Abstract
The ubiquitin-proteasome system is one of the major protein turnover mechanisms that plays important roles in the regulation of a variety of cellular functions. It is composed of E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 ubiquitin ligases that transfer ubiquitin to the substrates that are subjected to degradation in the 26S proteasome. The Skp1, Cullin, F-box protein (SCF) E3 ligases are the largest E3 gene family, in which the F-box protein is the key component to determine substrate specificity. Although the SCF E3 ligase and its F-box proteins have been extensively studied in the model yeast Saccharomyces cerevisiae, only limited studies have been reported on the role of F-box proteins in other fungi. Recently, a number of studies revealed that F-box proteins are required for fungal pathogenicity. In this communication, we review the current understanding of F-box proteins in pathogenic fungi.
Keywords: Cryptococcus neoformans, E3 ligase, F-box, Fungi, Virulence
The Ubiquitin-Proteasome System
Protein turnover is an important regulatory mechanism of cellular function in eukaryotes. During proteolysis, proteins are hydrolyzed to their constituent amino acids and used for synthesis of new proteins. Proteolysis is highly specific and a diverse group of enzymes designated proteases are involved in this process. In eukaryotic cells, there are two major protein degradation pathways mediating protein degradation: the lysosomal proteolysis and the ubiquitin-proteasome system [1].
Some cytosolic proteins are degraded in lysosomes after being engulfed in autophagotic vacuoles that fuse with lysosomes, which are accelerated by carbon or nitrogen starvation [2]. The proteins susceptible to degradation by this pathway are normally long-lived but dispensable proteins. However, the majority of intracellular proteins are degraded by the ubiquitin-proteasome pathway, in which the intracellular proteins are degraded with a high degree of specificity [1, 3].
The ubiquitin proteasome pathway uses ubiquitin as a marker that targets proteins for rapid proteolysis. Ubiquitin is a 76 amino acid regulatory protein that is highly conserved in all eukaryotes. Proteins are marked for degradation by the attachment of multiple ubiquitin molecules and are then recognized and degraded to small peptides by the 26S proteasome, a large multicatalytic protease complex, with the release of free and reusable ubiquitin [3]. Three enzymatic components are required to assemble chains of ubiquitin onto the substrates that are destined for proteolytic destruction. First, ubiquitin is activated by an E1 (ubiquitin-activating enzyme) in an ATP-dependent reaction and forms a thioester linkage between the C-terminal carboxyl group of the ubiquitin and the E1 cysteine sulfhydryl group. Ubiquitin is then transferred to an ubiquitin-conjugating enzyme E2 again via a thioesterification reaction. In the final step, an ubiquitin protein ligase E3 catalyzes the transfer of ubiquitin to specific substrates by creating an isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin (Fig. 1). Lysines on conjugated ubiquitin molecules can then be targeted for additional ubiquitination via isopeptide bond formation. Poly-ubiquitinated substrates are recognized and subsequently degraded by the 26S proteasome [4, 5].
Fig. 1.
The Skp1, Cullin, F-box protein (SCF) E3 ligase-mediated ubiquitin-proteasome system of protein degradation. Ubiquitin (Ub) is conjugated to substrate proteins through effort of three enzymes by an ATP-dependent process. First, ubiquitin is activated by an ubiquitin-actvating enzyme (E1) in an ATP-dependent way and transferred to its active site through formation of a thiol-ester bond between the ubiquitin and E1. Then the ubiquitin is passed to the second enzyme complex, the ubiquitin-conjugating enzyme (E2), through the same thiol-ester linkage. Finally, the target substrate is recognized by the third enzyme complex, ubiquitin ligase (E3), and labeled with the ubiquitin. This process can be repeated and a multiubiquitin chain can be formed, which usually targets the substrate for proteolysis by the 26S proteasome.
The importance of the ubquitin-proteasome system in higher eukaryotes has been recognized following the recent discovery that F-box proteins are receptors of several important plant hormones [6] and mutations in the E3 ubiquitin ligase pathway lead to human diseases [7]. The therapeutic potential of the ubiquitin-proteasome system for human diseases has been actively evaluated, and has resulted in the drug development such as bortezomib, a proteasomal inhibitor that has been used to treat human neoplasias [8].
E3 Ubiquitin Protein Ligases
The key enzymes in the ubiquitination process are the E3 ubiquitin ligases, because they function as the substrate recognition modules of the system and are capable of transferring the activated ubiquitin to the substrate. Specific E3s are responsible for the selectivity of ubiquitin-protein ligation and degradation by binding specific protein substrates that contain specific recognition signals [4].
Generally, E3s are divided into two broad structural classes: the HECT (homologous to E6-AP C-terminus) domain family, where the HECT domain from a monomeric E3 accepts the activated ubiquitin from E2 and transfers it to the substrate [9], and the RING (really interesting new gene) domain family [10].
The suppressor of kinetochore protein mutant (Skp1), Cullin (Cul1), F-box protein (SCF) E3 ligases are among the best-understood groups of the RING ligases. Three major components of the SCF complex, Skp1, Cul1, and ring-box protein (Rbx1), form an invariable core complex that associates with one of a number of F-box proteins that bind to specific substrates (Fig. 1). In cooperation with E1 and E2 enzymes, the SCF complex can transfer small ubiquitin proteins to the target protein [11-13].
The SCF E3 ligase was first identified in the budding yeast Saccharomyces cerevisiae, where it functions to catalyze the phosphorylation-dependent ubiquitination of G1 cyclins and Cdk inhibitors [14-16]. It also has important roles in regulating transcription factors [17] and regulators involved in G2 and M phases [18, 19], as well as having a role in the labeling of various other cellular proteins for degradation.
F-box Proteins
F-box proteins are proteins containing the F-box domain, a protein structural motif of approximately 50 amino acids initially identified in human cyclin F [14, 20]. F-box proteins commonly interact with Skp1 of the SCF and E3 ligase complexes to take part in protein ubiquitination and degradation. They are responsible for the substrate specificity of the SCF complexes by interacting with the substrates through their C-terminal protein binding domains, including leucine-rich repeats (LRR), WD40 repeats, as well as other protein-protein interaction domains such as carbohydrate-interacting (CASH), zinc-finger, and proline-rich domains [21-23], and then recruiting the substrates to the SCF core through the interaction of the F-box domain with Skp1 [14]. There are exceptions, in which an F-box protein does not interact with Skp1, such as Ela1 in S. cerevisiae [24]. To date, all known SCF substrates must be phosphorylated before being recognized by an F-box protein.
A large number of F-box proteins exist in all eukaryotic model organisms reported so far, and each of them likely binds to multiple substrates for ubiquitination. The fungal F-box proteins are important in regulating cellular functions including cell cycle, circadian clocks, transcription, development, signal transduction, and nutrient sensing [24]. The understanding of SCF E3 ligases has largely come from extensive studies in two model yeasts, S. cerevisiae and Schizosaccharomyces pombe. S. cerevesiae has at least 20 proteins containing an F-box domain and several have been well studied, including glucose repression resistant 1 (Grr1) [24]. Grr1 is part of the SCFGRR1 E3 ligase that interacts with Skp1 via its F-box domain and downstream targets via its LRR domain. Grr1 was originally identified as a primary element responsive for glucose repression [25], and was also found to control amino acid sensing [26] and cell cycle regulation [27]. S. pombe is another yeast in which a number of F-box proteins have been studied [24].
Despite extensive studies in both model yeasts, very limited studies of SCF E3 ligases have been reported in other fungi. Recent studies on the function of F-box proteins in pathogenic fungi have revealed that SCF E3 ligases are required for fungal virulence. Because of the proven therapeutic potential of the ubiquitin-proteasome pathway for human diseases [8], it would be very important to understand the molecular basis of how this pathway regulates fungal virulence, which may lead to novel approaches in developing new antifungal agents.
F-box Proteins in Fungal Pathogens
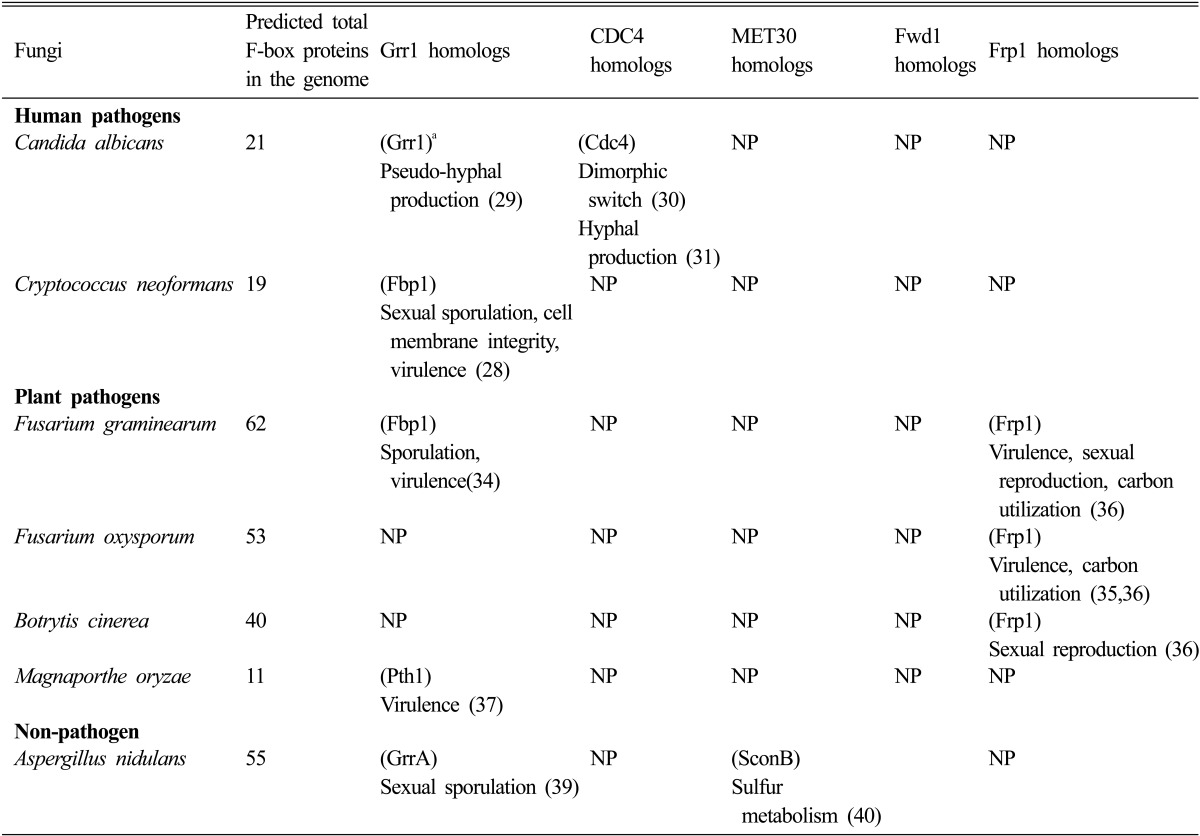
The function of F-box proteins and their potential as part of the ubiquitin-proteasome system (UPS) has been reported in a number of fungal pathogens, including those cause human infections (such as Cryptococcus neoformans and Candida albicans) or plant diseases (such as Fusarium species and Magnaporthe oryzae). A number of studies have focused on the fungal homologs of Grr1 in S. cerevisiae, and all the studies have demonstrated that these F-box proteins are important for fungal pathogenicity (Table 1).
Table 1.
F-box proteins reported in pathogenic fungi
NP, not reported.
aName of the F-box protein, followed by their major role in fungal development and virulence, and references.
C. neoformans is an encapsulated yeast causing fungal meningitis in immunocompromised patients. There are at least 19 proteins containing the F-box domain in C. neoformans. Among them, Fbp1 is identified as the only F-box protein with additional 12 LRR repeats, a common protein binding domain. Fbp1 shares sequence similarity with Grr1 in S. cerevisiae and physically interacts with Skp1 homologues in both S. cerevisiae and C. neoformans via its F-box domain, suggesting it may function as part of an SCF E3 ubiquitin ligase. Our recent studies also revealed that Fbp1 likely regulates multiple substrates (our unpublished data). Functional studies demonstrated that Fbp1 is essential for fungal virulence in a murine inhalation model. Fungal strains lacking Fbp1 protein produce normal virulence factors (capsule, melanin, and growth at 37℃), but fail to cause any disease symptom. Interestingly, the fungal burden in infected lungs remains at a consistent level from 3~60 days post-infection, which mimics a latent infection (our unpublished data). Fbp1 is also important for fungal sexual reproduction because basidiospore production is blocked in bilateral mating between fbp1Δ mutants, even though normal dikaryotic hyphae are observed during mating. Moreover, in a stress response assay, fbp1Δ mutants showed hypersensitivity to sodium dodecyl sulfate, but not to calcofluor white or Congo red, indicating that Fbp1 may regulate cell membrane integrity [28].
The dimorphic switch in C. albicans between yeast form, pseudohyphal form, and true hyphal form in response to different environmental stimuli is important for host invasion and establishment of disease, and is considered as an important virulence trait. Two F-box proteins, Cdc4 and Grr1, have been reported to be required for this morphological transition in C. albicans. Deletion of either CDC4 or GRR1 from the C. albicans genome results in a highly filamentous or pseudohyphal morphology under conditions that normally promote the yeast form of growth [29-31]. The suppression of pseudohyphal production by Grr1 is likely due to its negative regulation on cytokinesis via two G1 cyclins, Ccn1 and Cln3, in a similar way as that of the S. cerevisiae. These cyclin proteins are stabilized in a grr1Δ/Δ mutant background, which prevents cell separation after cytokinesis, indicating that they are potential substrates of Grr1. Furthermore, the cellular level of Hof1, a protein having a role in cytokinesis, is also significantly increased in grr1Δ/Δ cells [32]. Meanwhile, it is less clear how Cdc4 regulates cell morphology. One substrate of Cdc4, Sol1, was identified to play a role in C. albicans morphogenesis [30]. Additional efforts to identify Cdc4 associated proteins using in vitro affinity purification method revealed several novel proteins, including Gph1 (a putative glycogen phosphorylase) and Thr1 (a putative homoserine kinase). Both Gph1 and Thr1 are regulated by Tup1, a master regulator of hyphal production [33]. These studies suggest that ubiquitin-mediated protein degradation is involved in the regulation of the dimorphic switch of C. albicans. However, a direct role of these proteins in fungal virulence has not been established.
Interestingly, filamentous fungi such as Fusarium species contain much larger numbers of F-box proteins than yeast: 60~94 in Fusarium species compared to around 20 in yeasts (S. cerevisiae, C. albicans, or C. neoformans). This suggests that F-box proteins contribute to the regulation of a more complex developmental and metabolic processes occurring in filamentous fungi. Fusarium graminearum is a plant pathogen that infects wheat and barley to cause headblight disease, which not only leads to a significant loss of crop yield, but also produces a variety of harmful mycotoxins. By screening an insertional mutagenesis library, a homolog of Grr1 in S. cerevisiae, Fbp1, was identified to be important for fungal pathogenicity. Although Fbp1 is likely a part of the SCF complex since it can bind to S. cerevisiae Skp1 protein, Fbp1 can only partially complement several defects of a yeast grr1Δ deletion mutant. Functional analyses demonstrated that FBP1 is required for both sexual development and virulence in F. graminearum [34].
The same approach was used to identify another F-box protein required for pathogenicity (Frp1) in Fusarium oxysporum f. sp. Lycopersici, the causal agent of tomato wilt disease. Frp1 is filamentous fungi specific, only can be found in fungi belonging to Sordariomycetes, leotiomycetes, and Dothideomycetes. The frp1Δ mutant is avirulent due to its failure to colonize roots, confirming that the FRP1 gene is required for pathogenesis [35]. Interestingly, the homologs of Frp1 in different fungal pathogens that are closely related in evolution have different functions in sexual development, metabolism, and pathogenicity. Frp1 is essential for root infection in F. graminearum, but is dispensable for the infection on plant parts above ground in both F. graminearum and Botrytis cinerea, a common pathogen infecting wine grapes [36]. Frp1 is required for nonsugar carbon catabolism and regulates the growth of Fusarium species under a variety of conditions but does not play a role in the growth of B. cinerea, indicating a functional diversity on nonsugar carbon catabolism among these fungi.
Magnaporthe oryzae causes one of the most devastating diseases on rice. By screening an insertional mutagenesis library, an early study identified one F-box protein, Pth1, which is required for fungal pathogenicity both on rice and barley [37]. Pth1 is a homolog of Grr1 and is required for regulating maturation of the appressorium, a specialized cell structure for penetrating host cells. The pth1Δ mutant fails to penetrate the host leaf surface and to establish a successful host-pathogen interaction. Later studies demonstrated that Pth1 is critical for fungal carbohydrate metabolism and turgor pressure generation in appressoria, which could be the cause of producing defective appressoria in pth1Δ mutants [38].
There are several human and plant pathogens in Aspergillus species, such as Aspergillus fumigatus and A. flavus. There is no report of F-box proteins in these pathogenic Aspergillus species. A. nidulans is not a pathogen, but serves as a model system in understanding of other related species and has been extensively studied. Hence, we include the current understanding of F-box proteins in A. nidulans in this article.
The F-box protein GrrA identified in A. nidulans is a homolog of Grr1 in S. cerevisiae [39]. Functional studies have revealed that GrrA is dispensable in hyphal growth, asexual sporulation or development of asci-containing cleistothecia, but that the grrAΔ deletion mutant is unable to produce mature ascospores due to a block in meiosis [39]. The regulation of GrrA in meiosis is consistent with the role of other Grr1 homologs in pathogenic fungi, including C. neoformans, suggesting a conserved function of these F-box proteins among fungi. It will be interesting to analyze the potential role of GrrA in pathogenic Aspergillus species to understand whether they are also involved in regulation of fungal virulence.
The A. nidulans SconB is another F-box protein that shares sequence similarity with Met30 in S. cerevisiae and Scon2 in Neurospora crassa. SconB is required in regulation of sulphur metabolite repression. Besides the F-box domain, SconB also has seven WD repeats characteristic of the large WD40 family of eukaryotic regulatory proteins. The SCONB transcript is derepressed under sulphur limitation conditions and partly repressed by high methionine [40]. No homolog of conB has been reported in pathogenic fungi (Table 1).
Concluding Remarks and Future Directions
The recognition that the ubiquitin-proteasome system is a principle intracellular route for controlled protein turnover represents a major conceptual advancement in our understanding of cell biology. This protein degradation system has been proposed as the new generation therapeutic targets for human diseases [41]. Extensive studies in this field have been conducted in human [23, 42], as well as several model systems, such as S. cerevisiae [24], Arabidopsis thaliana [6, 43], and Caenorhabditis elegans [44]. The importance of the proteolytic function of the ubiquitin-proteasome system in regulating the virulence of pathogenic fungi has just been realized recently. Current studies of this system in pathogenic fungi are mostly focused on the F-box proteins. The substrates remain to be understood in most cases, which should be the focus of next step to understand the mechanism of the ubiquitin-proteasome system regulation in fungal diseases. An advanced understanding of the SCF E3 ligase-mediated ubiquitin-proteasome system in pathogenic fungi could potentially lead to innovative approaches to understand fungal virulence and novel strategies to explore therapeutic targets.
Acknowledgements
We thank Issar Smith and Hany Fahmy for critical reading of the manuscript and valuable comments. We acknowledge use of fungal genome sequences at the Broad Institute. This study is supported by UMDNJ institutional startup fund to C.X.
References
- 1.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 2.Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Spruck CH, Strohmaier HM. Seek and destroy: SCF ubiquitin ligases in mammalian cell cycle control. Cell Cycle. 2002;1:250–254. [PubMed] [Google Scholar]
- 6.Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto KM. Ubiquitin-dependent proteolysis: its role in human diseases and the design of therapeutic strategies. Mol Genet Metab. 2002;77:44–56. doi: 10.1016/s1096-7192(02)00146-4. [DOI] [PubMed] [Google Scholar]
- 8.Mitsiades CS, Mitsiades N, Hideshima T, Richardson PG, Anderson KC. Proteasome inhibitors as therapeutics. Essays Biochem. 2005;41:205–218. doi: 10.1042/EB0410205. [DOI] [PubMed] [Google Scholar]
- 9.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 11.Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 12.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 14.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 15.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 16.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 18.Bastians H, Topper LM, Gorbsky GL, Ruderman JV. Cell cycle-regulated proteolysis of mitotic target proteins. Mol Biol Cell. 1999;10:3927–3941. doi: 10.1091/mbc.10.11.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michael WM, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- 20.Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 24.Jonkers W, Rep M. Lessons from fungal F-box proteins. Eukaryot Cell. 2009;8:677–695. doi: 10.1128/EC.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flick JS, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernard F, Andre B. Ubiquitin and the SCF(Grr1) ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 2001;496:81–85. doi: 10.1016/s0014-5793(01)02412-7. [DOI] [PubMed] [Google Scholar]
- 27.Blondel M, Galan JM, Peter M. Isolation and characterization of HRT1 using a genetic screen for mutants unable to degrade Gic2p in Saccharomyces cerevisiae. Genetics. 2000;155:1033–1044. doi: 10.1093/genetics/155.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu TB, Wang Y, Stukes S, Chen Q, Casadevall A, Xue C. The F-Box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryot Cell. 2011;10:791–802. doi: 10.1128/EC.00004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler DK, All O, Goffena J, Loveless T, Wilson T, Toenjes KA. The GRR1 gene of Candida albicans is involved in the negative control of pseudohyphal morphogenesis. Fungal Genet Biol. 2006;43:573–582. doi: 10.1016/j.fgb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Atir-Lande A, Gildor T, Kornitzer D. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol Biol Cell. 2005;16:2772–2785. doi: 10.1091/mbc.E05-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shieh JC, White A, Cheng YC, Rosamond J. Identification and functional characterization of Candida albicans CDC4. J Biomed Sci. 2005;12:913–924. doi: 10.1007/s11373-005-9027-9. [DOI] [PubMed] [Google Scholar]
- 32.Li WJ, Wang YM, Zheng XD, Shi QM, Zhang TT, Bai C, Li D, Sang JL, Wang Y. The F-box protein Grr1 regulates the stability of Ccn1, Cln3 and Hof1 and cell morphogenesis in Candida albicans. Mol Microbiol. 2006;62:212–226. doi: 10.1111/j.1365-2958.2006.05361.x. [DOI] [PubMed] [Google Scholar]
- 33.Tseng TL, Lai WC, Jian T, Li C, Sun HF, Way TD, Shieh JC. Affinity purification of Candida albicans CaCdc4-associated proteins reveals the presence of novel proteins involved in morphogenesis. Biochem Biophys Res Commun. 2010;395:152–157. doi: 10.1016/j.bbrc.2010.03.162. [DOI] [PubMed] [Google Scholar]
- 34.Han YK, Kim MD, Lee SH, Yun SH, Lee YW. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol Microbiol. 2007;63:768–779. doi: 10.1111/j.1365-2958.2006.05557.x. [DOI] [PubMed] [Google Scholar]
- 35.Duyvesteijn RG, van Wijk R, Boer Y, Rep M, Cornelissen BJ, Haring MA. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol Microbiol. 2005;57:1051–1063. doi: 10.1111/j.1365-2958.2005.04751.x. [DOI] [PubMed] [Google Scholar]
- 36.Jonkers W, Van Kan JA, Tijm P, Lee YW, Tudzynski P, Rep M, Michielse CB. The FRP1 F-box gene has different functions in sexuality, pathogenicity and metabolism in three fungal pathogens. Mol Plant Pathol. 2011;12:548–563. doi: 10.1111/j.1364-3703.2010.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sweigard JA, Carroll AM, Farrall L, Chumley FG, Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol Plant Microbe Interact. 1998;11:404–412. doi: 10.1094/MPMI.1998.11.5.404. [DOI] [PubMed] [Google Scholar]
- 38.Silva MC. Signaling pathway in appressorium formation in Magnaporthe grisea [dissertation] College Station: Texas A&M University; 2004. [Google Scholar]
- 39.Krappmann S, Jung N, Medic B, Busch S, Prade RA, Braus GH. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol Microbiol. 2006;61:76–88. doi: 10.1111/j.1365-2958.2006.05215.x. [DOI] [PubMed] [Google Scholar]
- 40.Natorff R, Piotrowska M, Paszewski A. The Aspergillus nidulans sulphur regulatory gene sconB encodes a protein with WD40 repeats and an F-box. Mol Gen Genet. 1998;257:255–263. doi: 10.1007/s004380050646. [DOI] [PubMed] [Google Scholar]
- 41.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 43.Craig A, Ewan R, Mesmar J, Gudipati V, Sadanandom A. E3 ubiquitin ligases and plant innate immunity. J Exp Bot. 2009;60:1123–1132. doi: 10.1093/jxb/erp059. [DOI] [PubMed] [Google Scholar]
- 44.Kipreos ET. Ubiquitin-mediated pathways in C. elegans. WormBook; 2005. pp. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]