Abstract
A Mariannaea fungus was isolated during investigation of an elm tree infested with unidentified beetles. Based on morphological characteristics and molecular analysis of the internal transcribed spacer rDNA sequence, the fungus was identified as Mariannaea elegans var. elegans. Fungal growth was better on malt extract agar than on potato dextrose agar and oatmeal agar. Optimal temperature and pH for growth of the fungus were 30℃ and pH 7.0, respectively. The fungus was found to have the ability to produce extracellular enzymes such as amylase, β-glucosidase, cellulase, and protease. This is first report on M. elegans var. elegans in Korea.
Keywords: Elm Tree, ITS rDNA, Mariannaea elegans var. elegans
Introduction
The genus Mariannaea is comprised of eight species. Mariannaea species is widespread and has frequently been isolated from soil, decaying bark of pine, submerged wood in freshwater streams, and from insects [1-3]. M. elegans is the representative species of the genus Mariannaea. This species was described in 1974 by Samson as the type species of the genus [1]. M. elegans has two varieties, M. elegans var. elegans and M. elegans var. punicea. In 1991, the teleomorph of M. elegans was reported as Nectria mariannaeae by Samuels and Seifert [2]. In Korea, M. elegans was reported in 2004 as the anamorph stage of Cordyceps pruinosa, an entomopathogenic mushroom [4]. However, in 1991, the anamorph of C. pruinosa was reported as Mariannaea pruinosa by Liang [5]. In a recent study, Sung et al. [6] reported on their acceptance of M. pruinosa as the anamorph of C. pruinosa. Thus, currently, no official report has been published on M. elegans in Korea.
A species of Mariannaea was recently isolated during investigation of an elm tree infested with unidentified beetles. In this study, we report on identification of the isolated Mariannaea species and analysis of the growth properties and ability of its mycelia to produce extracellular enzymes.
Materials and Methods
Isolation of fungi
Several pieces of wood were taken as samples from an elm tree infested with unidentified beetles in Asan City in the Chungchungnamdo (province) of Korea in September, 2010 (Fig. 1A and 1B). Several unidentified beetle larvae were captured from the egg gallery of the beetle found in the sample wood pieces. One example is shown in Fig. 1C. For isolation of fungi, a few of the captured beetle larvae were washed with sterile water and dried for few minutes in a bio-safety cabinet. They were then placed on malt extract agar (MEA; Difco, Detroit, MI, USA) supplemented with streptomycin (200 µg/mL). Some of the captured beetle larvae were placed directly on MEA supplemented with streptomycin (200 µg/mL). All larvae containing MEA were incubated at 25℃ for 3~7 days. Using sterile needles, mycelia grown out from larvae containing MEA were transferred to new MEA plates, followed by incubation. Single spore isolates were obtained from fungi grown on the incubated MEA. Pure cultures of the isolates were maintained on MEA and 10% glycerol stock cultures were prepared. Following comparison of the colony pattern on MEA and microscopic examination of microstructure of the pure cultures, an isolate coded as DUCC400 with morphology resembling that of Mariannaea was used for subsequent study. The DUCC400 isolate was deposited in the Dankook University Culture Collection (DUCC), Cheonan, Korea.
Fig. 1.
Wilted and debarked elm tree branches infested with unidentified beetles (A), a sample piece of elm wood with the gallery and holes (B), and an unidentified beetle larva captured from a hole in the sample piece of elm wood (C). Morphology of colonies formed on potato dextrose agar (D), malt extract agar (E), and oatmeal agar (F) after growth of the isolate DUCC 400 obtained from the larva shown in (C) for 14 days at 25℃.
Microscopic analysis
A phase-contrast microscope (Axioskop 40; Carl Zeiss, Jena, Germany) and scanning electron microscope (SEM, Hitachi S-430; Hitachi, Tokyo, Japan) were used for observation of morphological characteristics of the isolate DUCC400. Examination of fungal structures was based on fresh materials prepared on MEA and potato dextrose agar (PDA) at 25℃ for 7~14 days. For observations using a SEM, culture agar blocks were cut from MEA plates and fixed with 2% glutaraldehyde in 0.1M cacodylate buffer for 12 hr and then 1% osmic acid for 1 hr. Fixed samples were washed with 0.05M cacodylate buffer, followed by dehydration in a series of different concentrations of ethanol from 50% to 100% for 20 min each. The samples were subsequently dried with a Hitachi critical point dryer and coated with platinum-palladium for 50 sec using a Hitachi E-1030 ion sputter, followed by observation using a SEM.
Molecular analysis
Using the drilling method described by Kim et al. [7], mycelia of the isolate DUCC400 freshly grown on PDA for 7 days were collected for DNA extraction. Fungal specific primers internal transcribed spacer (ITS) 1F and ITS4 were used for amplification of the ITS rDNA sequence of the isolate DUCC400 [8, 9]. PCR amplification was performed in a 50 µL reaction mixture containing 100 ng fungal genomic DNA, 20 pmol of each primer, 10 mM of the four deoxynucleotide triphosphates (dNTPs), 1 × PCR buffer (10 mM Tris-Cl [pH 8.0], 1.5 mM MgCl2, 50 mM KCl), and 1.0 unit Taq DNA polymerase (Promega, Madison, WI, USA). PCR conditions were as follows: denaturation at 94℃ for 10 min, followed by 30 cycles of denaturation at 94℃ for 1 min, annealing at 56℃ for 1 min, and extension at 72℃ for 1 min, and one final cycle of extension at 72℃ for 10 min. A PCR clean-up kit (Qiagen, Hilden, Germany) was used for purification of the PCR product, followed subsequently by ligation into T&A cloning vectors (RBC, Taipei, Taiwan). The ligated vectors were transformed into competent Escherichia coli DH5α cells, according to the manufacturer's instructions (RBC).
DNA Sequencing was performed at Macrogen Inc. (Seoul, Korea) using an ABI 3700 automated sequencer (Perkin-Elmer Inc, Waltham, MA, USA). The ITS sequence obtained was blasted in the GenBank database (http://www.ncbi.nlm.nih.gov). Phylogenetic analysis based on the ITS sequence was performed using the neighbor-joining method implemented in the PAUP* 4.0 program [10], and bootstrap values were determined through heuristic searches with 1,000 replicates. Nectria radicicola was used as an out-group.
Assessment of mycelial growth and the capacity for extracellular enzyme secretion
Mycelial growth on solid medium was assessed on PDA, MEA, and oatmeal agar (OA). Optimal temperature and pH for mycelial growth were tested on MEA. The capacity for secretion of extracellular enzymes was evaluated on chromogenic reaction medium containing 0.5% CM-cellulose (Sigma-Aldrich, St. Louis, MO, USA), D-cellobiose (Sigma-Aldrich), polygalacturonic acid (MP Biomedicals, Sanata Ana, CA, USA), starch (Sigma-Aldrich), xylan (Sigma-Aldrich), and Avicel (Fluka, Cork, Ireland) as a carbon source, 0.1% yeast nitrogen base without amino acid (BD, Franklin Lakes, NJ, USA) as a nitrogen source, 1.5% agar, and 0.5% Congo Red (Sigma-Aldrich) as a dye. Protease production was measured on skim milk (Fluka, Buchs, Switzerland) agar [11]. After cultivation for 14 days at 25℃, colony diameter and the clear zone formed by fungal growth were measured for determination of the capacity for extracellular enzyme production.
Results and Discussion
Morphology
Differences were observed in colony morphology of DUCC400 grown on MEA, PDA, and OA (Fig. 1D~1F). First, colony color on MEA was white, and later became yellow and yellow brown with age; the reverse plate was yellow brown due to diffusion of the exudate into the surrounding agar. Colonies on PDA were strongly zonate and mat like. Colonies on OA were velutinous in appearance but more weekly zonate than those on PDA. The reverse of colonies on PDA and OA were yellow brown to dark brown. Vegetative aerial hyphae of the isolate DUCC400 were hyaline and thin on MEA (Fig. 1D).
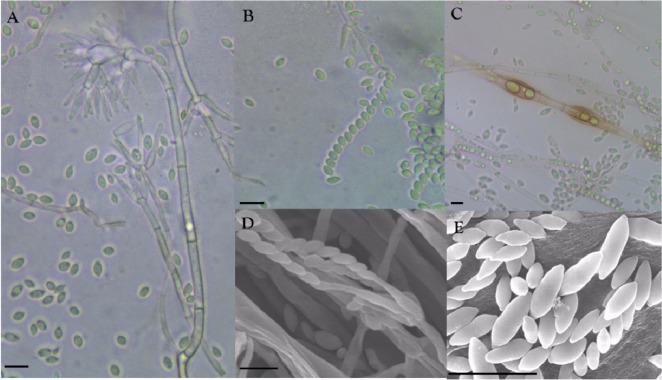
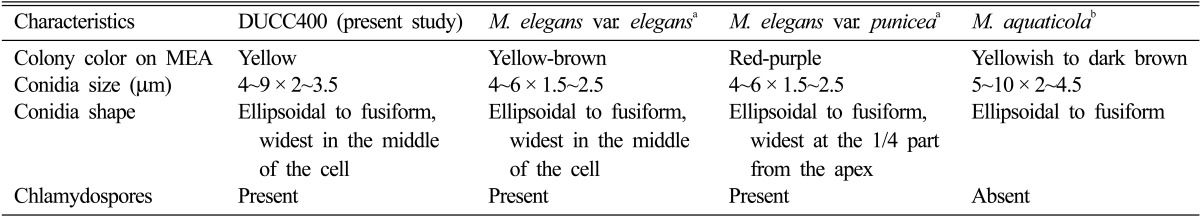
Conidiophores were erect and formed zonation on MEA, PDA, and OA. Conidiophore structures were irregular and complex, but usually verticillate, having a stalk that measured up to 300 µm in length and 3~7 µm in width, bearing short branches with whorls of 3~6 phialides (Fig. 2A and 2D). Phialides, which measured 9~25 × 2~3 µm, were flask shaped, hyaline, smooth walled, and sometimes inflated. Conidia, which measured 4~9 × 2~3.5 µm, were hyaline, ellipsoidal to fusiform, smooth walled, divergent in size, measuring from 4 × 2 to 9 × 3.5 µm, widest in the middle of the cell, and usually formed imbricate chains having contact with phialides (Fig. 2B, 2D and 2E). Chlamydospores, which were rarely present in intermediary chains, were thick walled, roughened, yellow-brown, and ellipsoidal in shape, and measured 30 × 10 µm (Fig. 2C). No microstructure of sexual stage was observed, indicating that the isolate DUCC400 was in anamorphic stage. These observed morphological properties were compared with those of M. elegans varieties and M. aquaticola (Table 1). The isolate DUCC400 was found to be most similar to M. elegans var. elegans.
Fig. 2.
Morphological features of the isolate DUCC400. Conidia and conidiophore structures (A), conidia imbricate chain (B), and chlamydospores (C) observed using a phase contrast light microscope. Structures of conidiophore (D) and conidia (E) observed by scanning electron microscope (scale bars: A~E = 10 µm).
Table 1.
Comparison of morphological characteristics of the isolate DUCC400 with three Mariannaea species
aData from Samson [1] in Mycobank (http://www.mycobank.org).
bData from Cai et al. [12] in Mycobank (http://www.mycobank.org).
Molecular characterization
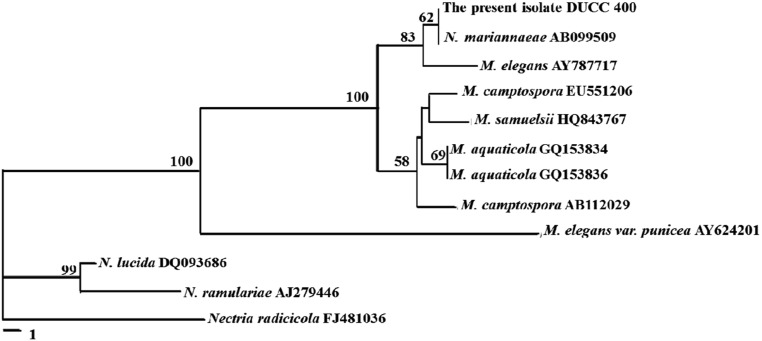
The ITS rDNA sequence of the isolate DUCC400 was determined as 601 bp and deposited in GenBank with the accession no. JQ690354. Sequence homology search through the Genbank DNA database revealed that the isolate DUCC400 showed 100% homology with Nectria mariannaeae (AB099509) and 99% homology with Marianaea elegans (AY787717). M. elegans is the anamorph of N. mariannaeae [2]; therefore, we could say that the ITS rDNA sequence of the isolate DUCC400, which did not show its telemorph in this study, is most homologous to that of M. elegans. Phylogenetic analysis of ITS rDNA sequences also positioned the isolate DUCC400 with M. elegans and its telemorph N. mariannaeae (Fig. 3). M. aquaticola, a species with morphological similarity to M. elegans, diverged from the isolate DUCC400. The molecular divergence of M. elegans from M. aquaticola is in agreement with the report by Cai et al. [12]. So far, two varieties of M. elegans, M. elegans var. elegans and M. elegans var. punicea, have been reported. These two varieties can be clearly separated based on their capacity for production of reddish-purple colonies on MEA. M. elegans var. punicea produced reddish-purple colonies on MEA, but M. elegans var. elegans did not [13, 14]. The isolate DUCC400 did not show a reddish-purple color, but rather a yellow color on MEA (Table 1, Fig. 1D). The isolate DUCC400 was well separated from M. elegans var. punicea in the phylogram (Fig. 4). These molecular data support that the isolate DUCC400 is not M. elegans var. punicea. Therefore, based on morphological (Table 1) and molecular data (Fig. 4), we identified the isolate DUCC400 as M. elegans var. elegans.
Fig. 3.
Phylogenetic position of DUCC400 among Nectira (N) and Mariannaea (M) species. The tree was generated by the neighbor-joining method based on internal transcribed spacer rDNA sequences. Numbers at nodes represent the percentage of bootstrap re-samplings based on 1,000 replicates. The bar represents the number of nucleotide substitutions per site. Nectria radicicola was used as an out-group.
Fig. 4.
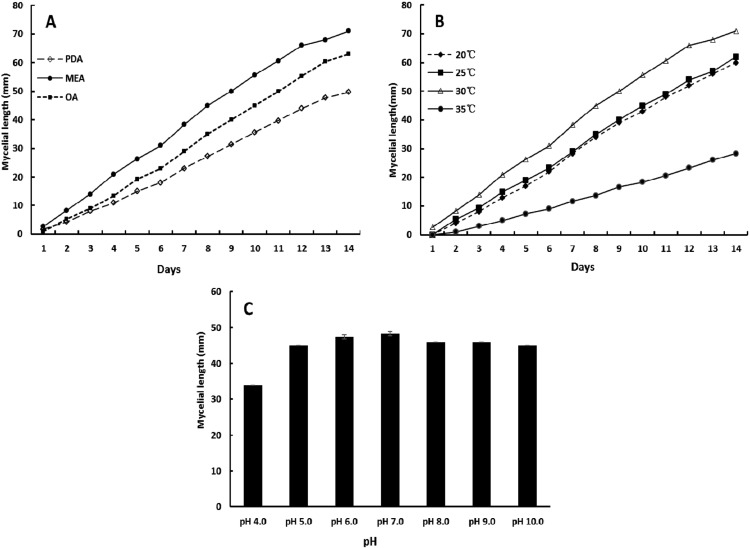
Mycelial growth of the isolate DUCC400 on different media at a temperature of 30℃ (A), on malt extract agar (MEA) at different temperature (B) and pH at 30℃ for 10 days (C). PDA, potato dextrose agar; OA, oatmeal agar.
Growth and biochemical properties
Results of the mycelia growth test showed that the isolate DUCC400 was moderately faster on MEA than on PAD and OA. The optimal temperature and pH for mycelial growth of the isolate DUCC400 on MEA was 30℃ and pH 7, respectively (Fig. 4). Fungal growth was slightly retarded at pH 4; however, it grew moderately well at pH 5 to 10.
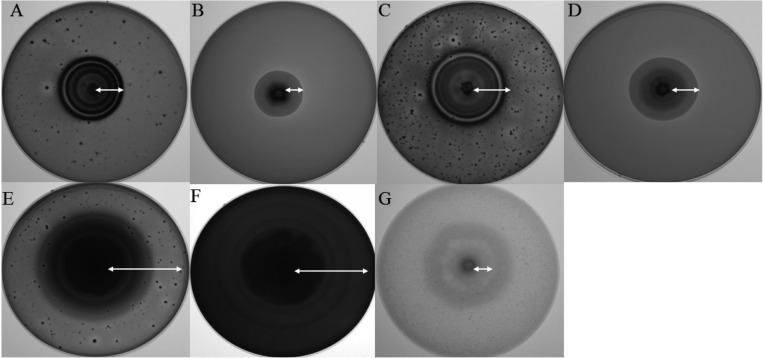
Results from assessment of the capacity for production of extracellular enzyme demonstrated that the isolate DUCC400 possessed diverse extracellular enzyme activity for all substrates used in the study. In particular, the isolate DUCC400 showed relatively higher β-glucosidase and amylase activity, when compared with other extracellular enzymes, such as CM-cellulase, Avicelase, and xylanase (Fig. 5). So far, there has been no report on the capacity for production of extracellular enzymes by M. elegans var. elegans. Considering that the isolate DUCC was isolated from a beetle's larva found in elm wood, it is agreeable that the fungus shows the ability to degrade wood components. Questions remain with regard to how M. elegans var. elegans can occur in an elm tree infested with beetle's larva, where the DUCC was isolated. Therefore, further study of the ecological role of M. elegans var. elegans in association with the elm tree is needed.
Fig. 5.
Extracellular enzyme activities of the isolate DUCC400 shown on chromogenic reaction medium containing each enzymatic substrate. A, CM-cellulase; B, xylanase; C, Avicelase; D, pectinase; E, β-glucosidase; F, amylase; G, protease.
In conclusion, by combining analyses of its morphological characteristics and phylogenetics, this study indentified and characterized the isolate DUCC as M. elegans var. elegans. This is first report on its description in Korea.
Acknowledgements
This study was supported by a grant from the National Institute of Biological Resources, the Ministry of Environment, and by a grant from the Next Generation BioGreen 21 program (Project No. PJ0081542011), Rural Development Administration, Republic of Korea.
References
- 1.Samson RA. Paecilomyces and some allied hyphomycetes. Stud Mycol. 1974;6:1–119. [Google Scholar]
- 2.Samuels GJ, Seifert KA. Two new species of Nectria with Stilbella and Mariannaea anamorphs. Sydowia. 1991;43:249–263. [Google Scholar]
- 3.Gräfenhan T, Schroers HJ, Nirenberg HI, Seifert KA. An overview of the taxonomy, phylogeny, and typification of Nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud Mycol. 2011;68:79–113. doi: 10.3114/sim.2011.68.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin JC, Shrestha B, Lee WH, Park YJ, Kim SY, Jeong GR, Kim HK, Kim TW, Sung JM. Distribution and favorable conditions for mycelial growth of Cordyceps pruinosa in Korea. Korean J Mycol. 2004;32:79–88. [Google Scholar]
- 5.Liang ZQ. Verification and identification of the anamorph of Cordyceps pruinosa Petch. Mycosystema. 1991;10:104–107. [Google Scholar]
- 6.Sung GH, Hywel-Jones NL, Sung JM, Luangsa-Ard JJ, Shrestha B, Spatafora JW. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 2007;57:5–59. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Uzunovic A, Breuil C. Rapid detection of Ophiostoma piceae and O. quercus in stained wood by PCR. Appl Environ Microbiol. 1999;65:287–290. doi: 10.1128/aem.65.1.287-290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardes M, Bruns TD. ITS primers with enhanced specificity for Basidiomycetes: application to identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 9.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Snindky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 10.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods). Version 4.0 b10. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- 11.Yoon JH, Hong SB, Ko SJ, Kim SH. Detection of extracellular enzyme activity in Penicillium using chromogenic media. Mycobiology. 2007;35:166–169. doi: 10.4489/MYCO.2007.35.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai L, Kurniawati E, Hyde KD. Morphological and molecular characterization of Mariannaea aquaticola sp. nov. collected from freshwater habitats. Mycol Prog. 2010;9:337–343. [Google Scholar]
- 13.Samson RA, Bigg WL. A new species of Mariannaea from California. Mycologia. 1988;80:131–134. [Google Scholar]
- 14.Okuda T, Yamamoto K. Materials for the fungus flora of Japan (56): Mariannaea camptospora and M. elegans var. punicea from Japan. Mycoscience. 2000;41:411–414. [Google Scholar]