Abstract
The tulip tree (Liriodendron chinense) has been widely cultivated in Korea as a street or garden tree for its large flowers, which have a superficial resemblance to tulips. Occurrence of anthracnose disease on the leaves of tulip trees growing on the campus of Gyeongsang National University, Jinju, Korea, has been observed. Based on mycological characteristics, pathogenicity, and internal transcribed spacer sequence, the causal fungus was identified as Colletotrichum gloeosporioides. This is the first report on anthracnose disease caused by C. gloeosporioides on tulip trees in Korea.
Keywords: Anthracnose, Colletotrichum gloeosporioides, Tulip tree
The tulip tree (Liriodendron chinense) has been widely cultivated in Korea as a street or garden tree for it large flowers, which have a superficial resemblance to tulips. Anthracnose is the name given to a group of diseases caused by several closely related fungi that attack many shade trees, including sycamore, white oak, elm, dogwood, and maple [1]. From July to October 2011, spot anthracnose disease was observed on leaves of tulip trees grown on the campus of Gyeongsang National University, Jinju, Korea.
Spot anthracnose disease was found to occur frequently on the leaves of tulip trees. Necrotic lesions became black as the spots expanded. Small to irregular with tan to black spots often have yellow to dark brown margins. Spot anthracnose does not lead to defoliation or death.
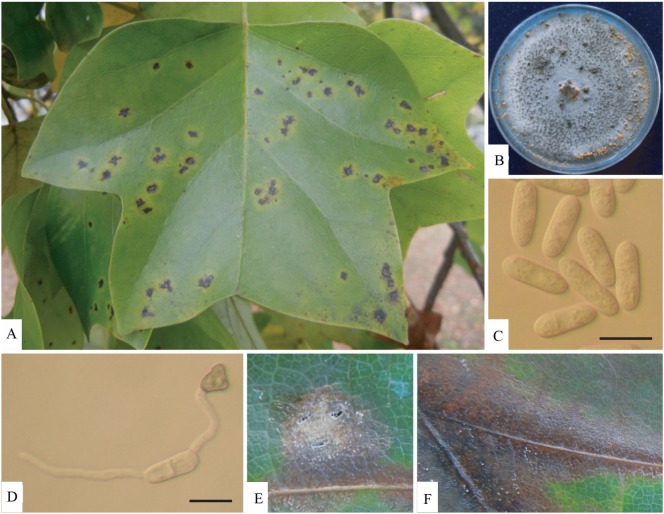
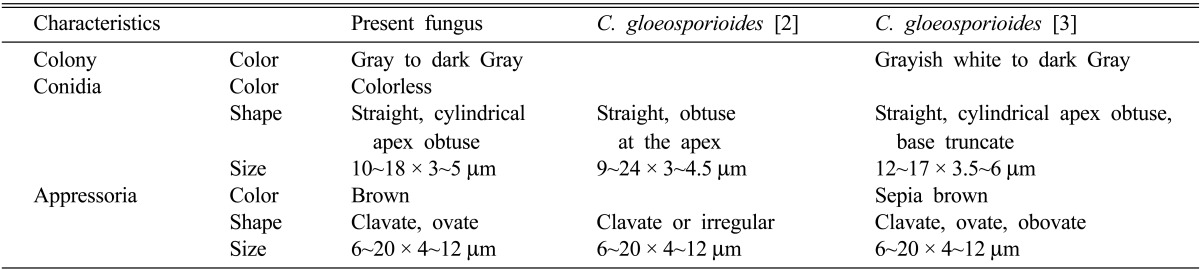
To isolate pathogens from infected leaves, small sections (5 mm square) from the margin of the infected lesion were disinfected for 1 min in 1% sodium hypochlorite solution, rinsed in sterile distilled water, dried on sterile absorbent paper, and placed on potato dextrose agar (PDA). Aseptic sub-culture of colonies of the pathogen was then performed for further study. A light microscope (Axioplan; Carl Zeiss, Jena, Germany) was used for detailed microscopic examination of a representative specimen. Fungus grown on PDA initially produced whitish mycelia, which became dark gray, with later formation of salmon-colored conidial masses (Fig. 1). The conidia, which were cylindrical and ovoid, measured 10~18 × 3~5 µm (Table 1, Fig. 1C) [2, 3]. Appressoria on water agar were one-celled, pale brown, thick-walled, ellipsoidal, or clavate, rarely irregular, and measured 6~20 × 4~12 µm (Table 1, Fig. 1D). Measurements and taxonomic characters coincided with those of Colletotrichum gloeosporioides (Penz.) Sacc. described by Sutton [2].
Fig. 1.
Symptoms of anthracnose of Colletotrichum gloeosporioides and its morphological characteristics. A, Symptoms of anthracnose on leaves of tulip trees; B, Mycelial colony grown on potato dextrose agar for 15 days; C, Un-germinated spore; D, Appressorium melanization; E, F, Symptoms after artificial inoculation on leaf (scale bars = 10 µm).
Table 1.
Comparison of mycological characteristics of Colletotrichum gloeosporioides and anthracnose fungus isolated from tulip tree
For pathogenicity testing, leaves of the tulip tree were surface sterilized with 1% sodium hypochlorite solution. A sterilized scalpel was used for wounding of leaves. A 10-µL drop of a conidial suspension of 104 conidia/mL obtained from PDA cultures was applied to injured leaves. As the control treatment, three leaves were inoculated in a similar manner with sterile water. Inoculated leaves were kept in a humid box with 95~100% relative humidity at 25℃ for 7 days. Necrotic spot symptoms were reproduced after 7 days of incubation. Of particular interest, infected areas were often found along the veins and midrib of artificially-inoculated leaves (Fig. 1F). Dead areas may merge until the whole leaf dies. To prove Koch's postulates, the fungus was re-isolated based on these symptoms. Development of symptoms was not observed in waster controls.
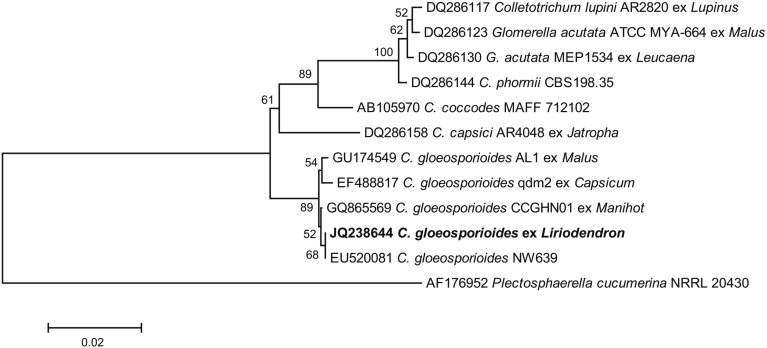
For molecular identification of the causal fungus, amplification of the complete internal transcribed spacer (ITS) rRNA gene sequence was performed according to the method previously described by White et al. [4]. The Exgene Plant-Fungal SV Mini kit (Geneall Biotechnology Co., Seoul, Korea) was used for isolation of total DNA, following the manufacturer's instructions. The PCR reaction was performed in a mixture containing 5 units Taq polymerase (Takara, Tokyo, Japan), 1 × PCR buffer, 0.5mM MgCl2, 0.2 mM of each dNTP, 5 pmol of each primer, and approximately 10~15 ng of fungal genomic DNA, with adjustment of the total volume to 50 µL with sterile water. PCR amplication was performed on an Astec PC 802 thermal cycler (Astec, Fukuoka, Japan), as follows: 98℃ for 2 min, 30 cycles of denaturation at 98℃ for 10 sec, annealing at 55℃ for 30 sec, and extension at 72℃ for 30 sec, with nal extension at 72℃ for 4 min. Primers used for amplification of the complete ITS region were ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3') [4]. Following agarose gel electrophoresis, extraction of PCR amplicons was performed using a Gel Extraction kit (Geneall Biotechnology Co.). Purified PCR products were cloned into a pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced with primers M13F (5'-GTAAAACGACGGCCAGT-3') and M13R (5'-GCGGATAACAATTTCACACAGG-3'). A Bigdye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) was used in performance of sequencing, following the manufacturer's instructions. The resulting 575-bp of the ITS rRNA gene sequence was deposited in GenBank (accession No. JQ238644). Blast of the DNA sequence showed 100% identity with accession No. EU520081, for C. gloeosporioides (Fig. 2). MEGA ver. 4 (www.megasoftware.net) with the neighbor-joining method and Tajima-Nei distance model was used in performance of phylogenetic analysis [5]. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. In the phylogenetic tree (Fig. 2), the representative isolate was placed within a clade comprising reference isolates of C. gloeosporioides. Previously published ITS sequences of C. gloeosporioides strains were included for reference.
Fig. 2.
Phylogenetic tree using internal transcribed spacer sequences showing the closest known relatives of Colletotrichum gloeosporioides, including anthracnose fungus infection of tulip trees. ClustalW was used in alignment of DNA sequences from the National Center for Biotechnology Information (NCBI) nucleotide database. Numbers above the branches indicate boots-trap values. Bars indicate the number of nucleotide substitutions per site. Our isolate from the infected tulip tree is shown in bold font.
Based on symptoms, mycological characteristics, molecular data, and pathogenicity to the host plant, this fungus was identified as Colletotrichum gloeosporioides. Anthracnose disease caused by Colletotrichum sp. has been previously reported; however, spot anthracnose disease caused by C. gloeosporioides has not been reported in Korea [6]. To the best of our knowledge, this is the first report on the occurrence of C. gloeosporioides in tulip trees in Korea. Incidence of this disease is highly dependent upon weather conditions. Recent occurrence of the disease suggests that C. gloeosporioides can be widespread. Although the economic importance of the disease in tulip trees is limited, the pathogen could present a risk to economically important fruit trees.
Acknowledgements
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ007345)" Rural Development Administration, Korea.
References
- 1.Farr DF, Rossman AY. Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA. Retrieved September 30, 2010 [Internet] Beltsville: USA Systematic Mycology and Microbiology Laboratory; 2010. [cited 2012 Jan 2]. Available from: http://nt.ars-grin.gov/fungaldatabases/ [Google Scholar]
- 2.Sutton BC. The Coelomycetes. Kew: Commonwealth Mycological Institute; 1980. [Google Scholar]
- 3.Bailey JA, Jeger MJ. Colletotrichum: biology, pathology and control. Wallingford: CAB International; 1992. [Google Scholar]
- 4.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 5.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 6.The Korean Society of Plant Pathology. List of plant diseases in Korea. 5th ed. Seoul: Korean Society of Plant Pathology; 2009. (in Korean) [Google Scholar]