosomal degradative compartments, suggesting its participation in both ST-IgG engulfment and degradation.63–65 Nevertheless, IgG-opsonization could be working only as a ‘bacterial number concentrator’, which facilitates both capture and degradation of ST. Moreover, our results suggest that FcγRs might not be responsible for mediating the capture and phagocytosis of opsonized bacteria.
Although our results suggest that DCs do not employ FcγRIII in the uptake of ST-IgG, this receptor seems to be required to restore the presentation of bacterial-derived antigens to specific T cells. In support of this observation are recent studies describing how only partial extracellular engagement of FcγRIII or FcγRI in the surface of an antigen-presenting cell is enough to trigger a strong intracellular degradative pathway, which renders host cells able to fuse early/late endosomes with lysosomal compartments.66 Hence, this observation unifies our findings and suggests that superficial IgGs, being present in ICs, could be engaging FcγRIII in the surface of the DCs and triggering a strong degradative pathway that promotes the fusion of already internalized Salmonella-ICs (engulfed without the assistance of FcγRs) with lysosomes.24,45 Interestingly, the degradation of ST-IgG within DCs was characterized as a function of PI3K.24 Given that phagocytic PI3K (class I PI3K) does not participate in the degradation of internalized ICs, the degradation of bacteria might depend on class III PI3K.47,67 Our findings corroborate this notion, because when we inhibited class III PI3K we observed a slight increment in the amount of GFP-containing DCs, which can be interpreted as decreased ability of DCs to degrade GFP protein associated with intracellular ST (Fig. S3). Hence, the action of degradative class III PI3K is independent of phagocytic class I PI3K, the function of which is spatially and temporally separated, and supports our results.
The degradation of intracellular ST-IgG restores antigen presentation both in vitro and in vivo (Fig. 6 and Fig. S7). Interestingly, both strains ST(OVA) and ST(OVA)-IgG caused acute splenomegaly (probably through the high degree of difference between PAMPs present in the pathogen). However, only IgG-coated bacteria were able to induce strong CD4+ T-cell activation (CFSE dilution). We showed similar data for in vitro experiments (Fig. 6). This increased activation was correlated with elevated numbers of CFUs in the spleen (Fig. S7E). Recently, it has been shown that DCs are responsible for transport of S. Typhimurium to the spleen.18 However, we did not observe a significant increment in the CD11c+ population in this organ (Fig. S7D). This observation suggests that in mice treated with ST(OVA)-IgG each DC could be transporting elevated numbers of intracellular bacteria when compared with ST(OVA)-treated mice, which supports our initials findings of enhanced ST-IgG capture.
In summary, here we provide data suggesting that IgG-opsonization of S. Typhimurium renders the pathogen unable to escape from DC capture. The process of uptake is class I PI3K/actin/dynamin/FcγRI/FcγRIIb/FcγRIII-independent. This suggests the existence of an alternative internalization mechanism still undisclosed for IgG ICs. This new mechanism of capture, in combination with engagement of FcγRIII, renders DCs highly degradative, hence restoring the capacity of antigen presentation to T cells of bacterially derived antigens, which triggers an anti-bacterial immune response.
Acknowledgments
This work was supported by grants from FONDECYT 1085281, 1070352, 3070018, SavinMuco-Path-INCO-CT-2006-032296, Millennium Nucleus on Immunology and Immunotherapy P04/030-F. AMK is a Chaire De La Région Pays De La Loire De Chercheur Étranger D’Excellence. SAR is a CONICYT-Chile fellow. In addition, we are grateful to Dr Jeffrey Ravetch for providing triple knockout FcγR mice.
Glossary
- APC
allophycocyanin
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- CFU
colony-forming unit
- CytD
cytochalasin D
- DC
dendritic cell
- FcγR
Fcγ receptor
- GFP
green fluorescent protein
- IC
immune complex
- MOI
multiplicity of infection
- OVA
ovalbumin
- PAMP
pathogen-associated molecular pattern
- PE
phycoerythrin
- PI3K
phosphatidylinositol 3-kinase
- SPI-1
Salmonella Pathogenicity Island 1
- SRBC
sheep red blood cell
- ST
Salmonella enterica serovar Typhimurium
- Wm
wortmannin
Disclosures
The authors declare no conflict of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Phenotype of DCs used in this study.
Figure S2. IgG opsonization ofSalmonella is highly effective.
Figure S3. Phagocytic PI3K (Class I PI3K) is not involved in ST(GFP)-mIgG1 internalization.
Figure S4. Neither PI3K nor Actin cytoskeleton is involved in the internalization of ST(GFP)-pIgG Serum in DCs.
Figure S5. Class I and II of Dynamin are not severely involved in the mechanism of internalization of mIgG1-coated Salmonella (GFP).
Figure S6. FcγRs (FcγRI,FcγRIIb and FcγRIII) are not involved in theinternalization of ST(GFP) opsonized with polyclonal IgG Serum.
Figure S7. IgG-opsonization of ST triggers TCD4+ proliferation in vivo.
Figure S8. Free antibiotic-containing medium does not contain lethal doses of antibiotics after replacement of old antibiotic-containing medium.
Data S1. Material and methods.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Ravetch JV. Fc receptors. Curr Opin Immunol. 1997;9:121–5. doi: 10.1016/s0952-7915(97)80168-9. [DOI] [PubMed] [Google Scholar]
- 3.Ravetch J. In vivo veritas: the surprising roles of Fc receptors in immunity. Nat Immunol. 2010;11:183–5. doi: 10.1038/ni0310-183. [DOI] [PubMed] [Google Scholar]
- 4.Restifo NP, Bacik I, Irvine KR, et al. Antigen processing in vivo and the elicitation of primary CTL responses. J Immunol. 1995;154:4414–22. [PMC free article] [PubMed] [Google Scholar]
- 5.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousso P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nat Rev Immunol. 2008;8:675–84. doi: 10.1038/nri2379. [DOI] [PubMed] [Google Scholar]
- 7.Karlsson C, Eliasson M, Olin AI, Morgelin M, Karlsson A, Malmsten M, Egesten A, Frick IM. SufA of the opportunistic pathogen Finegoldia magna modulates actions of the antibacterial chemokine MIG/CXCL9, promoting bacterial survival during epithelial inflammation. J Biol Chem. 2009;284:29499–508. doi: 10.1074/jbc.M109.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiology (Bethesda) 2005;20:326–39. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- 9.O’Loughlin JL, Spinner JL, Minnich SA, Kobayashi SD. Yersinia pestis two-component gene regulatory systems promote survival in human neutrophils. Infect Immun. 2010;78:773–82. doi: 10.1128/IAI.00718-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedoui S, Kupz A, Wijburg OL, Walduck AK, Rescigno M, Strugnell RA. Different bacterial pathogens, different strategies, yet the aim is the same: evasion of intestinal dendritic cell recognition. J Immunol. 2010;184:2237–42. doi: 10.4049/jimmunol.0902871. [DOI] [PubMed] [Google Scholar]
- 11.Bueno SM, Gonzalez PA, Schwebach JR, Kalergis AM. T cell immunity evasion by virulent Salmonella enterica. Immunol Lett. 2007;111:14–20. doi: 10.1016/j.imlet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85:112–8. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis GR. Type III secretion: a bacterial device for close combat with cells of their eukaryotic host. Philos Trans R Soc Lond B Biol Sci. 2000;355:681–93. doi: 10.1098/rstb.2000.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–74. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 15.Bogunovic M, Ginhoux F, Helft J, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–25. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alaniz RC, Cummings LA, Bergman MA, Rassoulian-Barrett SL, Cookson BT. Salmonella typhimurium coordinately regulates FliC location and reduces dendritic cell activation and antigen presentation to CD4+ T cells. J Immunol. 2006;177:3983–93. doi: 10.4049/jimmunol.177.6.3983. [DOI] [PubMed] [Google Scholar]
- 17.Albaghdadi H, Robinson N, Finlay B, Krishnan L, Sad S. Selectively reduced intracellular proliferation of Salmonella enterica serovar Typhimurium within APCs limits antigen presentation and development of a rapid CD8 T cell response. J Immunol. 2009;183:3778–87. doi: 10.4049/jimmunol.0900843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bueno SM, Wozniak A, Leiva ED, Riquelme SA, Carreno LJ, Hardt WD, Riedel CA, Kalergis AM. Salmonella pathogenicity island 1 differentially modulates bacterial entry to dendritic and non-phagocytic cells. Immunology. 2010;130:273–87. doi: 10.1111/j.1365-2567.2009.03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheminay C, Mohlenbrink A, Hensel M. Intracellular Salmonella inhibit antigen presentation by dendritic cells. J Immunol. 2005;174:2892–9. doi: 10.4049/jimmunol.174.5.2892. [DOI] [PubMed] [Google Scholar]
- 20.Wallet MA, Sen P, Tisch R. Immunoregulation of dendritic cells. Clin Med Res. 2005;3:166–75. doi: 10.3121/cmr.3.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bueno SM, Tobar JA, Iruretagoyena MI, Kalergis AM. Molecular interactions between dendritic cells and Salmonella: escape from adaptive immunity and implications on pathogenesis. Crit Rev Immunol. 2005;25:389–403. doi: 10.1615/critrevimmunol.v25.i5.40. [DOI] [PubMed] [Google Scholar]
- 22.Tobar JA, Carreno LJ, Bueno SM, Gonzalez PA, Mora JE, Quezada SA, Kalergis AM. Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect Immun. 2006;74:6438–48. doi: 10.1128/IAI.00063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobar JA, Gonzalez PA, Kalergis AM. Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fcγ receptors on dendritic cells. J Immunol. 2004;173:4058–65. doi: 10.4049/jimmunol.173.6.4058. [DOI] [PubMed] [Google Scholar]
- 24.Herrada AA, Contreras FJ, Tobar JA, Pacheco R, Kalergis AM. Immune complex-induced enhancement of bacterial antigen presentation requires Fcγ receptor III expression on dendritic cells. Proc Natl Acad Sci USA. 2007;104:13402–7. doi: 10.1073/pnas.0700999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Velden AW, Velasquez M, Starnbach MN. Salmonella rapidly kill dendritic cells via a caspase-1-dependent mechanism. J Immunol. 2003;171:6742–9. doi: 10.4049/jimmunol.171.12.6742. [DOI] [PubMed] [Google Scholar]
- 26.Gallo P, Goncalves R, Mosser DM. The influence of IgG density and macrophage Fcγ receptor cross-linking on phagocytosis and IL-10 production. Immunol Lett. 2010;133:70–7. doi: 10.1016/j.imlet.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson KE, Chessa TA, Davidson K, et al. PtdIns3P and Rac direct the assembly of the NADPH oxidase on a novel, pre-phagosomal compartment during FcR-mediated phagocytosis in primary mouse neutrophils. Blood. 2010;116:4978–4989. doi: 10.1182/blood-2010-03-275602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carreno LJ, Bueno SM, Bull P, Nathenson SG, Kalergis AM. The half-life of the T-cell receptor/peptide-major histocompatibility complex interaction can modulate T-cell activation in response to bacterial challenge. Immunology. 2007;121:227–37. doi: 10.1111/j.1365-2567.2007.02561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miki T, Okada N, Shimada Y, Danbara H. Characterization of Salmonella pathogenicity island 1 type III secretion-dependent hemolytic activity in Salmonella enterica serovar Typhimurium. Microb Pathog. 2004;37:65–72. doi: 10.1016/j.micpath.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Mallo GV, Espina M, Smith AC, et al. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J Cell Biol. 2008;182:741–52. doi: 10.1083/jcb.200804131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira OV, Botelho RJ, Rameh L, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou D, Galan J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 2001;3:1293–8. doi: 10.1016/s1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhou D, Chen LM, Hernandez L, Shears SB, Galan JE. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol Microbiol. 2001;39:248–59. doi: 10.1046/j.1365-2958.2001.02230.x. [DOI] [PubMed] [Google Scholar]
- 34.May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114:1061–77. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- 35.Ihle NT, Powis G. Take your PIK: phosphatidylinositol 3-kinase inhibitors race through the clinic and toward cancer therapy. Mol Cancer Ther. 2009;8:1–9. doi: 10.1158/1535-7163.MCT-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang ZY, Barreda DR, Worth RG, Indik ZK, Kim MK, Chien P, Schreiber AD. Differential kinase requirements in human and mouse Fcγ receptor phagocytosis and endocytosis. J Leukoc Biol. 2006;80:1553–62. doi: 10.1189/jlb.0106019. [DOI] [PubMed] [Google Scholar]
- 37.Link TM, Park U, Vonakis BM, Raben DM, Soloski MJ, Caterina MJ. TRPV2 has a pivotal role in macrophage particle binding and phagocytosis. Nat Immunol. 2010;11:232–9. doi: 10.1038/ni.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dell’Angelica EC, Mullins C, Caplan S, Bonifacino JS. Lysosome-related organelles. FASEB J. 2000;14:1265–78. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- 39.Harding CV. Class II antigen processing: analysis of compartments and functions. Crit Rev Immunol. 1996;16:13–29. doi: 10.1615/critrevimmunol.v16.i1.20. [DOI] [PubMed] [Google Scholar]
- 40.Geuze HJ. The role of endosomes and lysosomes in MHC class II functioning. Immunol Today. 1998;19:282–7. doi: 10.1016/s0167-5699(98)01269-9. [DOI] [PubMed] [Google Scholar]
- 41.Tierrez A, Garcia-del Portillo F. New concepts in Salmonella virulence: the importance of reducing the intracellular growth rate in the host. Cell Microbiol. 2005;7:901–9. doi: 10.1111/j.1462-5822.2005.00540.x. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-del Portillo F, Nunez-Hernandez C, Eisman B, Ramos-Vivas J. Growth control in the Salmonella-containing vacuole. Curr Opin Microbiol. 2008;11:46–52. doi: 10.1016/j.mib.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez PA, Carreno LJ, Coombs D, Mora JE, Palmieri E, Goldstein B, Nathenson SG, Kalergis AM. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc Natl Acad Sci USA. 2005;102:4824–9. doi: 10.1073/pnas.0500922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coombs D, Kalergis AM, Nathenson SG, Wofsy C, Goldstein B. Activated TCRs remain marked for internalization after dissociation from pMHC. Nat Immunol. 2002;3:926–31. doi: 10.1038/ni838. [DOI] [PubMed] [Google Scholar]
- 45.Riquelme SA, Wozniak A, Kalergis AM, Bueno SM. Evasion of host immunity by virulent Salmonella: implications for vaccine design. Curr Med Chem. 2011;18:5666–75. doi: 10.2174/092986711798347333. [DOI] [PubMed] [Google Scholar]
- 46.Araki N, Hatae T, Furukawa A, Swanson JA. Phosphoinositide-3-kinase-independent contractile activities associated with Fcγ-receptor-mediated phagocytosis and macropinocytosis in macrophages. J Cell Sci. 2003;116:247–57. doi: 10.1242/jcs.00235. [DOI] [PubMed] [Google Scholar]
- 47.Vieira OV, Bucci C, Harrison RE, et al. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol. 2003;23:2501–14. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Y, Kawamura I, Nomura T, et al. TLR2-MyD88-dependent PI3K and Rac1 activation facilitates the phagocytosis of Listeria monocytogenes by murine macrophages. Infect Immun. 78:2857–2867. doi: 10.1128/IAI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bohdanowicz M, Cosio G, Backer JM, Grinstein S. Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes. J Cell Biol. 2010;191:999–1012. doi: 10.1083/jcb.201004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nimmerjahn F, Ravetch JV. Fcγ receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Swanson JA, Hoppe AD. The coordination of signaling during Fc receptor-mediated phagocytosis. J Leukoc Biol. 2004;76:1093–103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- 52.Hazenbos WL, Heijnen IA, Meyer D, et al. Murine IgG1 complexes trigger immune effector functions predominantly via FcγRIII (CD16) J Immunol. 1998;161:3026–32. [PubMed] [Google Scholar]
- 53.Steinberg BE, Scott CC, Grinstein S. High-throughput assays of phagocytosis, phagosome maturation, and bacterial invasion. Am J Physiol Cell Physiol. 2007;292:C945–52. doi: 10.1152/ajpcell.00358.2006. [DOI] [PubMed] [Google Scholar]
- 54.Tse SM, Furuya W, Gold E, Schreiber AD, Sandvig K, Inman RD, Grinstein S. Differential role of actin, clathrin, and dynamin in Fcγ receptor-mediated endocytosis and phagocytosis. J Biol Chem. 2003;278:3331–8. doi: 10.1074/jbc.M207966200. [DOI] [PubMed] [Google Scholar]
- 55.Baudino L, Nimmerjahn F, Azeredo da Silveira S, et al. Differential contribution of three activating IgG Fc receptors (FcγRI, FcγRIII, and FcγRIV) to IgG2a- and IgG2b-induced autoimmune hemolytic anemia in mice. J Immunol. 2008;180:1948–53. doi: 10.4049/jimmunol.180.3.1948. [DOI] [PubMed] [Google Scholar]
- 56.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 57.Honko AN, Mizel SB. Effects of flagellin on innate and adaptive immunity. Immunol Res. 2005;33:83–101. doi: 10.1385/IR:33:1:083. [DOI] [PubMed] [Google Scholar]
- 58.Salazar-Gonzalez RM, Srinivasan A, Griffin A, Muralimohan G, Ertelt JM, Ravindran R, Vella AT, McSorley SJ. Salmonella flagellin induces bystander activation of splenic dendritic cells and hinders bacterial replication in vivo. J Immunol. 2007;179:6169–75. doi: 10.4049/jimmunol.179.9.6169. [DOI] [PubMed] [Google Scholar]
- 59.Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 60.McGreal EP, Miller JL, Gordon S. Ligand recognition by antigen-presenting cell C-type lectin receptors. Curr Opin Immunol. 2005;17:18–24. doi: 10.1016/j.coi.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JS, Nauseef WM, Moeenrezakhanlou A, et al. Monocyte p110α phosphatidylinositol 3-kinase regulates phagocytosis, the phagocyte oxidase, and cytokine production. J Leukoc Biol. 2007;81:1548–61. doi: 10.1189/jlb.0906564. [DOI] [PubMed] [Google Scholar]
- 62.Cambi A, Beeren I, Joosten B, Fransen JA, Figdor CG. The C-type lectin DC-SIGN internalizes soluble antigens and HIV-1 virions via a clathrin-dependent mechanism. Eur J Immunol. 2009;39:1923–8. doi: 10.1002/eji.200939351. [DOI] [PubMed] [Google Scholar]
- 63.Booth JW, Kim MK, Jankowski A, Schreiber AD, Grinstein S. Contrasting requirements for ubiquitylation during Fc receptor-mediated endocytosis and phagocytosis. EMBO J. 2002;21:251–8. doi: 10.1093/emboj/21.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 65.Ukkonen P, Lewis V, Marsh M, Helenius A, Mellman I. Transport of macrophage Fc receptors and Fc receptor-bound ligands to lysosomes. J Exp Med. 1986;163:952–71. doi: 10.1084/jem.163.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joller N, Weber SS, Muller AJ, Sporri R, Selchow P, Sander P, Hilbi H, Oxenius A. Antibodies protect against intracellular bacteria by Fc receptor-mediated lysosomal targeting. Proc Natl Acad Sci USA. 2010;107:20441–6. doi: 10.1073/pnas.1013827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


