Abstract
Microorganisms are significantly affected when the ambient pH of their environment changes. They must therefore be able to sense and respond to these changes in order to survive. Previous investigators have studied various fungal species to define conserved pH-responsive signaling pathways. One of these pathways, known as the Pal/Rim pathway, is activated in response to alkaline pH signals, ultimately targeting the PacC/Rim101 transcription factor. Although the central signaling components are conserved among divergent filamentous and yeast-like fungi, there is some degree of signaling specificity between fungal species. This specificity exists primarily in the downstream transcriptional targets of this pathway, likely allowing differential adaptation to species-specific environmental niches. In this review, the role of the Pal/Rim pathway in fungal pH response is discussed. Also highlighted are functional differences present in this pathway among human fungal pathogens, differences that allow these specialized microorganisms to survive in the various micro-environments of the infected human host.
Keywords: Aspergillus nidulans, Candida albicans, Cryptococcus neoformans, PacC, Rim101, Saccharomyces cerevisiae, Signal transduction, Yeast
In order to survive, microorganisms must be able to sense and respond to their surroundings. These cells are frequently exposed to rapidly changing environmental stresses. However, microorganisms are remarkably resilient and have developed mechanisms to efficiently adapt to changes in their environment. Ambient pH is one extracellular condition to which these cells must be able to respond to in order to survive. Changes in pH induce many stresses on cellular functions including altering micronutrient availability, protein function, and membrane potential.
Among microorganisms, fungi are often able to survive in a diverse range of environmental conditions. Those that live predominantly in the external environment must be able to propagate despite large variations in temperature, nutrient availability, and pH. Mammalian fungal pathogens are additionally able to survive the unique stresses of the various microenvironments within the infected host, where the pH can range from 2 to 10. Therefore, fungal signaling pathways responding to external pH signals are important components of their cellular machinery.
While many signaling pathways are directly or indirectly regulated by pH, one of the most specialized pH response pathways is the Pal/Rim alkaline response pathway. This pathway has been studied in multiple fungal species and is especially important for fungal pathogenesis. This review will focus primarily on how the Pal/Rim pathway stimulates changes in cellular processes as a response to alkaline ambient pH and how this pathway is utilized by fungal pathogens to colonize their host and cause disease.
The PacC/Rim101 Transcription Factors Mediate pH Responses in Aspergillus nidulans and Saccharomyces cerevisiae
Ambient pH response signaling in fungi was first elucidated in A. nidulans and S. cerevisiae. The major effector, PacC in A. nidulans and Rim101 in S. cerevisiae, is a three Cys2His2 zinc finger transcription factor that mediates changes in gene expression in response to neutral or alkaline pH. PacC/Rim101 is activated when a region of its C-terminus is proteolytically cleaved, allowing the processed protein to mediate gene regulation. This proteolysis is induced under alkaline or neutral conditions.
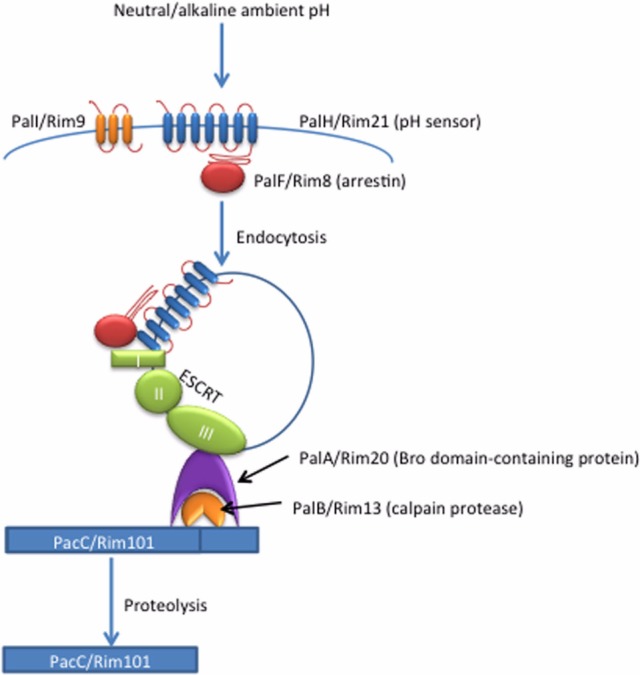
Activation of PacC/Rim101 is stimulated by a highly conserved signaling pathway, referred to as the Pal pathway in filamentous fungi, and the Rim pathway in yeast-like fungi, involving interactions between signaling complexes on the plasma membrane and on the endosomal membranes. The Pal/Rim signal is initiated at the plasma membrane where a complex of proteins first senses neutral/alkaline pH. This signal is transferred via non-canonical arrestin signaling to the endosomal membranes where PacC/Rim101 processing and activation ultimately occur (Fig. 1) [1-3].
Fig. 1.
PacC/Rim101 pH response pathway. Neutral/alkaline pH signals are sensed at the cell surface by the 7 transmembrane-domain protein PalH/Rim21. This extracellular signal leads to PalF/Rim8 ubiquitination and subsequent endocytosis of the PalH/PalF membrane complex. Prior to activation, PalI/Rim9 aids in the plasma membrane localization of the PalH/Rim21 protein. Once internalized to the endomembranes, the PalF/Rim8 arrestin-like protein interacts with the ESCRT complex, including ESCRT-I proteins that recruit and activate the ESCRT-II and -III proteins to this site. PalA/Rim20 is subsequently recruited to the ESCRT-III complex through its Bro domain, in turn recruiting both the calpain-like protease PalB/Rim13 and the transcription factor PacC/Rim101 into proximity. PalC also interacts with the ESCRT-III complex. Proteolysis of PacC/Rim101 by the PalB/Rim13 protease results in the activation of this transcriptional regulator, and the resulting induction of alkaline response genes.
Sensing the external environment: cell surface signaling events
In A. nidulans, the plasma membrane signaling complex is composed of a 7-transmembrane domain receptor, PalH, a 3-transmembrane domain protein, PalI, and an arrestin-related protein, PalF [1-3]. Orthologous proteins in S. cerevisiae have also been described (Fig. 1).
The arrestin protein function in this pathway helps to internalize the pH signal from the cell surface receptor. Classically, arrestins mediate the downregulation of G protein-coupled receptor (GPCR) signaling by interacting with the cytosolic regions of GPCRs, preventing G-protein interaction, and mediating receptor endocytosis and degradation by the lysosome. However, arrestins can also serve as signaling scaffolds that mediate signaling from an endocytosed vesicle [4]. Since a G-protein does not appear to be involved in the PacC/Rim101 signaling pathways, the arrestin-related PalF/Rim8 appears to be mediating transcription factor processing through the latter arrestin function. Also, the PalF and Rim8 arrestin proteins promote PacC/Rim101 processing, which is not characteristic of the typically inhibitory behavior of arrestins [5-7]. Furthermore, A. nidulans PalF appears to interact with the C-terminal, cytosolic region of PalH regardless of ambient pH [7]; this interaction would theoretically inhibit any PalH/Gprotein interaction. Therefore, the function of the PacF/Rim8 family of proteins is atypical for classical arrestinlike proteins.
The PalI protein is also a member of the plasma membrane signaling complex and functions by an unknown mechanism to promote proper PalH plasma membrane localization [8]. This hypothesized function is supported by the observation that PalH is located primarily on endosomal membranes in a palI strain, rather than its typical localization to the plasma membrane [8].
Much of the plasma membrane signaling complex is conserved between A. nidulans and S. cerevisiae; however, there are several differences. S. cerevisiae has two 7 TMD receptors, Dgf16 and Rim21, both of which are homologous to PalH, and both required for Rim101 processing [9, 10]. However, unlike PalF, Rim8 ubiquitination is not dependent on pathway activation. It appears that a certain proportion of Rim8 molecules are always ubiquitinated regardless of ambient pH, and this ubiquitinated population is in complex with the ESCRT-I subunit Vps23 [5]. Therefore, Rim8 may be recruited to Rim21 or Dgf16 in neutral to alkaline conditions, bringing together the ESCRT complex and the plasma membrane complex. However, it has not been shown if the interaction between Rim8 and Rim21 or Dgf16 is regulated by pH. This activation sequence appears to be different from the Pal pathway where PalF is already bound to PalH before pathway activation, and this activation induces PalF ubiquitination and phosphorylation to drive endosomal interactions [6, 7].
Internalization of the pH signal: endocytosis and endomembrane signaling
In neutral and alkaline conditions, PalF becomes phosphorylated and ubiquitinated, resulting in endocytosis of the PalH/PalF complex to endomembranes [6, 7]. The propagation of this signal at endomembranes requires the involvement of the ESCRT complex. The classical function of the ESCRT complex is to bind, sort, and transfer ubiquitinated cargo (often membrane proteins) into the multivesicular body for degradation [11]. The ESCRT complex is also involved in retroviral budding, and cytokinetic abscission [11]. Formation of the ESCRT complex is initiated by ESCRT-0 binding to ubiquitinated cargo on endosomal membranes and the sequential recruitment of the other 3 complexes (ESCRT-I, -II, -III) [11]. The ESCRT-I, -II, and -III subunits Vps20-Snf7 were found to be required for S. cerevisiae Rim101 processing, and many members of the Pal/Rim signaling pathway have proven or predicted ESCRT subunit interaction domains [12].
Once endocytosed, the PalH-PalF complex interacts with downstream signaling proteins, presumably by interacting with the ESCRT complex via a PalF Vps23 binding domain (ESCRT-I subunit). PalC could also mediate interaction between the plasma membrane signaling complex and the ESCRT machinery. PalC is required for PacC activation and was found to localize to PalF/PalH endocytic vesicles after endocytosis. While its true function is unknown, PalC contains a Vps20-Snf7 interaction domain that may help the plasma membrane complex interact with downstream Pal proteins that also contain Vps20-Snf7 interaction domains [13].
The assembly of the ESCRT complex may be induced by Rim8/PalF. Rim8, and possibly PalF, appears to be able to substitute for the ESCRT-0 protein Vps27 in binding to the ESCRT-I protein Vps23 to initiate ESCRT complex assembly [5, 13, 14]. Therefore, one possibility for activation of this pathway is that PalF/Rim8 mediated endocytosis may drive ESCRT complex assembly, bringing other ESCRT interacting signaling proteins into proximity to carry out PacC/Rim101 processing.
Transcription factor processing and activation
This pH-responsive signaling pathway ultimately results in the cleavage and activation of the PacC/Rim101 transcription factors. The recruitment of the remaining ESCRT complex components leads to the recruitment of the Vps20-Snf7 interacting protein PalA/Rim20 as well as the calpain-like protease PalB/Rim13. PalA/Rim20 interacts with both the C-terminus of PacC/Rim101 and PalB/Rim13, leading to the PalB/Rim13-mediated proteolysis and activation of PacC/Rim101. For PacC, this proteolysis removes ~180 C-terminal amino acids, allowing this transcription factor it to relocate to the nucleus. In contrast, Rim101 is able to localize to the nucleus without Rim13 processing, but it is not able to mediate gene regulation without the removal of ~70 C-terminal amino acids [15, 16]. One possibility for this observation is that Rim13-mediated cleavage may allow Rim101 to interact with other co-regulators that are necessary for gene regulation [15].
In A. nidulans once PacC is proteolytically processed, 245 C-terminal amino acids are further cleaved from this protein, presumably by the proteasome [17, 18]. This second cleavage does not occur in S. cerevisiae. It has been hypothesized that both fully cleaved PacC and intermediate PacC are able to mediate gene regulation, with one form possibly acting as an activator and the other acting as a repressor. This may explain why S. cerevisiae Rim101 is believed to act only as a repressor but A. nidulans PacC can act as both an activator and repressor. However, it has yet to be definitively demonstrated if the intermediate form of PacC is functional.
Transcriptional response to Pal/Rim pathway
In neutral to alkaline conditions, activated PacC represses acid response genes while also inducing the expression of alkaline response genes. Most genes regulated by this transcription factor encode secreted enzymes (proteases and phosphatases) and permeases that function at either acidic or alkaline pH. PacC also regulates genes involved in the synthesis of secreted metabolites, such as penicillin and toxins. PacC induces its own expression, thereby serving in a positive feedback loop [1-3, 19, 20].
In S. cerevisiae, Rim101 promotes alkaline growth by repressing the expression of the NRG1 gene. Negative regulator of glucose-repressed genes (Nrg1) is a transcription factor that inhibits the expression of ion transporters, such as the Na+-ATPase transporter Ena1, that are important for maintaining ion homeostasis during conditions of increased pH [15, 21]. Expression of the low zinc-induced glycosylphosphatidylinositol-anchored protein Zps1 is also repressed by Nrg1, and its expression is induced when Rim101 is activated [15, 21]. Smp1 is another transcription repressor that is direct target of Rim101, and is involved in the repression of haploid invasive growth, rough colony morphology, and sporulation. Its repression by Rim101 also induces CTW1 expression, which has been shown to promote cell wall integrity [15]. Along with promoting growth in alkaline pH, Rim101 activity is involved in invasive growth, sporulation, and ion homeostasis [15, 21].
Candida albicans
Candida albicans is a human commensal that commonly colonizes mucosal areas such as the oral-pharyngeal, gastrointestinal, and urogenital tracts. This organism is also able to cause invasive opportunistic infections at these mucosal surfaces, as well as systemic infections, potentially disseminating to almost any organ. The pH at these different anatomic areas ranges from quite acidic (gastrointestinal tract and vaginal tract) to slightly alkaline (oral-pharyngeal tract and bloodstream). To survive pH changes of this magnitude, C. albicans must be able to sense these changes and drastically change its cellular functions in response to new environments. It is not surprising therefore, that C. albicans requires pH-sensing pathways, such as the Rim101 pathway, to survive within the infected host [22-25].
C. albicans is able to grow in both a yeast and a hyphal form. Transition from acidic to neutral/alkaline conditions stimulates C. albicans to switch from yeast to filamentous growth [23-25]. This morphological change is important for virulence as filamentous growth has been found to be important for this organism's ability to cause systemic infections [26]. Conversely, growth in the yeast form is needed for colonizing acidic environments like the vaginal tract [27].
The Rim101 pathway is important in pH-responsive morphological transitions, and therefore for Candida pathogenesis [22-25]. Like in A. nidulans and S. cerevisiae, the Rim101 pathway in C. albicans is activated by neutral to alkaline ambient pH, resulting in the proteolytic processing of the Rim101 transcription factor [22, 23]. Many of the signaling components upstream of Rim101 activation are conserved in C. albicans, including homologs of PalH/Rim21, Dgf16; PalF/Rim8; PalA/Rim20; PalB/Rim13; and PalI/Rim9. All of these components were found to be required for proper alkaline response gene expression, proper processing of Rim101, and/or survival at high pH levels [10, 22, 23, 25, 28, 29].
While much of the activation pathway seems to be similar to the PacC/Rim101 pathways in A. nidulans and S. cerevisiae, the cleavage of C. albicans Rim101 is unique. In neutral to alkaline conditions, a ~10 kD C-terminal region is removed from the full length 85 kD Rim101p in a Rim13-dependent manner. However, Rim13 is also required for the cleavage of ~20 kD from the C-terminus of Rim101 in acidic conditions (pH 4). Rim8 and Rim20 were also found to be required for these processing events. It is hypothesized that the 75 kD form present in alkaline conditions functions to regulate alkaline response genes, while the 65 kD form present in acidic conditions regulates pH-independent genes [29]. C-terminally truncated forms of Rim101 are able to suppress mutations in genes encoding upstream components of Rim101 activation and restore alkaline gene expression. However, it has not been demonstrated whether reconstitution with the 65 kD-form of Rim101 present in acidic conditions would also rescue defects in alkaline-specific gene expression.
C. albicans Rim101 activation in alkaline conditions leads to the transcriptional induction of genes required for alkaline tolerance. Similar to S. cerevisiae, C. albicans Rim101 induces the expression of genes encoding ion pumps, such as ENA1, and iron acquisition genes, such as FRP2, FRE2, and ARN1. Along with increasing alkaline tolerance, the proteins encoded by these genes are important for high cation and low iron tolerance [22, 30]. Unlike S. cerevisiae, C. albicans Rim101 appears to be able to act as both a transcriptional activator and repressor. Also, C. albicans NRG1 is repressed under alkaline conditions, but in a Rim101-independent manner [22, 23, 31].
The C. albicans Rim101 pathway also regulates the expression of genes important for virulence. In addition to regulating genes necessary for hyphal growth, virulence-associated genes encoding adherence- and invasion-inducing proteins are also induced by the Rim101 pathway, such as the adhesin protein Als3 and the E-cadherin degrading protease Sap5p [32, 33]. The C. albicans Rim101 pathway also regulates cell wall composition [1, 22, 32, 33]. pH-regulated genes 1 (PHR1) and 2 (PHR2) were two of the first Rim101 regulated genes required for virulence. These β-1,3- and β-1,6-glucan crosslinking cell wall proteins are functionally homologous and are oppositely regulated by Rim101, with Rim101 inducing PHR1 expression at alkaline pH while suppressing PHR2 expression. Therefore, PHR1 is required for infection of neutral to alkaline areas inside the host, while PHR2 is required for infection of acidic areas [25, 27, 34]. In addition to controlling the expression of genes encoding cell surface proteins, Rim101 also regulates the expression of genes encoding cell wall modifying enzymes. These modification enzymes, including a chitinase and a glucosidase, are induced by Rim101 and are important for interactions with endothelial cells [32].
Analysis of the C. albicans Rim101 pathway revealed that the components upstream of the transcription factor function in much the same way as their homologues in the S. cerevisiae Rim101 pathway or the A. nidulans PacC pathway. However, major differences are evident in the way C. albicans Rim101 is processed, the relative levels of transcriptional repression or activation attributed to this transcription factor, and the types of genes regulated by Rim101. Therefore, C. albicans has customized this conserved pH-signaling pathway to adapt to surviving in specific regions within the human host. In this environment, pH changes are not only stressors but also clues that this commensal organism uses to adapt to different locations within the human body.
Cryptococcus neoformans
The ascomycete PacC/Rim101 signaling system has been studied in detail. Much less is known about this pathway in basidiomycetes. One such basidiomycete is the human opportunistic pathogen Cryptococcus neoformans. This organism causes fungal meningitis in immunocompromised individuals and has become a prevalent pathogen due to the human immunodeficiency virus (HIV) pandemic [35]. This organism grows in both yeast and hyphal form, and, similar to C. albicans, responds to pH signals to induce virulence factors. Unlike C. albicans, however, C. neoformans does not grow well above a pH of 8. It is able to grow inside macrophage phagolysosomes (pH 5) and in the slightly alkaline conditions of cerebrospinal fluid and serum (pH 7.4) [36, 37]. Melanin production, polysaccharide capsule, and titan cell formation are all virulence factors that C. neoformans induces during infection, and each is regulated to some degree by the C. neoformans Rim101 transcription factor.
C. neoformans Rim101 was first characterized in a mutant library screen in which the rim101 mutant was found to have a slight melanin defect and also to survive better in mice compared to wild type strains [38]. The Rim101 protein was also identified in a bioinformatic search for transcription factors potentially regulated by the protein kinase A (PKA) pathway [39]. Discovering that the C. neoformans Rim101 has a functional PKA phosphorylation site was unexpected since this type of regulation was not appreciated for other PacC or Rim101 homologues.
Some of the processes regulated by the C. neoformans Rim101 protein are characteristic of the classical Rim pathway, suggesting that it is a true Rim101 ortholog. For example, similar to what is observed in S. cerevisiae and C. albicans, the C. neoformans rim101 strain is more susceptible to ion stress and alkaline growth conditions [39]. Along with these predicted functions, CnRim101 is also involved in species-specific functions, such as capsule attachment and titan cell formation [39, 40]. The titan cell is a newly described C. neoformans morphotype that is 5~10 times larger than a regular cell, displaying a thickened capsule and increased resistance to oxidative and nitrosative stress [41, 42].
Activation of C. neoformans Rim101 appears to be dependent on both classical upstream Rim/Pal activators and also by the PKA pathway. As in other organisms, activation of CnRim101 also requires proteolytic processing. When C. neoformans is incubated in non-capsule inducing conditions (rich medium at 30℃), CnRim101 is processed from its predicted 140 kD form to 120 kD. It is further processed to 70 kD when the cells are incubated in capsule inducing conditions. This protein processing correlates with proper cellular localization. Rim101 is localized primarily to the nucleus in both capsule inducing and non-inducing conditions. This expected pattern of Rim101 localization is disrupted in conditions that disrupt proteolytic processing [39]. In the absence of activation by either Rim20 or PKA, CnRim101 remains in its full-length form and loses most of its nuclear localization, instead showing diffuse or punctate localization throughout the cell. Mutating the PKA phosphorylation site on CnRim101 also disrupts processing and nuclear localization [39].
Analysis of gene expression patterns regulated by CnRim101 under capsule inducing conditions reveal that this transcription factor primarily regulates genes associated with ion and metal homeostasis, cell wall modification, and capsule. For example, similar to S. cerevisiae and C. albicans, CnRim101 regulates the expression of the ENA1 gene encoding a sodium pump important for ion homeostasis at increased pH [39]. Likely in response to altered metal ion availability in alkaline pH, CnRim101 also induces the expression of genes encoding an iron transporter and an iron permease (SIT1 and CFT1), and a copper transporter (CTR4). Consistent with the capsule defect in the C. neoformans rim101 strain, CnRim101 appears to regulate genes known to be important for polysaccharide capsule biosynthesis, including, a glucose dehydrogenase (UGD1), a mannosyltransferase (CMT1), and a phosphomannomutase (PMM). Cell wall modification may also effect capsule attachment, which appears to be disrupted in the rim101 strain. Consistent with this hypothesis, CnRim101 regulates genes involved in cell wall production, including AGS1 encoding an α-1,3-glucan synthase and a glucan 1,3 beta-glucosidase protein [39]. C. albicans Rim101 was also found to regulate the expression of cell wall proteins, which were previously found to be important for virulence in an oral-pharyngeal infection model [32].
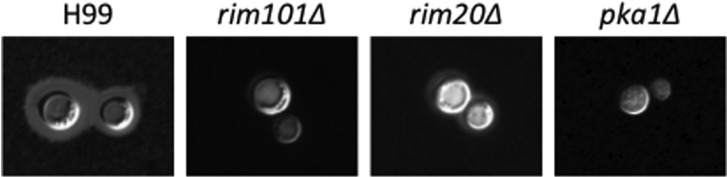
The C. neoformans rim101 mutant strain is hypersusceptible to elevated pH and iron deprivation, two conditions known to exist in the host. Moreover, this strain has a dramatic defect in attachment of capsule, the main virulence factor for this pathogenic fungus (Fig. 2). Therefore, one might have predicted that Rim101 would be required for pathogenesis. However, in two separate laboratories and two independent models of cryptococcosis, the Cnrim101 mutant displayed preserved/enhanced virulence [38, 39]. The mechanism behind this hypervirulence has yet to be elucidated. However, it was observed that the rim101 strain survived better than wild-type strains in macrophages, indicating that the mutant might be better adapted to grow in acidic conditions [39]. Therefore, CnRim101 may also be involved in the suppression of acid response genes. In a separate study, a rim101 mutant strain was noted to be engulfed by macrophages better than wild type [43]. However, because the level of phagocytosis was determined after 24 hr incubation with macrophages, it is unclear if the higher number of macrophage cells containing rim101 cells was due to increased phagocytosis or to better survival/replication inside the macrophages. Nevertheless, these data suggest that the rim101 strain may interact with its host's immune system differently than wild type C. neoformans, perhaps in part offering initial insight into the mechanism for the enhanced virulence of this mutant strain.
Fig. 2.
Cryptococcus neoformans Rim101 and Rim20 proteins control capsule expression on the cell surface. The indicated strains were incubated at 37℃ and 5% CO2 in Dulbecco's modified Eagle medium for 3 days and stained with India ink to visualize the polysaccharide capsule.
Studies on the Rim101 pathway in C. neoformans have already revealed significant differences in the way it is regulated. However, little is currently known about how CnRim101 is activated or the specific proteins mediating its activation. Genes encoding PalB/Rim13, PalI/Rim9, and PalC homologues are present in the C. neoformans genome. These homologues, along with a PacC/Rim101 and PalA/Rim20 homologues, were also found in the related basidiomycete Ustilago maydis. In this plant fungal pathogen, all of these Rim pathway components, except PalI/Rim9, are required for ion stress tolerance and growth in alkaline conditions [44, 45]. Interestingly, homologues of the cell surface signaling proteins PalH/Rim21, PalF/Rim8, or Dgf16 are not immediately recognizable in either the U. maydis or the C. neoformans genomes (unpublished) [44, 45]. Proteins with homologous functions could be present but without sufficient conservation of sequence to be detected by homology searches. For example, the predicted 7-transmembrane G-protein coupled receptor Grp5 was previously found to be important for titan cell formation. However, this protein did not readily interact with known G-alpha proteins in a modified yeast two-hybrid assay [39, 40]. Since Rim101 is also required for titan cell formation, it is interesting to speculate if Gpr5 or a related protein is acting as the pH-responsive sensor protein, similar to PalH/Rim21 or Dgf16. Alternatively, it is possible that the Pal/Rim pathway is regulated in a unique way in basidiomycetes, such as the unexpected PKA regulation of Rim101 in C. neoformans.
In addition to identifying the proteins responsible for activating Rim101 in C. neoformans, further studies will also define the active form of CnRim101. While this transcriptional regulator is minimally proteolytically processed in yeast-peptone-dextrose medium which has a pH of 6.5 [39], it is unknown how the protein is processed in more acidic conditions, such as a macrophage phagolysosome. Also, there appears to be a second processing even that occurs under host relevant conditions [39]. Does this protein processing occur in response to increases in pH or to another host-derived environmental cue, such as alterations in temperature, iron levels, or CO2 levels? Finally, defining the role of the Rim pathway in C. neoformans capsule attachment and immune activation will likely provide important insight into how this fungal pathogen avoids immune recognition and establishes long-lived infections in the human host.
Other Signaling Pathways Involved in the Fungal pH Response
The Pal/Rim pH response pathway acts together and in parallel with other signaling pathways to modulate cellular functions in response to changes in pH. Because pH changes create such widespread changes to cellular functions, these interacting signaling pathways are involved in diverse cellular processes, such as nutrient acquisition and carbonic anhydrase activity.
The calcineurin pathway acts in parallel with the Rim pathway in S. cerevisiae and C. albicans [46-48]. In response to alkaline pH signals, the cytoplasmic Ca2+ concentration increases, activating the calcium-responsive phosphatase calcineurin. This important signaling protein activates the transcription factor Crz1, which in turn controls the expression of pH response genes, such as that encoding the sodium pump Ena1 that is also regulated by the Rim101 pathway [46, 47].
The PKA pathway is also involved in fungal pH response. In S. cerevisiae, PKA activation correlates with decreased alkaline tolerance [49]. PKA activity may increase alkaline sensitivity through phosphorylation and inhibition of Crz1, as mentioned above. Therefore, PKA acts in opposition to calcineurin in the pH response [47]. In contrast, the C. neoformans PKA pathway is involved in increasing alkaline tolerance [39]. Therefore, the PKA pathway may be utilized for both inducing and repressing the alkaline response in different fungal species.
The concentration of intracellular bicarbonate (HCO3) changes with pH and CO2 levels. Carbonic anhydrase is the enzyme responsible for accelerating the conversion of CO2 into bicarbonate [50]. In C. neoformans, alterations in carbonic anhydrase activity and resulting shifts in bicarbonate levels regulate adenylyl cyclase activity in a pH-dependent manner [51]. Therefore, fungi such as C. neoformans use multiple, interacting proteins and signaling pathways (carbonic anhydrase, PKA, Rim101, calcineurin) to modulate the cellular response to the environmental pH signal.
Finally, changes in pH levels result in changes in nutrient availability. The nutrient sensing target of rapamycin (TOR) pathway negatively regulates the alkaline-induced yeast-to-hyphal transition in C. albicans. Mds3 is involved in inhibiting TOR in response to alkaline pH, allowing for morphologic changes [52]. Interestingly, C. albicans may be able to locally regulate is pH in low glucose conditions. Metabolism of amino acids during glucose starvation leads to the secretion of ammonia, which sufficiently increases the pH surrounding the C. albicans cells to induce a yeast-to-hyphal transition [53].
In summary, both pathogenic and non-pathogenic fungal species have adapted intricate mechanisms to respond to common environmental signals such as changes in ambient pH. Species-specific alterations in conserved signaling paradigms indicate ways in which these distantly related organisms have adapted these pathways to survive in their unique environmental niches. In particular, fungal pathogens of humans have linked the expression of virulence-associated phenotypes to host-derived pH cues in order to effectively colonize and survive within their host.
Acknowledgements
This work was supported by NIH grants R01 AI050128 and R01 AI074677 (JAA). KS is supported by an NIH T32 training grant to the Duke University Cellular and Molecular Biology Graduate Training Program.
References
- 1.Peñalva MA, Tilburn J, Bignell E, Arst HN., Jr Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Peñalva MA, Arst HN., Jr Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu Rev Microbiol. 2004;58:425–451. doi: 10.1146/annurev.micro.58.030603.123715. [DOI] [PubMed] [Google Scholar]
- 3.Arst HN, Peñalva MA. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 2003;19:224–231. doi: 10.1016/s0168-9525(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 4.Shenoy SK, Lefkowitz RJ. beta-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz S, Rodríguez JM, Bussink HJ, Sánchez-Ferrero JC, Arst HN, Jr, Peñalva MA, Vincent O. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci U S A. 2005;102:12141–12146. doi: 10.1073/pnas.0504776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hervás-Aguilar A, Galindo A, Peñalva MA. Receptor-independent ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J Biol Chem. 2010;285:18095–18102. doi: 10.1074/jbc.M110.114371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink HJ, Múnera-Huertas T, Stanton L, Hervás-Aguilar A, Espeso EA, Tilburn J, et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell. 2007;6:2365–2375. doi: 10.1128/EC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothfels K, Tanny JC, Molnar E, Friesen H, Commisso C, Segall J. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:6772–6788. doi: 10.1128/MCB.25.15.6772-6788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barwell KJ, Boysen JH, Xu W, Mitchell AP. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot Cell. 2005;4:890–899. doi: 10.1128/EC.4.5.890-899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Xu W, Smith FJ, Jr, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell. 2004;15:5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galindo A, Hervás-Aguilar A, Rodríguez-Galán O, Vincent O, Arst HN, Jr, Tilburn J, Peñalva MA. PalC, one of two Bro1 domain proteins in the fungal pH signalling pathway, localizes to cortical structures and binds Vps32. Traffic. 2007;8:1346–1364. doi: 10.1111/j.1600-0854.2007.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent O, Rainbow L, Tilburn J, Arst HN, Jr, Peñalva MA. YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue, AIP1/Alix. Mol Cell Biol. 2003;23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Mitchell AP. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Díez E, Alvaro J, Espeso EA, Rainbow L, Suárez T, Tilburn J, Arst HN, Jr, Peñalva MA. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002;21:1350–1359. doi: 10.1093/emboj/21.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hervás-Aguilar A, Rodríguez JM, Tilburn J, Arst HN, Jr, Peñalva MA. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J Biol Chem. 2007;282:34735–34747. doi: 10.1074/jbc.M706723200. [DOI] [PubMed] [Google Scholar]
- 19.Peñalva MA, Arst HN., Jr Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol Mol Biol Rev. 2002;66:426–446. doi: 10.1128/MMBR.66.3.426-446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caddick MX, Brownlee AG, Arst HN., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 21.Lamb TM, Xu W, Diamond A, Mitchell AP. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem. 2001;276:1850–1856. doi: 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 22.Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol. 2009;12:365–370. doi: 10.1016/j.mib.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Davis D. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet. 2003;44:1–7. doi: 10.1007/s00294-003-0415-2. [DOI] [PubMed] [Google Scholar]
- 24.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell AP. Dimorphism and virulence in Candida albicans. Curr Opin Microbiol. 1998;1:687–692. doi: 10.1016/s1369-5274(98)80116-1. [DOI] [PubMed] [Google Scholar]
- 27.De Bernardis F, Mühlschlegel FA, Cassone A, Fonzi WA. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornet M, Richard ML, Gaillardin C. The homologue of the Saccharomyces cerevisiae RIM9 gene is required for ambient pH signalling in Candida albicans. Res Microbiol. 2009;160:219–223. doi: 10.1016/j.resmic.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Martin SJ, Bruno VM, Mitchell AP, Davis DA. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot Cell. 2004;3:741–751. doi: 10.1128/EC.3.3.741-751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell. 2008;7:1168–1179. doi: 10.1128/EC.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 32.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, Mitchell AP, Filler SG. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10:2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, Hube B. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol. 2007;63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- 34.Baek YU, Martin SJ, Davis DA. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall beta-glycosidase Phr2. Eukaryot Cell. 2006;5:1550–1559. doi: 10.1128/EC.00088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. Cryptococcus: from human pathogen to model yeast. Washington, D.C.: ASM Press; 2010. [Google Scholar]
- 36.Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, Perfect JR. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyberg K, Johansson U, Johansson A, Camner P. Phagolysosomal pH in alveolar macrophages. Environ Health Perspect. 1992;97:149–152. doi: 10.1289/ehp.9297149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okagaki LH, Wang Y, Ballou ER, O'Meara TR, Bahn YS, Alspaugh JA, Xue C, Nielsen K. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot Cell. 2011;10:1306–1316. doi: 10.1128/EC.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010;6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chun CD, Madhani HD. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One. 2010;5:pii: e12503. doi: 10.1371/journal.pone.0012503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cervantes-Chávez JA, Ortiz-Castellanos L, Tejeda-Sartorius M, Gold S, Ruiz-Herrera J. Functional analysis of the pH responsive pathway Pal/Rim in the phytopathogenic basidiomycete Ustilago maydis. Fungal Genet Biol. 2010;47:446–457. doi: 10.1016/j.fgb.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Aréchiga-Carvajal ET, Ruiz-Herrera J. The RIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryot Cell. 2005;4:999–1008. doi: 10.1128/EC.4.6.999-1008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kullas AL, Martin SJ, Davis D. Adaptation to environmental pH: integrating the Rim101 and calcineurin signal transduction pathways. Mol Microbiol. 2007;66:858–871. doi: 10.1111/j.1365-2958.2007.05929.x. [DOI] [PubMed] [Google Scholar]
- 47.Ariño J. Integrative responses to high pH stress in S. cerevisiae. OMICS. 2010;14:517–523. doi: 10.1089/omi.2010.0044. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Liang Y, Zhang B, Zheng W, Xing L, Li M. Alkaline stress triggers an immediate calcium fluctuation in Candida albicans mediated by Rim101p and Crz1p transcription factors. FEMS Yeast Res. 2011;11:430–439. doi: 10.1111/j.1567-1364.2011.00730.x. [DOI] [PubMed] [Google Scholar]
- 49.Casado C, González A, Platara M, Ruiz A, Ariño J. The role of the protein kinase A pathway in the response to alkaline pH stress in yeast. Biochem J. 2011;438:523–533. doi: 10.1042/BJ20110607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahn YS, Mühlschlegel FA. CO2 sensing in fungi and beyond. Curr Opin Microbiol. 2006;9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Mogensen EG, Janbon G, Chaloupka J, Steegborn C, Fu MS, Moyrand F, Klengel T, Pearson DS, Geeves MA, Buck J, et al. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot Cell. 2006;5:103–111. doi: 10.1128/EC.5.1.103-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zacchi LF, Gomez-Raja J, Davis DA. Mds3 regulates morphogenesis in Candida albicans through the TOR pathway. Mol Cell Biol. 2010;30:3695–3710. doi: 10.1128/MCB.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2:e00055–e00011. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]