Abstract
Four Trichoderma strains (CH2, SH16, PQ34, and TN42) were isolated from soil samples collected from Quang Tri and Thua Thien Hue provinces in Vietnam. The strains exhibited high chitinolytic secretion. Strain PQ34 formed the largest zone of chitinase-mediated clearance (> 4 cm in diameter) in agar containing 1% (w/v) colloidal chitin. Analysis of the internal transcribed spacer regions of these strains indicated that they were Trichoderma asperellum. The molecular weights of the chitinases were approximately 42 kDa. Chitinase genes (chi42) of T. asperellum strains TN42, CH2, SH16, and PQ34 were 98~99% homologous to the ech42 gene of T. harzianum CB-Pin-01 (accession No. DQ166036). The deduced amino acid sequences of both T. asperellum strains SH16 and TN42 shared 100% similarity.
Keywords: Chi42 gene, Internal transcribed spacer region, Trichoderma asperellum, 42 kDa chitinase
Chitinase (EC 3.2.1.14) is a glycosyl hydrolase that catalyzes the hydrolytic degradation of chitin, and is found in a wide variety of organisms including bacteria, fungi, invertebrates, plants, and animals [1]. Chitin, a (1,4)-β-linked homopolysaccharide composed of N-acetylglucosamine residues, is considered the second most abundant biopolymers in nature after cellulose. It is present in the cell walls of bacteria and fungi, mushrooms, exoskeleton of crustaceans and insects, the microfilarial sheath of parasitic nematodes, and the lining of the digestive tracts of many insects [2-4].
Chitinases are produced by a variety of organisms and may play important physiological and ecological roles. Numerous genes encoding chitinase have been cloned from a variety of organisms. Chitinases have been classified into two glycoside hydrolase families, 18 and 19, based on amino acid sequence similarity [5]. Family 18 chitinases cleave the β-1,4-glycosidic linkage of not only GlcNAc-GlcNAc but also GlcNAc-GlcN, whereas family 19 chitinases cleave GlcNAc-GlcNAc and GlcN-GlcNAc linkages [6, 7]. Therefore, it is generally accepted that chitinases cannot cleave the β-1,4-glycosidic linkage of GlcN-GlcN [4].
Trichoderma asperellum is a well-known mycoparasite and effective biocontrol agent of several plant pathogens. A complex system of more than six chitinolytic enzymes that degrades chitin (two are N-acetylhexosaminidases and others are endochitinases) has been described in Trichoderma strains [8]. These mycoparasites utilize chitinolytic enzymes along with β-1,3-glucanases and proteases to degrade the cell walls of host pathogens and thus reduce the disease level [4]. However, recent studies on these enzymes have focused in T. harzianum [9, 10], T. reesei [4], T. atroviride [11-14], T. virens [15, 16], and T. aureoviride [17].
The present work deals with the isolation and identification of the chitinase producing Trichoderma strains and isolation of their chitinase genes.
Materials and Methods
Isolation and identification of Trichordema strains
Fifty-six soil samples were collected from the surface layer (15~20 cm) from eight different sites of agricultural fields in Thua Thien Hue and Quang Tri provinces, Vietnam. One gram of each sample was dissolved in 50 mL water by mixing for 10 min at room temperature. The supernatant was diluted 10-2 and 0.5 mL was spread on the surface of Trichoderma isolation medium [18], supplemented with propamocarb [19]. Cultures were incubated at 28℃ until fungi colonies become visible. To induce chitinase activities, all selected isolates were inoculated on solid Czapeck-Dox without glucose, supplemented with 1% (w/v) colloidal chitin at 28℃ for 36 hr [20]. Hydrolysis of colloidal chitin was detected by Lugol's solution [21].
The Trichoderma strains showing the highest chitinase activity was transferred onto potato dextrose broth medium [22] at 28℃ for 2 days for biomass production. Total DNA from fungi biomass was isolated by the phenol-chloroform method [23]. PCR was performed by internal transcribed spacer (ITS)1 and ITS4 primers of fungi rDNA [24]. PCR components in a 25 µL volume included 2.5 µL Taq buffer, 1 unit Taq polymerase, 10 pmol each primer, 20 µM deoxynucleotides (dNTP), 1.5 mM MgCl2, and 20 ng genomic DNA. PCR was carried out using the following program: 95℃ for 5 min; followed by 32 cycles of 95℃ for 30 sec, 55℃ for 30 sec, and 72℃ for 1 min; and a final extension at 72℃ for 10 min. PCR products were sequenced by the method of fluorescent dideoxy-terminator on 3130 ABI system (Applied Biosystems, Foster City, CA, USA).
Substrate electrophoresis of chitinase
Crude chitinase from the broth after 4 days of culture of Trichoderma strains on liquid Czapeck-Dox medium without glucose was precipitated by ammonium sulphate (70% saturation) at 4℃ for 2 hr and centrifuged at 15,000 rpm at 4℃ for 10 min. The pellet was then resuspended in suitable buffer (100 mM sodium acetate, pH 5) and dialyzed overnight against 0.05 M acetate buffer (pH 5). Molecular weight of enzyme was determined by 12% SDS-PAGE including 0.1% (w/v) colloidal chitin following a standard procedure [25]. Electrophoresis was performed at 4℃ for 3 hr and the gel was then incubated at 37℃ for 2 hr with gentle shaking in 100 mM sodium acetate buffer (pH 5.0) containing 1% (v/v) Triton X-100 to remove SDS [26]. The gel was stained in Lugol's solution [21] and the image was analyzed by Quality One software, ver. 4.1 (BioRad Laboratories, Richmond, CA, USA).
DNA isolation and PCR amplification of chitinase gene
Genomic DNA of T. asperellum was isolated by a phenol extraction method [23]. Then, genomic DNA was used as template in PCR amplification with the forward primer CHI-F (5'-ATG TTG GGC TTC CTC GGA-3') and the reverse primer CHI-R (5'-TTC GGG ATG GTT GTC ATA CTG-3'), which were specifically designed to detect the chitinase gene based on the nucleotide sequence of chitinase gene from T. harzianum CB-Pin-01 (accession No. DQ166036). The PCR mixture consisted of 1.25 unit Taq DNA polymerase (Fermentas, Maryland, CA, USA), 5 µL 10× Taq DNA polymerase buffer, 200 µM deoxynucleotides (dNTP mixture), 20 pmol of each primer, 1.5 mM MgCl2, 40 ng genomic DNA, and distilled water to a final volume of 50 µL. After 5 min of genomic DNA denaturation at 95℃, 30 cycles of 1 min for denaturation at 95℃, 1 min for annealing at 55℃ and 1 min for polymerization at 72℃ were carried out in an iCycler thermocycler (BioRad Laboratories, Richmond, CA, USA). In the final cycle, the temperature of 72℃ was held for an additional 5 min. PCR products were detected by electrophoresis on 0.8% agarose gel and stained with ethidium bromide.
Cloning and sequencing of the chitinase gene
PCR products were cut from the gel and purified by Wizard® SV Gel and PCR Clean-Up Kit (Promega, Madison, WI, USA). They were then ligated to a pGEM®-T easy vector (Invitrogen, Carlsbad, CA, USA). The ligation reaction consisted of 5 µL ligase buffer, 1 µL vector, 1 µL PCR product, 1 µL ligase, and 2 µL distilled water. The reaction mixture was incubated at 16℃ overnight and then was transformed into Escherichia coli TOP10 cells by the heat-shock method. Positive colonies were cultured on 5mL LB medium with 50 mg/mL ampicillin and recombinant plasmids were isolated using Quick Plasmid Miniprep Kit (Invitrogen). The presence of the insert was determined by PCR followed by 0.8% agarose gel electrophoresis. For determination of chitinase gene, PCR products were fluorescent dideoxy-terminator sequenced on 3130 ABI system (Applied Biosystem, Foster City, CA, USA).
Results and Discussion
Isolation and identification of Trichoderma strains
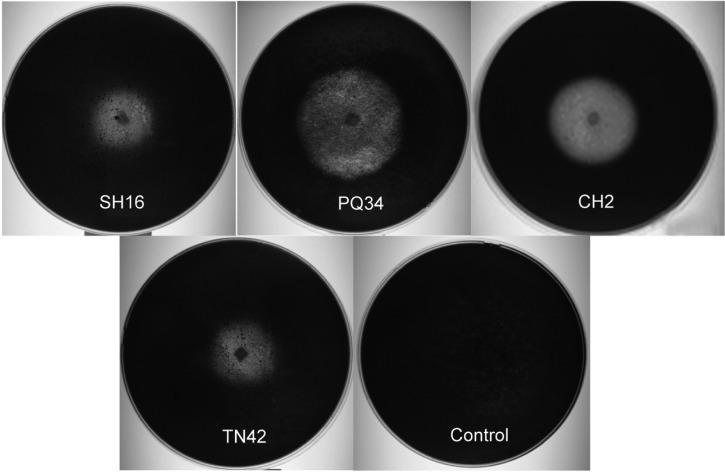
Eighty-one Trichoderma strains were isolated from soil samples collected from Quang Tri and Thua Thien Hue provinces. Chitinase activity was detected in 41 strains. However, only four Trichoderma strains (CH2, SH16, PQ34, and TN42) exhibited high chitinolytic secretion in the assay of colloidal chitin hydrolytic activity on agar plates (Fig. 1). Among these four chitinase producing fungi isolates, strain PQ34 formed the largest zone of clearance, exceeding 4 cm in diameter.
Fig. 1.
Determination of action of chitinase from Trichoderma asperellum strains on assayed plate containing 0.1% (w/v) colloidal chitin. CH2, SH16, PQ34, and TN42, all four Trichoderma strains exhibited high chitinolytic secretion; Control, without Trichoderma.
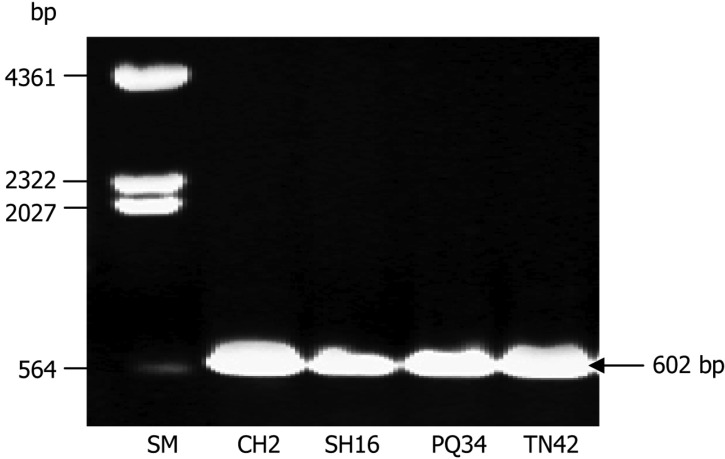
The ITS regions of these strains were amplified by PCR (Fig. 2), then sequenced and used to search BLAST in Genbank database for identification. They were deposited on NCBI (accession Nos. HM545079, HM545080, HM545081, and HM545083). Nucleotide comparison of ITS regions showed 100% identity of both these strains and T. asperellum strain ZJPH0810 (accession No. GU318216). Hence, these strains were classified as T. asperellum. Among them, T. asperellum strain SH16 has characterized chitinase properties [27].
Fig. 2.
Agarose gel electrophoresis (0.8% w/v) of PCR products from ITS region of Trichoderma strains CH2, SH16, PQ34, and TN42. SM, DNA size marker (λDNA/Hind III).
Molecular weight determination of chitinase
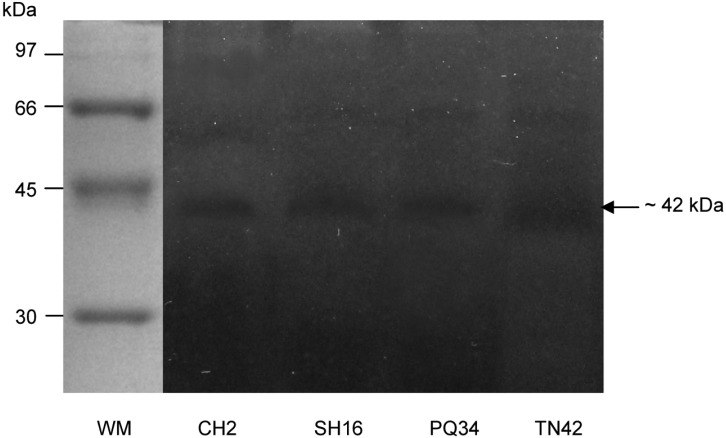
The chitinase activity and molecular weight were determined by SDS-PAGE with colloidal chitin as substrate after ammonium precipitation at 4℃ for 2 hr and dialysis overnight in acetate buffer. One chitinase band, confirming enzyme activity, clearly occurred in the dark-colored gel after staining in Lugol's solution. The molecular weights of enzyme were found to be approximately 42 kDa (Fig. 3).
Fig. 3.
SDS-PAGE (12%) with 0.1% (w/v) colloidal chitin as a substrate. WM, protein weight markers (97~14.4 kDa). CH2, SH16, PQ34 and TN42, chitinase enzyme of four Trichoderma asperellum strains. A clear band on the gel confirmed the chitinolytic activity of enzyme with an estimated molecular weight of 42 kDa.
Chitinase of 42 kDa is a common enzyme in a complex chitinase system of Trichoderma species. For example, 42 kDa chitinase has been demonstrated in T. harzianum [28-30], T. atroviride [31], T. hamatum [32]), and T. reesei [33]. The present substrate SDS-PAGE in this work showed that 42 kDa chitinase in T. asperellum is dominant of complex chitinase system.
Chitinase gene of T. asperellum
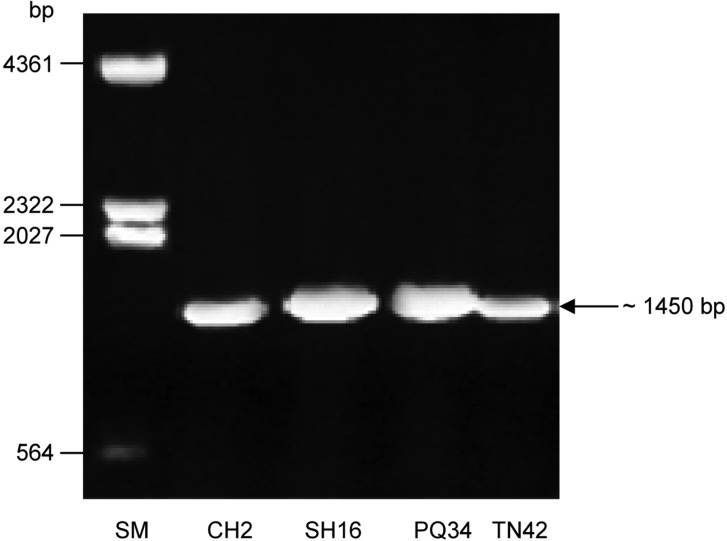
As shown in Fig. 4, DNA fragments of approximately 1,450 bp were amplified from the genome of T. asperellum strains CH2, SH16, PQ34, and TN42, suggesting that a chitinase gene might be present in them. These results also showed that two CHI-F and CHI-R primers were specific for this chitinase gene. To verify the amplification of the chitinase gene from T. asperellum strains (called chi42), PCR products were cloned into pGEM®-T easy vector, and then their sequence was analyzed. Sequencing profiles indicated that all these sequences had the same 1,459 nucleotides. The chi42 genes of T. asperellum strains TN42, CH2, SH16, and PQ34 (accession Nos. HM208346, HM191682, HM191683, and HM191684, respectively) exhibited 98~99% homology to the ech42 gene of T. harzianum CB-Pin-01 (accession No. DQ166036).
Fig. 4.
Agarose gel electrophoresis (0.8% w/v) of PCR products of CHI-F and CHI-R primers from Trichoderma strains CH2, SH16, PQ34 and TN42. SM, DNA size marker (λDNA/Hind III).
Jinzhu (2005) suggested that the gene coding for a 42 kDa chitinase of Trichoderma species may be strongly conserved because of the high homology (more than 90%) [17]. Our sequences also showed high similarity (more than 98%).
Chi42 genes encode proteins of 424 amino acids; the molecular weight calculated from deduced amino acid sequence was approximately 42 kDa (Fig. 5). The deduced amino acid sequences of both T. asperellum SH16 and TN42 strains showed 100% similarity. Using the ScanProsite tool (http://expasy.org/tools/scanprosite), we found an active site of chitinase family 18 in chi42 genes of these T. asperellum strains.
Fig. 5.
Multiple alignment of deduced amino acid sequence of the chitinase gene from Trichoderma harzianum CB-Pin-01 (ECH42, GenBank DNA sequence, accession No. DQ166036) and four T. asperellum strains SH16, TN42, CH2, and PQ34 (CHI42-). Amino acid residues blocked in black differing from the corresponding one of ECH42.
Chitinases have been demonstrated to have several importance applications, especially in biological control and medicinal industry [34-36]. However, the large-scale production of chitinase using the wild type may be disadvantageous, due to the limitation of enzyme secretion and the difficulty of purification. Therefore, cloning and expression of chitinase genes in the optimal heterologus expression system is a potential means of overproduction of these enzymes. This study is the first report of the discovery of genetic material to express chi42 gene in an appropriate host. This processing will be investigated.
Acknowledgements
This study was supported by Vietnam Ministry of Education and Training (2009~2011).
References
- 1.Gooday GW. The ecology of chitin decomposition. Adv Microb Ecol. 1990;11:387–430. [Google Scholar]
- 2.Duo-Chuan L. Review of fungal chitinases. Mycopathologia. 2006;161:345–360. doi: 10.1007/s11046-006-0024-y. [DOI] [PubMed] [Google Scholar]
- 3.Krajewska B. Application of chitin- and chitosan-based materials for enzyme immobilizations: a review. Enzyme Microb Technol. 2004;35:126–139. [Google Scholar]
- 4.Ike M, Nagamatsu K, Shioya A, Nogawa M, Ogasawara W, Okada H, Morikawa Y. Purification, characterization, and gene cloning of 46 kDa chitinase (Chi46) from Trichoderma reesei PC-3-7 and its expression in Escherichia coli. Appl Microbiol Biotechnol. 2006;71:294–303. doi: 10.1007/s00253-005-0171-y. [DOI] [PubMed] [Google Scholar]
- 5.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitsutomi M, Ohtakara A, Fukamizo T, Goto S. Action pattern of Aeromonas hydrophila chitinase on partially N-acetylated chitosan. Agric Biol Chem. 1990;54:871–877. [PubMed] [Google Scholar]
- 7.Ohtakara A, Matsunaga H, Mitsutomi M. Action pattern of Streptomyces griseus chitinase on partially N-acetylated chitosan. Agric Biol Chem. 1990;54:3191–3199. [PubMed] [Google Scholar]
- 8.Haran S, Schickler H, Oppenheim A, Chet I. New components of the chitinolytic system of Trichoderma harzianum. Mycol Res. 1995;99:441–446. [Google Scholar]
- 9.Sandhya C, Adapa LK, Nampoothiri KM, Binod P, Szakacs G, Pandey A. Extracellular chitinase production by Trichoderma harzianum in submerged fermentation. J Basic Microbiol. 2004;44:49–58. doi: 10.1002/jobm.200310284. [DOI] [PubMed] [Google Scholar]
- 10.Limón MC, Lora JM, García I, de la Cruz J, Llobell A, Benítez T, Pintor-Toro JA. Primary structure and expression pattern of the 33-kDa chitinase gene from the mycoparasitic fungus Trichoderma harzianum. Curr Genet. 1995;28:478–483. doi: 10.1007/BF00310819. [DOI] [PubMed] [Google Scholar]
- 11.Gruber S, Vaaje-Kolstad G, Matarese F, López-Mondéjar R, Kubicek CP, Seidl-Seiboth V. Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology. 2011;21:122–133. doi: 10.1093/glycob/cwq142. [DOI] [PubMed] [Google Scholar]
- 12.Arriaga S, Acosta-Munguía JA, Pérez-Martínez AS, León-Rodríguez AD, Barba de la Rosa AP. Coupling aerobic biodegradation of methanol vapors with heterologous protein expression of endochitinase Ech42 from Trichoderma atroviride in Pichia pastoris. Bioresour Technol. 2010;101:9661–9665. doi: 10.1016/j.biortech.2010.06.092. [DOI] [PubMed] [Google Scholar]
- 13.Klemsdal SS, Clarke JL, Hoell IA, Eijsink VG, Brurberg MB. Molecular cloning, characterization, and expression studies of a novel chitinase gene (ech30) from the mycoparasite Trichoderma atroviride strain P1. FEMS Microbiol Lett. 2006;256:282–289. doi: 10.1111/j.1574-6968.2006.00132.x. [DOI] [PubMed] [Google Scholar]
- 14.Hoell IA, Klemsdal SS, Vaaje-Kolstad G, Horn SJ, Eijsink VG. Overexpression and characterization of a novel chitinase from Trichoderma atroviride strain P1. Biochim Biophys Acta. 2005;1748:180–190. doi: 10.1016/j.bbapap.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Shah JM, Raghupathy V, Veluthambi K. Enhanced sheath blight resistance in transgenic rice expressing an endochitinase gene from Trichoderma virens. Biotechnol Lett. 2009;31:239–244. doi: 10.1007/s10529-008-9856-5. [DOI] [PubMed] [Google Scholar]
- 16.Abd-Aziz S, Fernandez CC, Salleh MM, Illias RM, Hassan MA. Effect of agitation and aeration rates on chitinase production using Trichoderma virens UKM1 in 2-l stirred tank reactor. Appl Biochem Biotechnol. 2008;150:193–204. doi: 10.1007/s12010-008-8140-4. [DOI] [PubMed] [Google Scholar]
- 17.Jinzhu S, Qian Y, Beidong L, Dianfu C. Expression of the chitinase gene from Trichoderma aureoviride in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2005;69:39–43. doi: 10.1007/s00253-005-1957-7. [DOI] [PubMed] [Google Scholar]
- 18.Elad Y, Chet I, Henis Y. A selective medium for improving qualitative isolation of Trichoderma spp. from soil. Phytoparasitica. 1981;9:59–67. [Google Scholar]
- 19.Askew DJ, Laing MD. An adapted selective medium for the quantitative isolation of Trichoderma species. Plant Pathol. 1993;42:686–690. [Google Scholar]
- 20.Shanmugaiah V, Mathivanan N, Balasubramanian N, Manoharan PT. Optimization of cultural conditions for production of chitinase by Bacillus laterosporous MML2270 isolated from rice rhizosphere soil. Afr J Biotechnol. 2008;7:2562–2568. [Google Scholar]
- 21.Orpin CG. The occurrence of chitin in the cell walls of the rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J Gen Microbiol. 1977;99:215–218. doi: 10.1099/00221287-99-1-215. [DOI] [PubMed] [Google Scholar]
- 22.Atlas RM. Handbook of microbiological media. 3rd ed. Boca Raton: CRC Press; 2004. [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Manter DK, Vivanco JM. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J Microbiol Methods. 2007;71:7–14. doi: 10.1016/j.mimet.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Trudel J, Asselin A. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem. 1989;178:362–366. doi: 10.1016/0003-2697(89)90653-2. [DOI] [PubMed] [Google Scholar]
- 27.Hung NB, Quang HT, Ha TT, Thao TN, Chau TT, Loc NH. Isolation and characterization of 42 kDa chitinase from Trichoderma asperellum. Vietnam J Biotechnol. 2010;8:1651–1657. [Google Scholar]
- 28.Lienemann M, Boer H, Paananen A, Cottaz S, Koivula A. Toward understanding of carbohydrate binding and substrate specificity of a glycosyl hydrolase 18 family (GH-18) chitinase from Trichoderma harzianum. Glycobiology. 2009;19:1116–1126. doi: 10.1093/glycob/cwp102. [DOI] [PubMed] [Google Scholar]
- 29.Carsolio C, Gutiérrez A, Jiménez B, Van Montagu M, Herrera-Estrella A. Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc Natl Acad Sci U S A. 1994;91:10903–10907. doi: 10.1073/pnas.91.23.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García I, Lora JM, de la Cruz J, Benítez T, Llobell A, Pintor-Toro JA. Cloning and characterization of a chitinase (chit42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr Genet. 1994;27:83–89. doi: 10.1007/BF00326583. [DOI] [PubMed] [Google Scholar]
- 31.Mach RL, Peterbauer CK, Payer K, Jaksits S, Woo SL, Zeilinger S, Kullnig CM, Lorito M, Kubicek CP. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl Environ Microbiol. 1999;65:1858–1863. doi: 10.1128/aem.65.5.1858-1863.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steyaert JM, Stewart A, Jaspers MV, Carpenter M, Ridgway HJ. Co-expression of two genes, a chitinase (chit42) and proteinase (prb1), implicated in mycoparasitism by Trichoderma hamatum. Mycologia. 2004;96:1245–1252. [PubMed] [Google Scholar]
- 33.Seidl V, Huemer B, Seiboth B, Kubicek CP. A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J. 2005;272:5923–5939. doi: 10.1111/j.1742-4658.2005.04994.x. [DOI] [PubMed] [Google Scholar]
- 34.Aam BB, Heggset EB, Norberg AL, Sørlie M, Vårum KM, Eijsink VG. Production of chitooligosaccharides and their potential applications in medicine. Mar Drugs. 2010;8:1482–1517. doi: 10.3390/md8051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JK, Shen CR, Liu CL. N-acetylglucosamine: production and applications. Mar Drugs. 2010;8:2493–2516. doi: 10.3390/md8092493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neeraja C, Anil K, Purushotham P, Suma K, Sarma P, Moerschbacher BM, Podile AR. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit Rev Biotechnol. 2010;30:231–241. doi: 10.3109/07388551.2010.487258. [DOI] [PubMed] [Google Scholar]