Natural killer (NK) cells are lymphocytes that serve as components of the early, innate, immune system, in contrast to B and T lymphocytes, that provide acquired specific immune responses. First described on the basis of capacity to kill cellular targets, such as tumor and infected cells, NK cells also can be activated to secrete cytokines that influence the subsequent development of acquired immunity (1). With respect to target recognition, NK cells appear to express two functional types of receptors, one for activation and another for inhibition (2). Activation receptors remain less well characterized but include receptors such as NKR-P1 in rodents (3) that can presumably recognize target cell ligands and stimulate NK cell release of perforin- and granzyme-containing granules that mediate target apoptosis. Inhibitory NK cell receptors, specific for major histocompatibility complex (MHC; HLA in humans) class I molecules, have been the subject of intense investigation over the last few years, providing major advances in understanding NK cell specificity (4). Such studies have also provided support for the “missing-self” hypothesis that was proposed to explain the effect of target cell MHC class I molecules in providing protection from NK cell killing (5). NK cells are hypothesized to survey tissues for the presence of normal MHC class I molecules that are ubiquitously expressed. When tissues express mutant MHC class I or lack expression, such as during tumorigenesis or infection, this inhibitory influence is released, permitting NK cell-mediated target lysis. In recent issues of the Proceedings, two articles provide significant insight into the physiologic mechanisms by which this inhibition occurs (6, 7).
The MHC class I-specific inhibitory NK cell receptors fall into two distinct structural groups of molecules containing a single transmembrane domain (8). The Ig-like receptors are (usually monomeric) type I integral membrane proteins that include the human killer inhibitory receptors (KIR), whereas the C-type lectin-like receptors are disulfide-linked dimeric, type II integral membrane proteins. Among the lectin-like receptors are the mouse Ly49 family and human CD94 and NKG2 family. Despite their structural differences, both types of receptors are polymorphic at several levels; the receptors belong to several families of highly related molecules that also display allelic forms. Furthermore, many of these molecules are expressed on subsets of NK cells, and an individual NK cell may express several different receptors. Both types of inhibitory receptors mediate their effects through cytoplasmic sequences termed immunoreceptor tyrosine-based inhibitory motifs (ITIMs) which can be phosphorylated and recruit intracellular tyrosine phosphatase, SHP-1, that presumably dephosphorylates components in the activation cascade (9). In addition, both types of receptors also include related isoforms that do not display ITIMs and may instead activate.
Given their highly related properties (selective NK cell expression, MHC class I specificity, inhibitory signaling), it was surprising that initial descriptions of each structural type of NK cell receptor indicated that human NK cells expressed the Ig-like receptors, whereas rodent NK cells expressed the lectin-like molecules (8). This raised the speculation that each species evolved structurally divergent NK cell receptors to deal with missing-self. However, recent studies have indicated that NK cells in humans, rats, and mice express receptors belonging to either structural type. Interestingly, all known members of each structural type and their isoforms are encoded by genes that form separate genetic complexes, and, at least for the case of the lectin-like receptors, the corresponding syntenic genetic regions have been identified (10). Moreover, it is now clear that an individual NK cell can express multiple different isoforms of either structural type and that there is considerable variability among individual NK cells with respect to the repertoire of receptors expressed.
CD94, however, appears to be expressed on all human NK cells (11) and its corresponding orthologue has been identified in rodents, where it is less well studied (12–14). On human NK cells, CD94 forms disulfide-linked heterodimers with molecules belonging to the NKG2 family that require CD94 for surface expression (15). Genomic studies indicate that NKG2C–F are products of distinct genes, whereas NKG2A and -B appear to be alternatively spliced from the same gene (16). The cytoplasmic domain of CD94 is only 7 amino acids; therefore, the signal transduction capacity of the heterodimer is derived from the cytoplasmic domain of the NKG2 partner chain (17). NKG2A/B contains ITIMs and is inhibitory, whereas NKG2C does not contain an ITIM and is stimulatory.
Previous studies on the HLA class I specificity of CD94/NKG2A were confusing. Its specificity appeared to be promiscuous; interactions with classical HLA class I molecules (class Ia) such as HLA-A, -B, or -C, and nonclassical (class Ib) molecules such as HLA-G have been described (15, 18–23). For example, transfection of a classical HLA class I molecule into a susceptible target cell could confer resistance to killing by an NK cell clone that expressed CD94/NKG2A but not the KIR molecules. Moreover, this inhibition could be overcome by a monoclonal antibody (mAb) reactive with most if not all HLA class I molecules (such as mAb W6/32; specific mAbs for individual HLA molecules are less readily available) or a mAb specific for CD94. However, several closely related alleles were apparently not recognized and there were discrepancies in reproducibility. Nevertheless, it had become generally accepted that CD94/NKG2A specifically recognized individual HLA-A, -B, -C, and/or -G molecules.
In this issue of the Proceedings, however, there is a deus ex machina concerning HLA-E, providing an unexpected explanation for discrepancies in previous findings and a new role for nonclassical MHC class I molecules (6). HLA-E is a class Ib molecule that is widely expressed and has limited polymorphism (24–26). Similar to other MHC class I molecules, the HLA-E heavy chain is expressed on the cell surface as a noncovalent complex with β2-microglobulin (light chain) and a peptide that occupies the peptide-binding (α1 and α2) domain of the heavy chain (27). For most MHC class I molecules, the peptide is an octamer or nonamer fragment derived from degradation of cytoplasmic proteins, such as housekeeping enzymes, or viral proteins in infected cells, by the proteasome, a multisubunit catalytic complex (Fig. 1A). These peptides are then translocated across the endoplasmic reticulum (ER) into the ER lumen by TAP (transporter for antigen presentation) for assembly with nascent class I heavy chain and β2-microglobulin. The assembled complex is then exported and displayed on the cell surface. In the absence of an adequate peptide supply, such as when the cell lacks functional TAP molecules, MHC class I molecules fail to form stable, intact complexes at the cell surface. However, such molecules apparently still cycle to the surface, where they can be stabilized as intact complexes by experimentally providing exogenous peptides if the peptides have the capacity to bind the α1/α2 domains of the heavy chain. The endogenous peptides bound to the heavy chain are a complex mixture because the major constraints for peptide binding are only two or three anchor residues that are oriented into the peptide binding groove with smaller contributions from interactions with side pockets of the groove. The anchor residues are relatively invariant between peptides that have the capacity to bind a given MHC class I molecule but many other amino acids can be accommodated in the other positions of the octamer/nonamer, permitting a large number of different peptides to bind. For HLA-E, however, there are additional constraints. Although HLA-E expression is TAP-dependent (28, 29), its peptide repertoire is largely comprised of the leader sequences from other class I molecules, a situation previously noted for the mouse Qa-1 molecule that shares other features with HLA-E (30, 31). Furthermore, the leader sequences for HLA-E or Qa-1 and certain other MHC class I molecules do not bind HLA-E or Qa-1, respectively. Interestingly, HLA-E and Qa-1 share an unusual feature involving the replacement of otherwise highly conserved Thr-143 and Trp-147 by serines (31). This could change one of the side pockets (F) in the peptide binding cleft, perhaps accounting for the restricted repertoire of peptides (29, 32). Hence, expression of HLA-E or Qa-1 requires not only the normal production of these molecules but also synthesis of certain classical MHC class I molecules (Fig. 1B). Importantly, therefore, these findings on HLA-E provide another possible interpretation of previous studies involving the specificity of NK cell receptors.
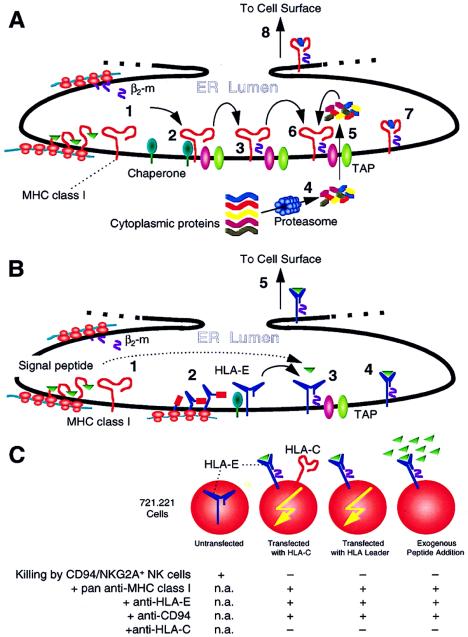
Figure 1.
(A) Synthesis and assembly of classical MHC class I molecules. Nascent MHC class I heavy (MHCI) chains and β2-microglobulin (β2-m) are generated in the endoplasmic reticulum (ER) lumen (step 1). The heavy chain initially associates with several different chaperone molecules (not shown for simplicity) (step 2). Upon assembly with β2-m, the MHCI-β2-m complex dissociates from the chaperones and associates with TAP molecules (step 3), which facilitate the transport of proteasome-degraded (step 4) small peptides across the ER membrane (step 5). TAP also has other assembly functions. The peptides then bind to the peptide binding cleft of MHCI (step 6), forming stable complexes that dissociate from TAP (step 7) and are transported to the cell surface (step 8). If assembly is disrupted, such as with TAP deficiency (as in the TAP-deficient RMA-S mouse cell), MHCI-β2-m molecules do not form intact stable complexes on the cell surface. The complexes can be stabilized by addition of exogenous peptides, but only if these peptides can bind MHCI (see ref. 27). (B) Synthesis and assembly of HLA-E (and Qa-1) molecules. In contrast to classical MHC class I molecules, HLA-E and Qa-1 bind peptides that are derived from the signal peptides of classical MHC class I molecules. The details of HLA-E synthesis and assembly are forthcoming but presumably should otherwise resemble those of classical MHC class I production, as initial studies indicate similarities (28, 29). Nascent MHCI and β2-m are generated in the ER lumen (step 1). In addition, nascent HLA-E heavy chains are also produced that associate with chaperones (step 2). Upon assembly with β2-m, MHCI-β2-m complex dissociates from chaperones and associates with TAP molecules (step 3) that are required, perhaps because of chaperone/assembly functions other than peptide transport. Fragments of signal peptides from classical MHC class I heavy chains then bind to the peptide binding cleft of HLA-E, forming stable complexes that dissociate from TAP (step 4) and are transported to the cell surface (step 5). If assembly is disrupted, such as with a deficiency in peptide supply because of lack of the classical MHC class I signal peptides, HLA-E-β2-m molecules do not form stable complexes on the cell surface. (Such is the case for 721.221 cells.) These complexes can be stabilized by addition of exogenous peptides only if these peptides can bind HLA-E, such as leader peptides from classical MHC class I molecules. Thus, HLA-E expression requires normal HLA-E and classicalHLA class I molecules and intact assembly pathways. (C) Transfection of 721.221 cells with leader sequences of HLA class I molecules renders resistance to killing by CD94/NKG2A+ NK cells. 721.221 cells express HLA-E transcripts but no stable HLA-E molecules on the surface because the cells lack classical HLA class I molecules due to a mutation. When HLA-C (and its leader) is transfected, HLA-E is also expressed, resulting in resistance to killing by CD94/NKG2A+ NK cells (6, 32, 35). The resistance is reversed (killing occurs) with antibodies that cross-react with HLA-E or CD94/NKG2A. If the leader sequence alone is transfected (as part of HLA-E, for example), resistance still occurs despite absence of HLA-C. Moreover, addition of exogenous peptides, representing the leader sequence of classical HLA class I molecules, stabilizes otherwise unstable HLA-E-β2m complexes and renders resistance. An anti-HLA-C specific antibody does not reverse resistance in any situation. Finally, soluble HLA-E molecules bind CD94/NKG2A molecules (not depicted) (32). The data therefore indicate a specific and direct interaction between HLA-E and CD94/NKG2A. n.a., Not applicable.
Indeed, most previous studies on CD94/NKG2A specificity were based on analysis of 721.221 cells that harbor HLA-E transcripts (Fig. 1C). 721.221 cells do not express intact HLA-E molecules on the cell surface because they lack an adequate peptide supply due to deficient expression of other HLA molecules. Intact HLA-E molecules can be expressed on the cell surface by transfection of cDNAs encoding HLA molecules or by addition of peptides representing the leader sequences of HLA molecules (28, 29, 33). The former is exactly the situation previously used to describe the specificity of CD94/NKG2A for HLA-A, -B, -C, or -G—i.e., transfection of an HLA molecule into 721.221 cells may result in expression not only of the transfected HLA molecule but also of endogenous HLA-E. Moreover, there was a good correlation between capacity of a specific HLA molecule to confer resistance and the ability of its leader peptide to bind HLA-E. Hence, the paper by Lee et al. (6) evaluates the possibility that HLA-E is directly recognized by CD94/NKG2A.
Lee et al. (6) demonstrate that transfection of 721.221 cells with a chimeric gene encoding a molecule with the leader sequence of HLA-A2 resulted in expression of HLA-E and resistance to lysis by CD94/NKG2A-expressing NK cell clones (6). Killing was restored by an anti-HLA-E-specific or anti-CD94-specific mAb. Furthermore, expression of HLA-Cw4 conferred HLA-E-mediated protection through CD94/NKG2A recognition as evidenced by restoration of lysis by anti-HLA-E or anti-CD94/NKG2A but not anti-HLA-C mAbs. In addition, stabilization of HLA-E on the cell surface with exogenous peptides from appropriate leader sequences of class I molecules conferred resistance to killing. By contrast, HLA-E is not apparently recognized by the Ig-like NK cell receptors. Therefore, HLA-E appears to be directly recognized by CD94/NKG2A receptors on NK cells, leading to inhibition by phosphorylation of the ITIM and SHP-1 recruitment (34).
These observations and interpretations are further substantiated by recent work from two other groups (32, 35). Related approaches were utilized by Borrego et al. (35), who provided additional information. In particular, addition of peptides capable of binding HLA-E to TAP-deficient, HLA-E-transfected, mouse RMA-S cells conferred protection against human CD94/NKG2A-expressing NK cell clones. Furthermore, the capacity of peptides to confer protection could be correlated with leader sequences of HLA molecules, such as HLA-G, previously known to protect by transfection. In addition, Braud et al. (32) utilized a soluble form of HLA-E in the form of tetramers and demonstrated binding to CD94/NKG2A transfectants. Interestingly, the HLA-E tetramer also bound to CD94/NKG2C, raising the possibility that HLA-E could activate NK cells through this ITIM-less isoform that can deliver stimulatory signals. Furthermore, the HLA-E tetramer was derived from constructs expressed in bacteria and therefore devoid of glycosidic residues. Although CD94 and NKG2 molecules are homologous to the C-type lectin superfamily, which contains other members that bind carbohydrates, and MHC class I molecules have conserved Asn-linked glycosylation sites, the capacity of CD94/NKG2A to bind MHC class I molecules is not carbohydrate-dependent. Similar conclusions have been recently reported for another lectin-like NK cell receptor, mouse Ly-49A (36). Taken together, there are abundant data to substantiate the conclusion that CD94/NKG2A directly recognizes HLA-E molecules rather than direct engagement of HLA-A, -B, -C, or -G.
These data also suggest a conceptual framework with relevance to the missing-self hypothesis in which at least two different MHC class I molecules, HLA-E, and the MHC class I molecule that provides the leader peptide, contribute to simultaneous recognition by a single NK cell receptor, CD94/NKG2A. Inasmuch as Qa-1 has features similar to those of HLA-E and rodent NK cells express transcripts for CD94 and NKG2 molecules, related studies should be forthcoming in rodent NK cell systems because this “two for one” system appears to be capable of gauging the health of a cell (at least of the MHC class I assembly pathway) as determined by HLA-E expression. Interestingly, other C-type lectin-like NK cell receptors, such as mouse Ly-49A, have no peptide specificity, but peptide is required for recognition of its MHC class I ligand, again providing a means to evaluate the health of the cell, as previously proposed (37, 38). On the other hand, the KIR molecules appear to have some specificity for the peptides themselves (39). It is noteworthy that viruses manifest several strategies to subvert the MHC class I assembly and antigen presentation pathway at different steps, resulting in interference in the capacity of antigen presentation to MHC class I-restricted cytotoxic T lymphocytes (40). However, this interference also will result in down-regulation of HLA-E and other MHC class I molecules, enhancing NK cell activity. NK cells therefore appear to have different receptor systems to evaluate missing-self, such as in the context of viral infections. How these receptor systems provide a integrated signal to the NK cell needs to be explored.
The HLA-E story also provides a cautionary note. In the April 14 issue of the Proceedings, Paul et al. (7) suggest that malignant melanoma may be correlated with HLA-G expression that is not normally expressed, and metastatic lesions have enhanced HLA-G expression. This would provide an inhibitory mechanism for tumor cells to evade NK cell-mediated effects, an important conceptual advance. However, given the new information with regard to HLA-E, of which Paul et al. were unaware, the findings could be due to enhanced HLA-E expression as a result of HLA-G up-regulation. Furthermore, the role of HLA-G in conferring resistance of fetal tissues to maternal NK cells could be due to a similar mechanism (41, 42). Perhaps this HLA-E-based mechanism explains the capacity of HLA-G2 molecules to confer protection even though it lacks the α2 domain. Although it is still plausible that these results could be explained by direct recognition of HLA-G, a reevaluation is required to explore whether HLA-E and HLA class I leader peptide loading is involved in these systems. The expression of HLA-E is generally not taken into account in most experiments to date, and anti-HLA-specific mAbs may cross-react with HLA-E, since mAbs specific for only HLA-E have only recently been developed (28). Likewise, other examples of NK cell specificity for MHC class I may require further examination and understanding of whether the putative NK cell receptor is directly recognizing the transfected MHC class I molecule or its peptide in the context of another MHC class I molecule, such as HLA-E or Qa-1. On the other hand, the KIR molecules appear not to bind HLA-E and may directly engage HLA-G (6, 32, 43), but further evaluation may be necessary.
In any event, the capacity to inhibit NK cell activity with a small repertoire of peptides that bind HLA-E raises the possibility that high-affinity peptide mimetics could be developed to displace endogenous peptides bound to HLA-E. If such peptides result in HLA-E destabilization or an HLA-E conformation that is not recognized by CD94/NKG2A, the possibility exists to exploit the potent killing activity of NK cells in new therapeutic strategies.
Finally, another remarkable aspect of these findings (6, 32, 35) is the significant influence of a signal peptide on an important immunologic recognition event. After completing its mission, this portion of a protein was generally thought of as being useless and jettisoned as waste analogous to a rocket booster once it had inserted a multistage spacecraft into space. Moreover, some of the issues raised by the Qa-1 and HLA-E investigations relate to other questions that should be of interest to cell biologists. Are these leader peptides also important for other cellular functions? Why are the Qa-1- and HLA-E-associated leader peptides TAP-dependent when they would presumably be in the ER already? Do the peptides efflux into the cytosol and then are transported back? Are there other TAP functions besides transport that are being revealed by these experiments, such as a chaperone-like property (folding) or requirement for TAP in peptide loading (29, 44)? The myriad questions generated by these findings should generate enthusiasm from a wide audience of immunologists and cell biologists, yet another two-for-one from these interesting studies.
Acknowledgments
The author thanks Andrew Brooks, Lewis Lanier, and Miguel López-Botet for sharing prepublication manuscripts and Hamish Smith, Hubert Chuang, and other members of his laboratory for stimulating discussions. Work in the author’s laboratory is supported by grants from the National Institutes of Health and the Barnes-Jewish Research Foundation. The author is an investigator of the Howard Hughes Medical Institute.
Footnotes
The companions to this commentary are published in page 4510 in issue 8 and page 5199 of this issue.
References
- 1.Trinchieri G. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama W M. Curr Opin Immunol. 1993;5:67–73. doi: 10.1016/0952-7915(93)90083-5. [DOI] [PubMed] [Google Scholar]
- 3.Ryan J C, Niemi E C, Nakamura M C, Seaman W E. J Exp Med. 1995;181:1911–1915. doi: 10.1084/jem.181.5.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama W M. Proc Natl Acad Sci USA. 1995;92:3081–3085. doi: 10.1073/pnas.92.8.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kärre K. In: Mechanisms of Cytotoxicity by NK Cells. Herberman R B, Callewaert D M, editors. Orlando, FL: Academic; 1985. pp. 81–103. [Google Scholar]
- 6.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, López-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal F A, Avril M F, Dausset J, Guillet J G, Carosella E D. Proc Natl Acad Sci USA. 1998;95:4510–4515. doi: 10.1073/pnas.95.8.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parham P. Immunol Rev. 1997;155:1–221. doi: 10.1111/j.1600-065x.1997.tb00948.x. [DOI] [PubMed] [Google Scholar]
- 9.Leibson P J. Immunity. 1997;6:655–661. doi: 10.1016/s1074-7613(00)80441-0. [DOI] [PubMed] [Google Scholar]
- 10.Brown M G, Scalzo A A, Matsumoto K, Yokoyama W M. Immunol Rev. 1997;155:53–65. doi: 10.1111/j.1600-065x.1997.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 11.López-Botet M, Perezvillar J J, Carretero M, Rodriguez A, Melero I, Bellon T, Llano M, Navarro F. Immunol Rev. 1997;155:165–174. doi: 10.1111/j.1600-065x.1997.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 12.Dissen E, Berg S F, Westgaard I H, Fossum S. Eur J Immunol. 1997;27:2080–2086. doi: 10.1002/eji.1830270836. [DOI] [PubMed] [Google Scholar]
- 13.Vance R E, Tanamachi D M, Hanke T, Raulet D H. Eur J Immunol. 1997;27:3236–3241. doi: 10.1002/eji.1830271222. [DOI] [PubMed] [Google Scholar]
- 14.Ho, E., Heusel, J. W. , Brown, M. G., Matsumoto, K., Scalzo, A. A. & Yokoyama, W. M. (1998) Proc. Natl. Acad. Sci. USA 95, in press. [DOI] [PMC free article] [PubMed]
- 15.Lazetic S, Chang C, Houchins J P, Lanier L L, Phillips J H. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 16.Plougastel B, Trowsdale J. Eur J Immunol. 1997;27:2835–2839. doi: 10.1002/eji.1830271114. [DOI] [PubMed] [Google Scholar]
- 17.Houchins J P, Lanier L L, Niemi E C, Phillips J H, Ryan J C. J Immunol. 1997;158:3603–3609. [PubMed] [Google Scholar]
- 18.Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, Barbaresi M, Pende D, Ciccone E, López-Botet M, et al. J Exp Med. 1994;180:545–555. doi: 10.1084/jem.180.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pazmany L, Mandelboim O, Valesgomez M, Davis D M, Reyburn H T, Strominger J L. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 20.Sivori S, Vitale M, Bottino C, Marcenaro E, Sanseverino L, Parolini S, Moretta L, Moretta A. Eur J Immunol. 1996;26:2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 21.Phillips J H, Chang C W, Mattson J, Gumperz J E, Parham P, Lanier L L. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 22.Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D, Le Bouteiller P, Moretta L, Moretta A. Eur J Immunol. 1997;27:1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Villar J J, Melero I, Navarro F, Carretero M, Bellon T, Llano M, Colonna M, Geraghty D E, López-Botet M. J Immunol. 1997;158:5736–5743. [PubMed] [Google Scholar]
- 24.Wei X H, Orr H T. Hum Immunol. 1990;29:131–142. doi: 10.1016/0198-8859(90)90076-2. [DOI] [PubMed] [Google Scholar]
- 25.Koller B H, Geraghty D E, Shimizu Y, DeMars R, Orr H T. J Immunol. 1988;141:897–904. [PubMed] [Google Scholar]
- 26.Ulbrecht M, Honka T, Person S, Johnson J P, Weiss E H. J Immunol. 1992;149:2945–2953. [PubMed] [Google Scholar]
- 27.York I A, Rock K L. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 28.Lee, N., Goodlett, D. R., Ishitani, A., Marquardt, H. & Geraghty, D. E. (1998) J. Immunol. in press. [PubMed]
- 29.Braud V M, Allan D S J, Wilson D, McMichael A J. Curr Biol. 1998;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 30.Aldrich C J, DeCloux A, Woods A S, Cotter R J, Soloski M J, Forman J. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 31.Soloski M J, DeCloux A, Aldrich C J, Forman J. Immunol Rev. 1995;147:67–89. doi: 10.1111/j.1600-065x.1995.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 32.Braud V M, Allen D S J, O’Callaghan C A, Söderström K, D’Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, McMichael A J. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 33.Ulbrecht M, Kellermann J, Johnson J P, Weiss E H. J Exp Med. 1992;176:1083–1090. doi: 10.1084/jem.176.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carretero, M., Palmieri, G., Llano, M., Tullio, V., Santoni, A., Geraghty, D. E. & López-Botet, M. (1998) J. Exp. Med., in press. [DOI] [PubMed]
- 35.Borrego F, Ulbrecht M, Weiss E H, Coligan J E, Brooks A G. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto N, Ribaudo R K, Abastado J-P, Margulies D H, Yokoyama W M. Immunity. 1998;8:245–254. doi: 10.1016/s1074-7613(00)80476-8. [DOI] [PubMed] [Google Scholar]
- 37.Correa I, Raulet D H. Immunity. 1995;2:61–71. doi: 10.1016/1074-7613(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 38.Orihuela M, Margulies D H, Yokoyama W M. Proc Natl Acad Sci USA. 1996;93:11792–11797. doi: 10.1073/pnas.93.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malnati M S, Peruzzi M, Parker K C, Biddison W E, Ciccone E, Moretta A, Long E O. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 40.Spriggs M K. Annu Rev Immunol. 1996;14:101–130. doi: 10.1146/annurev.immunol.14.1.101. [DOI] [PubMed] [Google Scholar]
- 41.Rouas-Freiss N, Marchal R E, Kirszenbaum M, Dausset J, Carosella E D. Proc Natl Acad Sci USA. 1997;94:5249–5254. doi: 10.1073/pnas.94.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Söderström K, Corliss B, Lanier L L, Phillips J H. J Immunol. 1997;159:1072–1075. [PubMed] [Google Scholar]
- 43.Munz C, Holmes N, King A, Loke Y W, Colonna M, Schild H, Rammensee H G. J Exp Med. 1997;185:385–391. doi: 10.1084/jem.185.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehner P J, Cresswell P. Curr Opin Immunol. 1996;8:59–67. doi: 10.1016/s0952-7915(96)80106-3. [DOI] [PubMed] [Google Scholar]