Abstract
The objective of this study was to find useful fungi with α-amylase activity from the Korean traditional nuruk for the quality of traditional Korean alcoholic beverage. In this study, 165 samples of traditional nuruk were collected from 170 regions throughout Korea and the fungi were isolated to a total of 384 strains. In order to investigate the effect of microflora on nuruk, α-amylase activity, saccharogenic power (SP), starch hydrolysis activity and acid producing activity were evaluated. Ten strains were selected by α-amylase activity, which ranged from 458.47 to 1,202.75 U/g. The size of the discolored zone for the starch hydrolysis activity of each fungus ranged from 0.3 to 2 cm. The SP of the 10 strains ranged from 228.8 to 433.4 SP. Of the 10 stains, three were identified as Aspergillus oryzae, two as Aspergillus flavus, two as Lichtheimia sp., one as Rhizopus oryzae and two as other strains. The total aflatoxins present in the nuruks were examined using enzyme-linked immunosorbent assay. The 10 nuruks had less than 1.11 ppb of aflatoxins.
Keywords: Aflatoxins, α-Amylase activity, Fungi, Nuruk, Saccharogenic power
Introduction
Traditional Korean liquor is obtained through a parallel fermentation process in brewing [1]. The fermentation involves an initial stage, in which the microorganisms, usually fungi, produce a number of enzymes that break down substrate components. Following this, yeast species produce alcohol from the sugar that was previously produced by activity of mold on starch [2, 3]. The use of nuruk in this process is responsible for many of the characteristic features of this traditional Korean liquor. In contrast, koji, which is comparable to malts used for beer brewing, is used for the saccharification of starch and the decomposition of the protein contained in the raw material, rice grains [4]. Nuruk and koji are often confused in the literature, with "koji" sometimes called "modified nuruk" [5]. Koji, however, is artificially inoculated, while nuruk combines both a saccharification and starter by yeast, and so is used to fabricate the liquor without pre-fermentation. In addition, this process can be done at a low temperature due to the slightly lower saccharogenic power (SP) and fermentative activity of nuruk; as a result, various flavors of the liquor produced by a number of these microorganisms are retained [6].
During the production of the nuruk, it is possible to produce secondary metabolites of the fungi, including Aspergillus flavus, Aspergillus parasiticus, and Aspergillus nomius, that contain aflatoxins B1, B2, G1, and G2 [7-9]. Aflatoxins are both acutely and chronically toxic to animals and humans, and are classified as hazardous substances [10]. Hence, the development of traditional Korean liquor must be improved to minimize contamination with the aforementioned pathogenic bacteria, and to produce modified nuruk that has excellent SP and fermentative activity.
In this study, fungi samples were collected from various regions throughout Korea and the samples with outstanding α-amylase activity and SP were isolated and identified. The presence of aflatoxins in these identified strains was examined by an enzyme-linked immunosorbent assay (ELISA).
Materials and Methods
Preparation of nuruk
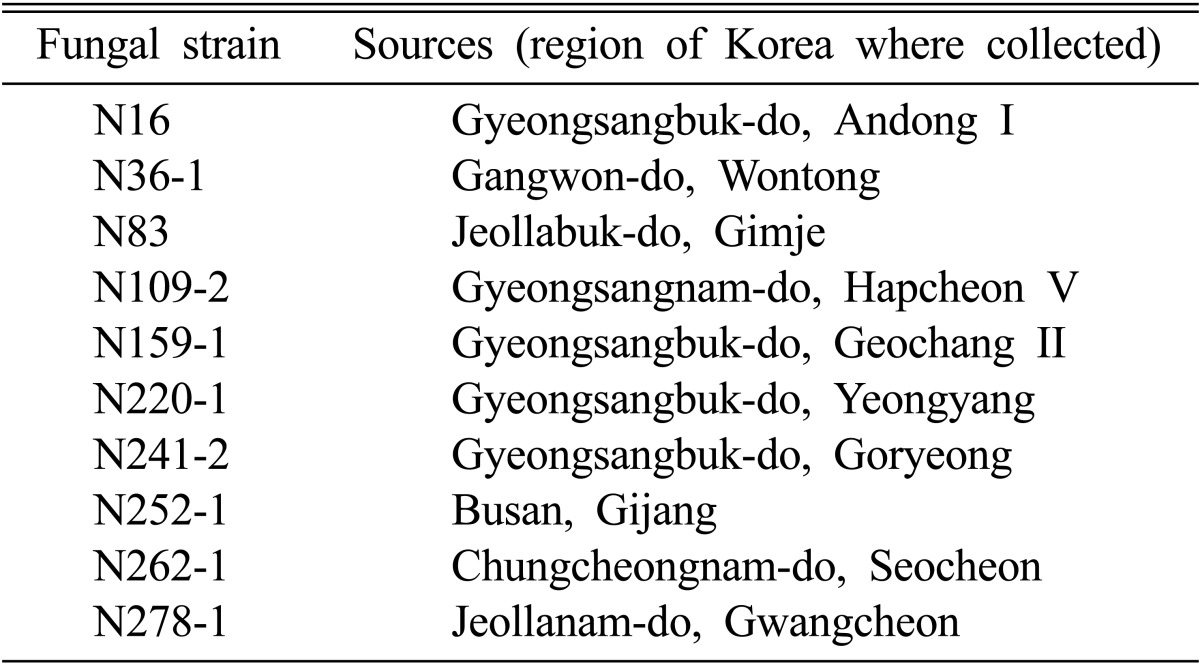
The samples used in this study were collected from 170 regions throughout Korea. Then Three hundred eighty four strains of fungi were isolated from 165 nuruks. The samples used in this study were collected at traditional markets in 23 regions of Chungcheongnam-do, 30 regions of Gyeongsangbuk-do, 18 regions of Gyeongsangnam-do, 12 regions of Chungcheongbuk-do, 20 regions of Gyeonggi-do, 13 regions of Jeollanam-do, 16 regions of Jeollabuk-do, 34 regions of Gangwon-do and four regions of Jeju-do. The collected nuruk from these 170 regions included 165 different varieties that were isolated and kept at 4℃. Fungi were grown on potato dextrose agar medium (0.4% [w/v] potato starch, 2% [w/v] dextrose; Difco, Sparks, MD, USA) at 30℃ for 4 days.
α-Amylase activity and SP
The samples were made from the wheat using selected strains of nuruk. The nuruk was inoculated in spore suspension (1.0 × 107 spores/mL) and shaken in 0.5% (w/v) NaCl at 20℃ for 3 hr. The extraction solution was centrifuged at 8,000 rpm for 10 min at 4℃. A clear supernatant was used in the enzyme solution. The substrate solution contained an acetate buffer (40 mM, pH 5.0) and 1% (w/v) soluble starch (preheated at 40℃ for 5 min). The enzyme solution (0.1 mL) was added, and the mixture was incubated at 40℃ for 30 min. Thereafter, a 0.00025 N iodine solution was added, and the transmittance (%T) was determined at 670 nm. Sample, substrate, and α-amylase blank determinations were undertaken under the same conditions. One unit of enzyme activity was defined as the amount of glucose released from 1 g of nuruk in 30 min [11]. SP of the final nuruk was measured using a 2% (w/v) soluble starch solution at a substrate, according to the methods of National Tax Service [12].
Starch hydrolysis and acid producing activity
To measure the starch hydrolysis activity of the isolated fungi in the nuruk, the starch medium was made using 0.1% (w/v) yeast nitrogen base without amino acids (Difco) as a nitrogen source and 0.2% (w/v) soluble starch (Difco) as a carbon source. Isolated strains were cultured on the starch medium, after which the strains were stained with an iodine solution (2% [w/v] KI, 1% [w/v] I2, and distilled water) and the size of their clear zone was measured [13]. To measure the acid-producing activity of the isolated fungi, a Czapek solution agar medium (Difco) was made using rose bengal (0.005% [w/v]; Kanto Chemical, Tokyo, Japan), bromocresol green (0.004% w/v as a pH indicator; Sigma-Aldrich, St. Louis, MO, USA), and filter-sterilized chloramphenicol (0.1% v/v; Sigma-Aldrich). The isolated colonies were then cultured and selected according to the size of their visible discolored zones [14]. Unless otherwise specified, all the chemicals were of analytical grade.
Microorganism identification
The microorganisms were sent to Macrogen (Seoul, Korea) for identification via PCR. PCR was performed using a model PTC-225 peltier thermal cycler (MJ Research, Reno, NV, USA) after DNA extraction (99℃, 10 min). The primers used were: ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'TCCTCCGCTTATTGATATGC-3') [15]. The amplification conditions were as follows: 95℃ for 5 min, 35 cycles at 94℃ for 45 sec, 75℃ for 1 min, 72℃ for 10 sec; and 72℃ for 5 min. PCR fragments were purified using ABI PRISM BigDyeTM terminator cycle sequencing kits (Applied Biosystems, Foster City, CA, USA). 18S rRNA sequencing was performed using an ABI PRISM 3730XL DNA analyzer (Applied Biosystems) with the same PCR primers. The sequences were identified using the BLAST program (http://www.ncbi.nlm.nih.gov).
Total aflatoxins
ELISA analysis was performed according to the instructions in the Neogen Veratox aflatoxin procedure. Using a blender, 5 g of a ground sample was shaken vigorously for 3 min in 25 mL of 70% methanol. The extract was filtered using Whatman No. 1 filter paper (Whatman, Maidstone, England), and the filtrate was collected. The concentration of total aflatoxins in parts per billion was recorded from a 650 nm-filter ELISA reader (Molecular Devices, Sunnyvale, CA, USA) that was calibrated using aflatoxin standards, after which all the data were read and calculated using Neogen's Veratox software ver. 2.3.4 (Neogen, Lansing, MI, USA).
Results and Discussion
Isolation of useful fungi
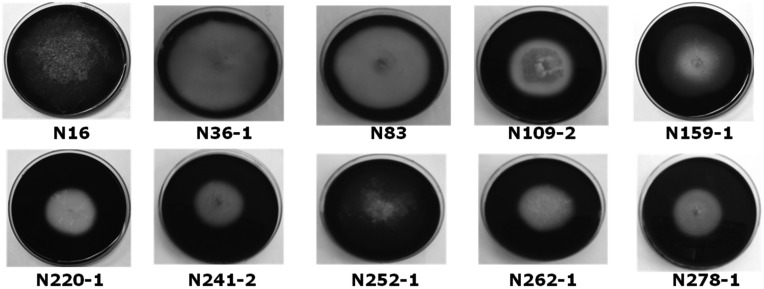
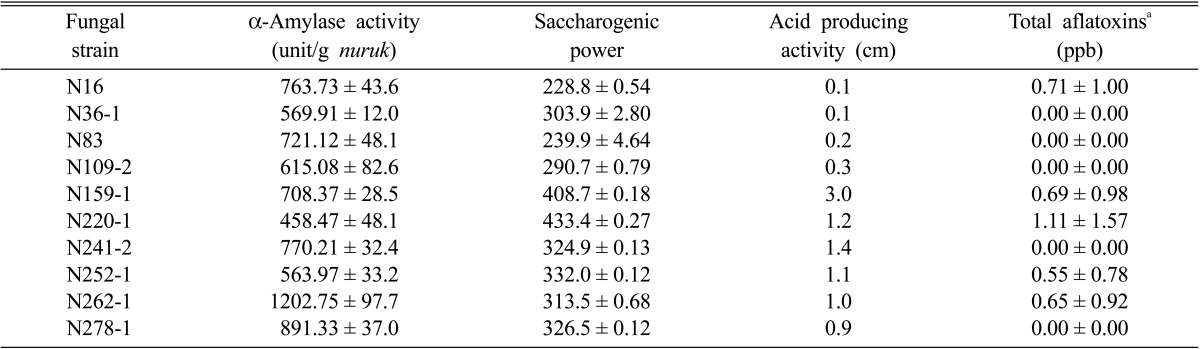
Three hundred and eighty four strains of fungi were isolated. Of these, 40 strains were isolated according to their morphology and cultured on starch media, after which the samples were stained with an iodine solution. Finally, 10 strains were selected according to their α-amylase activity, which ranged from 458.47 ± 48.1 to 1,202.75 ± 97.7 U/g) (Table 1). α-Amylase activity of nuruk made with strain N262-1 was the highest, and nuruk made with strain N220-1 was the lowest. The 10 selected strains, were N16 (2.0 cm), N36-1 (0.3 cm), N83 (0.3 cm), N109-2 (0.3 cm), N159-1 (0.8 cm), N220-1 (0.6 cm), N241-2 (0.3 cm), N252-1 (0.4 cm), N262-1 (0.5 cm), and N278-1 (0.3 cm), according to the size of the radius of the visible discolored zone, which ranged from 0.3~2.0 cm (Fig. 1). The SP of the 10 strains ranged from 228.8 ± 0.54 to 433.4 ± 0.27 SP (Table 2). The SP of nuruk made with N220-1 was the highest, and nuruk made with N16 was the lowest. The existence of acid-producing activity in fungi is important to prevent contamination with various microorganisms and abnormal fermentation in alcohol beverages made using nuruk. In this study, 10 fungi were examined using the size of their visible discolored zone via acid production. The visible discolored zones of the isolated strains ranged from 0.1~3.0 cm (Table 2). The systematic manufacture of liquor using useful strains with high acid-producing and starch hydrolysis activities must be carried out to globalize these traditional liquors, including makgeolli.
Table 1.
Sources of the selected fungi from traditional nuruk
Fig. 1.
Starch hydrolysis of selected fungi from traditional nuruk. N16, Rhizopus oryzae; N36-1, Talaromyces spectabilis; N83, Paecilomyces variotii; N109-2 and N278-1, Lichtheimia sp.; N159-1, N241-2, and N252-1, Aspergillus oryzae; N220-1 and N262-1, Aspergillus flavus.
Table 2.
Characteristics of the selected fungi from traditional nuruk
aTotal aflatoxins (B1, B2, G1, G2).
Identification of selected fungi
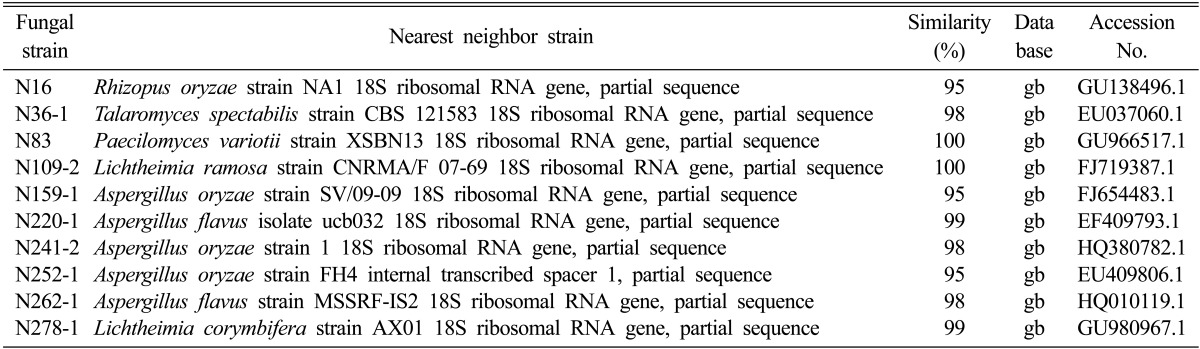
The 10 selected fungi were identified. N159-1 (KCTC 11927BP), N241-2, and N252-1 were Aspergillus oryzae; N220-1 and N262-1 were A. flavus, N36-1 was Talaromyces spectabilis, N83 was Paecilomyces variotii, and N109-2 and N278-1 were Lichtheimia sp. (Table 3). A. oryzae has previously been isolated from traditional Korean nuruk [6, 7]. This strain shows high amylase activity for starch degradation [16, 17] and also has a high level of acid-producing activity and a unique flavor [14, 18]. A. flavus has also been reported in Korean nuruk and was reported to produce aflatoxin [19].
Table 3.
Identification of the selected fungi from traditional nuruk
gb, genebank.
Total aflatoxins
Aflatoxin was detected in the nuruk made with 10 strains, A. oryzae (three strains), A. flavus (two strains), and others (five strains) at less than 1.11 ± 1.57 ppb (Table 2). The results were tested in triplicate via an ELISA. The current maximum levels set by the European commission are 2~8 mg/kg for aflatoxin B1 and total aflatoxins for groundnut, nuts, dried fruits, cereals, and processed products [20]. The action levels of the United States Food and Drug Administration [21] are 20 mg/kg of total aflatoxins in foods, peanuts, peanut products, and pistachio nuts. In the Codex Alimentarius [22], the guideline levels of total aflatoxins are 15 mg/kg for peanuts intended for further processing, almonds, hazelnuts, and pistachios. The current regulations in Korea for aflatoxin B1 are 10 mg/kg for cereals, bean products, peanuts, processed products, meju, doenjang, and gochujang [23, 24]. Kim et al. [25] reported that Aspergillus sp., Penicillium sp. and Rhizopus sp. isolated from nuruk cultured in raw wheat bran were devoid of aflatoxins. Yang et al. [26] noted that A. flavus, A. parasiticus, A. niger, and A. oryzae in doenjang were negative for aflatoxins, using multiplex PCR and direct-competitive-ELISA assays. Zheng et al. [27] compared the results of ELISA and high-performance liquid chromatography in the detection of total aflatoxins. ELISA was effective in measuring total aflatoxins (B1 + B2 +G1 +G2) in several appropriate commodities in quantitative ranges of 4~40 ppb. The ELISA method, due to its high variation in replicates, was found to be applicable as a screening method [8]. Therefore, the results indicated that ELISA is suitable for comparison and screening of a large number of samples.
It was verified that the useful fungi that were identified and isolated from nuruk produced only a small amount of aflatoxins (0~1.11 ppb). These may be a good material for manufacturing traditional Korean liquor in the near future.
Acknowledgements
This study was supported by a grant from the Traditional Food Globalization Research and Development Projects (Characterization of microorganism isolated from traditional Korean fermented foods and industrial use of starter through technology development of the fermentation process) of the Korea Food Research Institute.
References
- 1.Lee SR. Korean fermentation foods. 2nd ed. Seoul: Ewha Women's University Press; 1992. pp. 210–229. [Google Scholar]
- 2.Garbutt J. Essentials of food microbiology. London: Hodder Arnold; 1997. pp. 203–204. [Google Scholar]
- 3.Djien KS. Indigenous fermented foods. In: Rose AH, editor. Economic microbiology. Vol. 7. Fermented foods. London: Academic Press; 1982. pp. 27–29. [Google Scholar]
- 4.Kodama K, Yoshizawa K. Saké. In: Rose AH, editor. Economic microbiology. Vol. 1. Alcoholic beverages. Orlando: Academic Press; 1977. pp. 423–425. [Google Scholar]
- 5.Bae SM. Processing technology of traditional liquor. 3rd ed. Seoul: Woo-Gok Press; 2002. pp. 107–109. [Google Scholar]
- 6.Yu TS, Kim HS, Hong J, Ha HP, Kim TY, Yoon IW. Bibliographical study on microorganisms of nuruk (until 1945) J Korean Soc Food Sci Nutr. 1996;25:170–179. [Google Scholar]
- 7.Yu TS, Kim J, Kim HS, Hyun JS, Ha HP, Park MG. Bibliographical study on microorganisms of traditional Korean nuruk (since 1945) J Korean Soc Food Sci Nutr. 1998;27:789–799. [Google Scholar]
- 8.Nilüfer D, Boyacıoğlu D. Comparative study of three different methods for the determination of aflatoxins in Tahini. J Agric Food Chem. 2002;50:3375–3379. doi: 10.1021/jf020005a. [DOI] [PubMed] [Google Scholar]
- 9.Zaika LL, Buchanan RL. Review of compounds affecting the biosynthesis or bioregulation of aflatoxins. J Food Prot. 1987;50:691–708. doi: 10.4315/0362-028X-50.8.691. [DOI] [PubMed] [Google Scholar]
- 10.Butler WH, Barnes JM. Toxic effects of groundnut meal containing aflatoxin to rats and guinea pigs. Br J Cancer. 1963;17:699–710. doi: 10.1038/bjc.1963.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Japan Sake Brewers Association. A book with notes National Tax Service methods of analysis. 4th ed. Tokyo: JSB Association; 1993. pp. 213–221. [Google Scholar]
- 12.National Tax Service Technical Service Institute. Textbook of alcoholic beverage-brewing. Seoul: NTSTS Institute; 1997. pp. 368–370. [Google Scholar]
- 13.Williams PC, Kuzina FD, Hlynka I. A rapid colorimetric procedure for estimating the amylose content of starches and flours. Cereal Chem. 1970;47:411–420. [Google Scholar]
- 14.Kim HS, Hyun JS, Kim J, Ha HP, Yu TS. Enzymological characteristics and identification of useful fungi isolated from traditional Korean nuruk. Korean J Appl Microbiol Bioeng. 1998;26:456–464. [Google Scholar]
- 15.White TJ, Bruns T, Lee S, Tailor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 16.Park JW, Lee KH, Lee CY. Identification of filamentous molds isolated from Korean traditional nuruk and their amylolytic activities. Korean J Appl Microbiol Bioeng. 1995;23:737–746. [Google Scholar]
- 17.Lee SH, Jung HJ, Yeo SH, Kim HS, Yu TS. Characteristics of α-amylase of, a new species, Aspergillus coreanus NR 15-1. Korean J Biotechnol Bioeng. 2004;19:301–307. [Google Scholar]
- 18.Lee TS, Han EH. Volatile flavor components in mash of takju prepared by using Aspergillus oryzae nuruks. Korean J Food Sci Technol. 2001;33:366–372. [Google Scholar]
- 19.Lee SS, Park DH, Sung CK, Yoo JY. Studies on the yellow fungal isolates (Aspergillus species) inhabiting at the cereals in Korea. Korean J Mycol. 1997;25:35–45. [Google Scholar]
- 20.European Food Safety Authority. Parma: European Food Safety Authority; 2010. [cited 2010 Jun 7]. Commission Regulation (EU) No. 165 [Internet] Available from: http://ec.europa.eu/food/food/chemicalsafety/contaminants/aflatoxins_en.htm. [Google Scholar]
- 21.Guidance for industry: action levels for poisonous or deleterious substances in human food and animal feed [Internet] College Park: U. S. Food and Drug Administration; 2010. [cited 2010 Jun 4]. Available from: http://www.fda.gov/Food/GuidanceComplianceRegulatoryInformation/GuidanceDocuments/ChemicalContaminantsandPesticides/ucm077969.htm#afla. [Google Scholar]
- 22.Codex general standard for contaminants and toxins in food and feed [Internet] Rome: Codex Alimentarius; 2010. [cited 2010 Jun 7]. Available from: http://www.codexalimentarius.net/download/standards/17/CXS_193e.pdf. [Google Scholar]
- 23.A outline of the mycotoxins [Internet] Cheongwon: Korea Food and Drug Administration; 2009. [cited 2009 Aug 31]. Available from: http://www.kfda.go.kr/index.kfda?mid=69&pageNo=30.No.1055.hwp. [Google Scholar]
- 24.Park SK, Jang JI, Ha KT, Kim SD, Kim OH, Choi YH, Seung HJ, Kim SJ, Lee KA, Jo HB, et al. A survey of the presence of aflatoxins in herb medicines. J Food Hyg Saf. 2009;24:169–173. [Google Scholar]
- 25.Kim HS, Hyun JS, Ha HP, Yu TS. Characteristics of useful fungi isolated from traditional Korean nuruk. J Korean Soc Food Sci Nutr. 1997;26:767–774. [Google Scholar]
- 26.Yang ZY, Shim WB, Kim JH, Park SJ, Kang SJ, Nam BS, Chung DH. Detection of aflatoxin-producing molds in Korean fermented foods and grains by multiplex PCR. J Food Prot. 2004;67:2622–2626. doi: 10.4315/0362-028x-67.11.2622. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Z, Humphrey CW, King RS, Richard JL. Validation of an ELISA test kit for the detection of total aflatoxins in grain and grain products by comparison with HPLC. Mycopathologia. 2005;159:255–263. doi: 10.1007/s11046-004-8666-0. [DOI] [PubMed] [Google Scholar]