Abstract
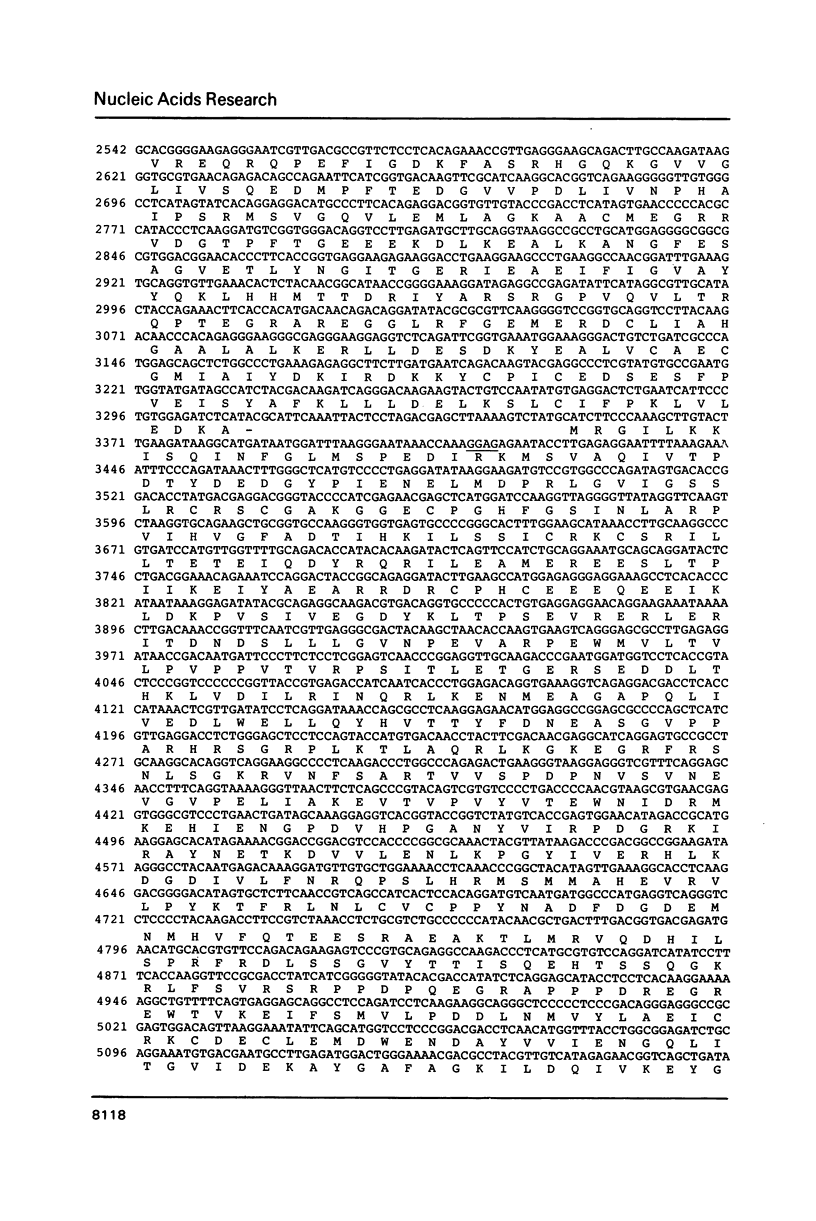
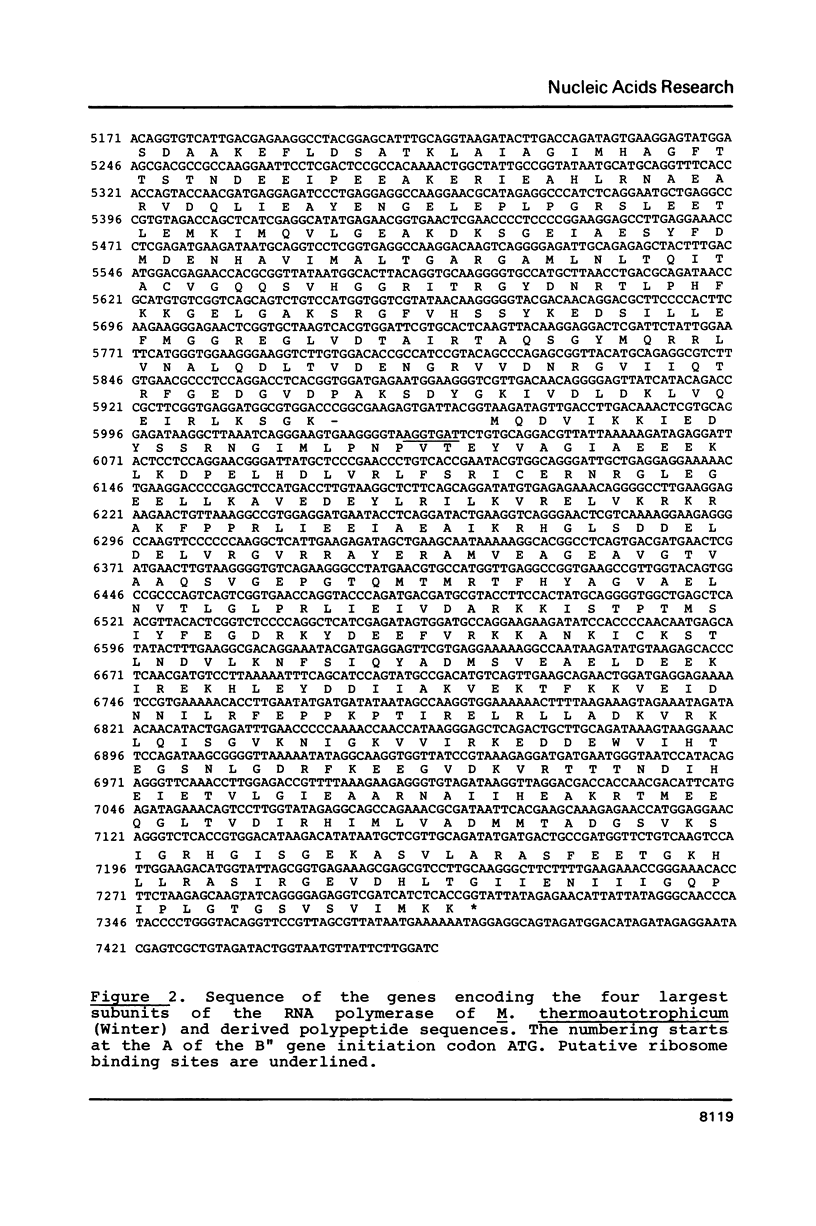
The sequence of the genes encoding the four largest subunits of the RNA polymerase of the archaebacterium Methanobacterium thermoautotrophicum was determined and putative translation signals were identified. The genes are more strongly homologous to eukaryotic than to eubacterial RNA polymerase genes. Analysis of the polypeptide sequences revealed colinearity of two pairs of adjacent archaebacterial genes encoding the B" and B' or A and C genes, respectively, with two eubacterial and two eukaryotic genes each encoding the two largest RNA polymerase subunits. This difference in sequence organization is discussed in terms of gene fusion in the course of evolution. The degree of conservation is much higher between the archaebacterial and the eukaryotic polypeptides than between the archaebacterial and the eubacterial enzyme. Putative functional domains were identified in two of the subunits of the archaebacterial enzyme.
Full text
PDF













Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Bokranz M., Klein A. Nucleotide sequence of the methyl coenzyme M reductase gene cluster from Methanosarcina barkeri. Nucleic Acids Res. 1987 May 26;15(10):4350–4351. doi: 10.1093/nar/15.10.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Rikhter V. A., Rabinov I. V. Lokalizatsiia ostatkov lizina v oblasti sviazyvaniia initsiiruiushchego substrata RNK-polimerazy E. coli. Bioorg Khim. 1987 Apr;13(4):552–555. [PubMed] [Google Scholar]
- Guo L. H., Wu R. Exonuclease III: use for DNA sequence analysis and in specific deletions of nucleotides. Methods Enzymol. 1983;100:60–96. doi: 10.1016/0076-6879(83)00046-4. [DOI] [PubMed] [Google Scholar]
- Hartmann M., Schnurbus R., Henkes H., Kubicki S., Bienzle U. Veränderungen des EEG-Grundrhythmus und des Hyperventilations-Effektes in verschiedenen Stadien der HIV-Infektion. EEG EMG Z Elektroenzephalogr Elektromyogr Verwandte Geb. 1988 Jun;19(2):101–105. [PubMed] [Google Scholar]
- Marsh J. L., Erfle M., Wykes E. J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984 Dec;32(3):481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Matzura B. Regulation of biosynthesis of the DNA-dependent RNA polymerase in Escherichia coli. Curr Top Cell Regul. 1980;17:89–136. doi: 10.1016/b978-0-12-152817-1.50008-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Salomatina I. S., Shuvaeva T. M., Lipkin V. M., Sverdlov E. D. The primary structure of E. coli RNA polymerase, Nucleotide sequence of the rpoC gene and amino acid sequence of the beta'-subunit. Nucleic Acids Res. 1982 Jul 10;10(13):4035–4044. doi: 10.1093/nar/10.13.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Chertov OYu, Modyanov N. N., Grinkevich V. A., Makarova I. A., Marchenko T. V., Polovnikova I. N. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the beta-subunit. Eur J Biochem. 1981 Jun 1;116(3):621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- Panka D., Dennis D. RNA polymerase. Direct evidence for two active sites involved in transcription. J Biol Chem. 1985 Feb 10;260(3):1427–1431. [PubMed] [Google Scholar]
- Schallenberg J., Moes M., Truss M., Reiser W., Thomm M., Stetter K. O., Klein A. Cloning and physical mapping of RNA polymerase genes from Methanobacterium thermoautotrophicum and comparison of homologies and gene orders with those of RNA polymerase genes from other methanogenic archaebacteria. J Bacteriol. 1988 May;170(5):2247–2253. doi: 10.1128/jb.170.5.2247-2253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R., Thomm M., Gerardy-Schahn R., Zillig W., Stetter K. O., Huet J. Structural homology between different archaebacterial DNA-dependent RNA polymerases analyzed by immunological comparison of their components. EMBO J. 1983;2(5):751–755. doi: 10.1002/j.1460-2075.1983.tb01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K., Beck E., Strohmaier K., Schaller H. Characterization of foot-and-mouth disease virus gene products with antisera against bacterially synthesized fusion proteins. J Virol. 1986 Mar;57(3):983–991. doi: 10.1128/jvi.57.3.983-991.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]



