Abstract
BACKGROUND:
Outer inflammatory protein A (OipA) is an outer membrane protein of Helicobacter pylori that is involved in inducing IL-8 and intracellular signaling. In this study, we have predicted exposure amino acid sequences of OipA for insertion in permissive sites of CstH subunit of Eschierchia coli CS3 pilli for bacterial surface display.
MATERIALS AND METHODS:
Databases: National Center for Biotechnology Institute and Protein Data Bank. Servers: PHD, SABLE, GOR 4, SignalP3.0, TBBpred, PRODIV-TMHMM, TMRPres2D, CPH Models, PHYRE, GETAREA, VADAR, Pep state and pep window. Software: Swiss PDB viewer and Discovery studio.
RESULTS:
In silico prediction of exposure amino acid sequences of OipA led to detection of six sequences of amino acid, 76-87, 106-112, 170-182, 222-230, 242-258, and 278-290. These sequences inserted between amino acid sequences 66-67, 100-101, and 109-110 of CstH that were predicted by Eskandari et al. as permissive sites of CstH.
CONCLUSION:
OipA has the ability to induce IL-8 from gastric epithelial cells and some papers are mentioned that this outer membrane protein involve to attachment and intracellular signaling. Receptor of OipA and adhesion motifs on this protein is unknown. Detection of exposure motifs aids to recognition of adhesion motifs and receptor of OipA on gastric epithelial cells. In this study, we have predicted exposure amino acid sequences for insert to subunit CstH of CS3 pilli E. coli for surface display.
Keywords: CstH, in silico prediction, OipA, surface sequences
Introduction
Helicobacter pylori is a gram negative spiral bacterium that colonizes stomach for long time, and this colonization can lead to gastritis, peptic ulcer, and gastric cancer.[1] World Health Organization and International Agency for Research on Cancer have classified this bacterium as type one carcinogen for gastric cancer.[2,3] H. pylori colonizes half of the world population and this colonization reaches up to 90% in developing country.[4] Outer membrane proteins have pivotal role in pathogenesis of this bacterium.[5] Most of these outer membrane proteins increase attachment to gastric epithelial cell and some of them induce host inflammatory responses.[5] Outer inflammatory protein A (OipA) is an outer membrane protein that is involved to increase pathogenesis of H. pylori.[6] Presence of this protein is linked to proinflammatory signaling of gastric epithelial cell, duodenal ulceration, gastric cancer, increase of H. pylori density, and neutrophil infiltration.[6,7] OipA mutant of H. pylori revealed that this outer membrane protein associated with bacterial adhesion to gastric epithelial cell. Inducing IL-8 secretion by this protein is controversial; some reports showed that oipA mutants had not reduced IL-8 secretion but the others have reported about main role of OipA in induction of IL-8.[5,6,8]
The aim of this research is computational studies for prediction of OipA exposure sequences of amino acid. Results of this study apply for insert in permissive sites of CstH subunit of Eschierichia coli CS3 pilli for bacterial surface display of outer inflammatory protein A on E. coli for detection adhesion motif and role of OipA in induction of IL-8 and proinflammatory signaling.
Materials and Methods
Databases: Amino acid sequences of OipA and CstH were retrieved from National Center for Biotechnology Institute and Uni-prot KB/Swiss-Prot. Accession number of CstH in NCBI and Uni-prot KB/Swiss-Prot were X16944 and P15488.[9,10] Tertiary structure of CstH presents in Protein Data Bank.[11]
Servers: Secondary structure of proteins predicted by PHD, SABLE, and GOR 4 servers.[12,13] SignalP3.0 server detected signal sequence of OipA and CstH.[14] TBBpred, PRODIV-TMHMM, and TMRPres2D servers used for transmembrane topology prediction in bilayer lipid and external loops of OipA. CPH Models and PHYRE servers applied for finding of homology modeling and tertiary structure prediction.[15–18] GETAREA and VADAR servers applied for prediction of accessible surface area amino acids among sequences of external loops. Pep state and pep window used for probability of expression in inclusion bodies and hydropathy plot of two proteins.
Software: Swiss PDB viewer and Discovery studio applied for data analysis.[19,20]
Results
In silico prediction of exposure amino acid sequences of OipA led to detection of six sequences of amino acid; 76-87, 106-112, 170-182, 222-230, 242-258, and 278-290. These sequences inserted between amino acid sequences 66-67, 100-101 and 109-110 of CstH were predicted by Eskandari et al. as permissive sites of CstH.
Discussion
Protein engineering is a branch of bioinformatics that is useful for progress of biological research.[21] In this study, we used from some tools of protein engineering for process of outer inflammatory protein A and prediction of exposure amino acid sequences for detection of adhesion motifs and sequences involve to proinflammatory signaling after bacterial surface display on E. coli.
Carrier protein for OipA on surface of E. coli is CstH subunit of CS3 pili that previously was analyzed by Eskandari et al. for permissive sites. Prediction for secondary, tertiary structure, and hydropathy plot showed three sites for insertion of our sequences.[21]
We applied three servers for prediction of secondary structure and compared these results with together [Figures 1–3].
Figure 1.

Secondary structure prediction of OipA by PHD server E: beta sheet, H: alpha helices, C: random coil
Figure 3.

Secondary structure prediction of OipA by SABLE server E: beta sheet, H: alpha helices, C: random coil
Figure 2.

Secondary structure prediction of OipA by GOR4 server E: beta sheet, H: alpha helices, C: random coil
SignalP3.0 server predicted seventeen N-terminal amino acids of OipA which do not present in mature OipA on the cell surface and this sequence was deleted before further analysis by other tools [Figure 4].
Figure 4.
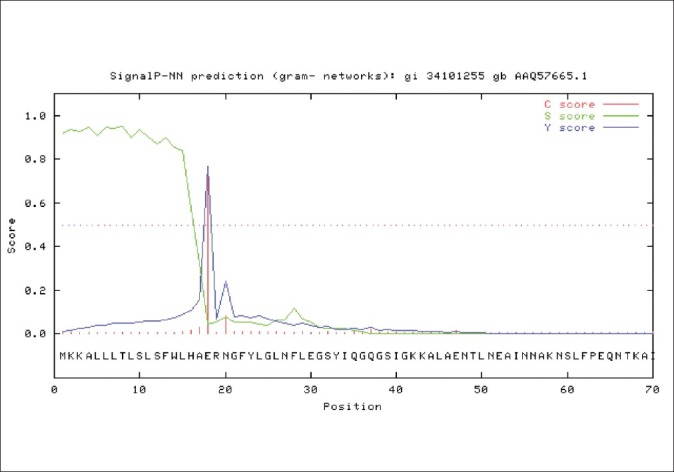
SignalP3.0 server for prediction of signal sequence of OipA
Secondary structure of transmembrane proteins in prokaryotes has a little difference from eukaryotes. In outer membrane of prokaryotes, hydrophobic beta barrel integrate in bilayer lipid and famous tool for prediction of transmembrane topology is TBBpred server [Figure 5].[22]
Figure 5.

Prediction of beta barrel of OipA in H. pylori outer membrane by TBBpred server “n” refers to residues in non barrel regions and “b” refers to residues in barrel region
Tertiary structure of protein causes that some sequences of external loops get surface and the others get inside of protein.[22] Next step was tertiary structure prediction of OipA and detection of exposure amino acid sequences in external loops [Figures 6–8].
Figure 6.
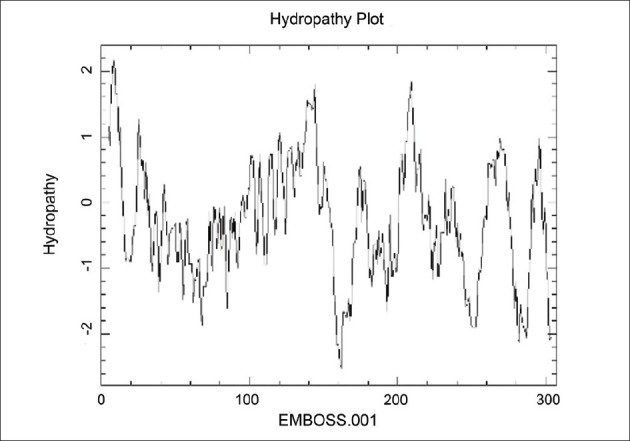
Hydropathy plot of OipA by pepwindow server
Figure 8.

Tertiary structure prediction of OipA by CPH model server and shown with Discovery Studio Visualizer
Figure 7.
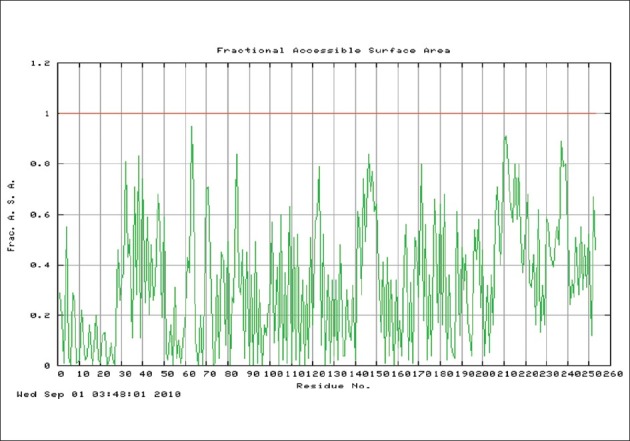
Prediction of accessible surface area amino acids by vadar server
The CPH model server revealed that tertiary structure of OipA, same as VacA, was most likely an auto display protein and its insertion in the outer membrane was carried out by type V secretion system (T5SS). N-terminal long hydrophilic region confirms this finding [Figures 6–10].
Figure 10.
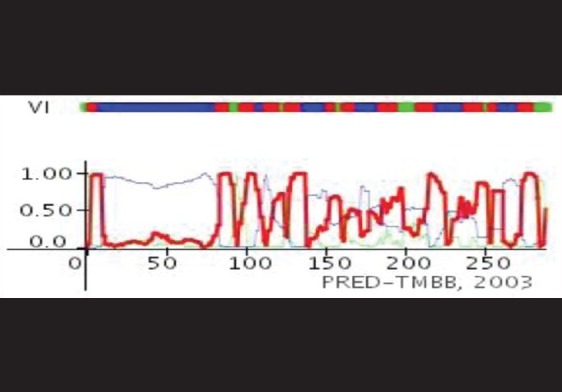
Prediction of external and internal loops sequence of OipA by TMRPres2D server
Figure 9.
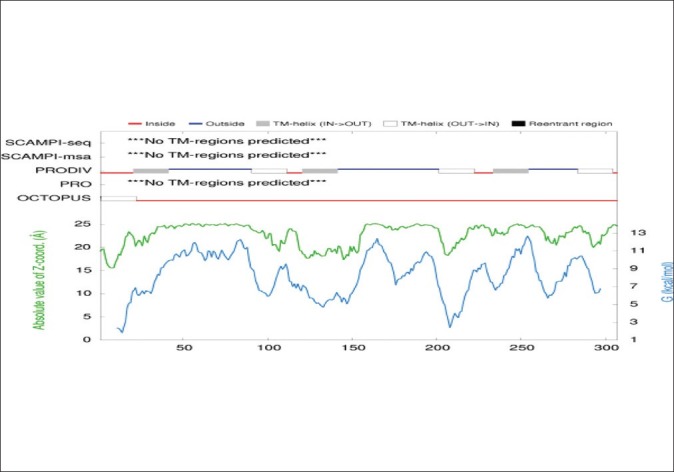
Prediction of external and internal loops sequence of OipA by PRODIV-TMHMM server
Conclusion
Exposure amino acid sequences of OipA and other outer membrane proteins have important role in interaction with stomach epithelial cell, and detection of these sequences is very useful for finding of receptors on surface of Gastric epithelial cell.[5]
Receptor of OipA and adhesion motifs on this protein is unknown. Detection of exposure motifs aids to recognition of adhesion motifs and receptor of OipA on gastric epithelial cells.[23] In this study, we have predicted exposure amino acid sequences for insert to subunit CstH of CS3 pilli E. coli for surface display.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Wei J, O’Brien D, Vilgelm A, Piazuelo MB, Correa P, Washington MK, et al. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology. 2008;134:1412–23. doi: 10.1053/j.gastro.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Fang DC, Wang RQ, Yang SM, Liu HF, Luo YH. Effect of Helicobacter pylori infection on expression of Bcl-2 family members in gastric adenocarcinoma. World J Gastroenterol. 2004;10:227–30. doi: 10.3748/wjg.v10.i2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–87. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang ZW, Farthing MJ. Molecular mechanisms of H. pylori associated gastric carcinogenesis. World J Gastroenterol. 1999;5:369–74. doi: 10.3748/wjg.v5.i5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dossumbekova A, Prinz C, Mages J, Lang R, Kusters JG, Van Vliet AH, et al. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: Genetic and functional genomic analysis of hopH gene polymorphisms. J Infect Dis. 2006;194:1346–55. doi: 10.1086/508426. [DOI] [PubMed] [Google Scholar]
- 6.Clyne M, Dolan B, Reeves EP. Bacterial factors that mediate colonization of the stomach and virulence of Helicobacter pylori. FEMS Microbiol Lett. 2007;268:135–43. doi: 10.1111/j.1574-6968.2007.00648.x. [DOI] [PubMed] [Google Scholar]
- 7.Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci U S A. 2000;97:7533–8. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao SH, Wang H, Chai SG, Liu LM. Research progress on Helicobacter pylori outer membrane protein. World J Gastroenterol. 2005;11:3011–3. doi: 10.3748/wjg.v11.i20.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bairoch A. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–74. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jalajakumari MB, Thomas CJ, Halter R, Manning PA. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: Novel regulation of pilus production by bypassing an amber codon. Mol Microbiol. 1989;3:1685–95. doi: 10.1111/j.1365-2958.1989.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 11.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rost B, Sander C, Schneider R. PHD-An automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 13.Adamczak R, Porollo A, Meller J. The SABLE. Server, which was rigorously. Bioinformatics. 2004 [Google Scholar]
- 14.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: Signal P 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–25. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 16.Krieger E, Nabuurs SB, Vriend G. Homology modeling. Methods Biochem Anal. 2003;44:509–23. doi: 10.1002/0471721204.ch25. [DOI] [PubMed] [Google Scholar]
- 17.Sujatha MS, Balaji PV. Fold-recognition and comparative modeling of human alpha2,3-sialyltransferases reveal their sequence and structural similarities to CstII from Campylobacter jejuni. BMC Struct Biol. 2006;6:9. doi: 10.1186/1472-6807-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardesty J. Protein Simulation, Function and Prediction. Computational Structure Biology. 2005 [Google Scholar]
- 19.Kaplan W, Littlejohn TG. Swiss-PDB Viewer (Deep View) Brief Bioinform. 2001;2:195–7. doi: 10.1093/bib/2.2.195. [DOI] [PubMed] [Google Scholar]
- 20.Niemeyer EA. Application Guide tools and methods used in Discovery Studio for the visualization. Nucleic Acids Res. 2000;28:235–42. [Google Scholar]
- 21.Eskandari V, Yakhchali B, Minuchehr Z. In-silico prediction of permissive sites of CstH subunit of E. coli CS3 pili for insertion of foreign peptides. Iran J Biol. 2010;1:73–84. [Google Scholar]
- 22.Xiong J. Protein Secondary Structure Prediction. In: Xiong J, editor. Essential Bioinformatics. 1st ed. New York: Cambridge University Press; 2006. pp. 200–13. [Google Scholar]
- 23.Yamaoka Y, Alm RA. Helicobacter pylori outer membrane proteins. In: Yamaoka Y, editor. Helicobacter pylori Molecular Genetics and Cellular Biology. 1st ed. Norfolk: Caister Academic Press; 2008. pp. 37–61. [Google Scholar]


