Abstract
Background & objectives:
Intermittent cholera outbreaks are major problem in many of the states of India. It is essential to identify cholera at the earliest for timely mobilization of public health responses and to abort the outbreaks. The present study was a part of a diarrhoeal outbreak investigation in Secunderabad, India, during May 2009 where the usefulness of Crystal VC rapid dipstick kit was assessed for detecting the aetiologic agent of the outbreak.
Methods:
Stool specimens were collected from 15 hospitalized patients with acute watery diarrhoea and analyzed for detection of cholera vibrios using Crystal VC rapid dipstick kit and the usefulness of the kit was determined by comparative analysis of the same set of specimens using both microbiological and real-time PCR (RT-PCR) based assays.
Results:
Detection of Vibrio cholerae O1 from 10 of 15 specimens was recorded using dipstick assay. Microbiological methods detected V. cholerae O1 positivity among 11 specimens. However, RT-PCR based assay showed all 15 specimens positive for the presence of V. cholerae O1. In addition, the same assay showed that the pathogen load in the dipstick as well as RT-PCR positive specimens ranged from 106 colony forming units (cfu)/ml or more.
Interpretation & conclusions:
Crystal VC kit had the potential to identify cholera cases in 10 min in field conditions without having good laboratory support. Therefore, dipstick kit may be considered as cholera detecting tool in diarrhoeal outbreak investigations. Specimens from clinically typical cholera cases, if negative by dipstick, should be reanalyzed by culture based methods.
Keywords: Cholera, diarrhoea, dipstick test, outbreak, rapid test, Vibrio cholerae
Among the diarrhoeal diseases, cholera is one of the most significant causal elements in terms of severity of the disease and outcomes. Several epidemics of cholera have been reported from different parts of India and abroad1–8. Vibrio cholerae strains belonging to O1 and O139 serogroups are responsible to cause cholera in the form of epidemics and pandemics9,10. Despite the fact that the pathophysiology and transmission of cholera have been well understood and several advances have been made in modern health systems, cholera remains as a dreadful disease due to its rapid onset, severity to the extent of death, if left untreated, and potential to cause outbreaks that easily break through the public health systems in impoverished settings11,12. Outbreaks of cholera are related to limited access to safe drinking water and poor sanitation12,13. Earlier reports have pointed out the fact that high case fatalities in cholera outbreaks were due to inadequate use of oral rehydration therapy and in some cases inadequate experience in managing severe cholera by the health workers13. In a recent report, lack of clean water has been attributed for an exacerbation of cholera outbreaks in Haiti14. Strengthening of health services infrastructure in rural areas along with enhanced public education can significantly reduce cases fatalities15. Case fatality rates for untreated cholera cases can reach to 50 per cent, while implementation of good case management can reduce this figure below 1 per cent12,15. Outbreaks often start in fringes in very low number but without effective and efficient healthcare system it can become threat to wider communities. The reported cholera cases to WHO ranged from 178,000 to 237,000 with 4000-6300 deaths12. There are numerous evidences, which show that without having timely intervention, delayed diagnosis of cholera has turned handful of cases into large epidemics16–19. Therefore, prompt, rapid and sufficiently sensitive diagnosis of cholera is the key factors to prevent the disease from being an epidemic.
Currently, cholera diagnosis relies on the microbiological identification of the pathogen V. cholerae from the diarrhoeal stool specimens followed by serological characterization into either O1 or O139 serogroups. Culture confirmed cholera case detection requires 2~ 3 days in a good laboratory set up with trained manpower. Therefore, rapid diagnostic tests consuming least time, requiring minimum laboratory infrastructure and technical skill, will be the best choice in diagnosing cholera, in an outbreak situation. These rapid tests are based on the chromatographic changes due to the presence of O1 and/or O139 antigens in the test specimen. Studies have already been carried out on the applicability of such rapid tests in cholera endemic areas like Madagascar and Bangladesh; the specificity of O1 and O139 dipsticks ranged between 84-100 per cent and the sensitivity ranged between 94.2-100 per cent20.
National Institute of Cholera and Enteric Diseases (NICED), Kolkata, conducts diarrhoeal outbreak investigations, as and when required, in different parts of the country. This study was a part of an outbreak investigation carried out NICED, Kolkata, at Bholakpur, Secunderabad, India.
This study also evaluated the potential of using rapid dipstick test to identify the aetiologic agent of the outbreak.
Material & Methods
Affected area and specimen collection: The NICED team collected clinical and epidemiological data of patients admitted to the Gandhi Medical College and Quarantine Hospital at Secunderabad, Adhra Pradesh, India. The diarrhoeal outbreak broke out at Bholakpur, between May 3-11, 2009. Bholakpur area consists of mainly of slum population of about 65,000 residing an average of 4-5 families per premise. The community first faced the brunt of the outbreak on May 3. The daily number of cases increased to 600 by May 5. On May 7, the numbers went down to 500 and since then decreased to 150 cases on May 10. There have been 16 deaths reported from the area of whom 10 were children below 5 yr. Four medical camps were set up in the community manned by physicians and ancillary health staff and patients were referred to different hospitals. Patients started pouring since May 3, 2009, mainly into two local hospitals, namely Gandhi Medical College Hospital and Quarantine Hospital which were about 2 km away from the affected area. All cases presented with acute watery diarrhoea and were mostly admitted within 6~12 h of initiation of symptom. Patients were admitted in the paediatric disaster and other special wards open to combat the situation. NICED team clinically assessed hospitalized diarrhoeal patients, collected relevant epidemiological information and made microbiological assessment on diarrhoeal stool specimens. The team clinically assessed diarrhoeal patients for likelihood of cholera according to the WHO guidelines21, appropriate management was advised, their dehydration status and deaths were recorded. Between May 3 and 11, 2009, 515 and 173 diarrhoeal patients were admitted at Gandhi hospital and Quarantine hospital, respectively. In Gandhi hospital about 70 per cent cases were in the paediatric age group while most of the adult cases were admitted in the Quarantine hospital. Patients were admitted with severe dehydration caused by acute watery diarrhoea and vomiting, with episodes of diarrhoea being 10 per adult subjects and 6-20 in cases of children. The mean duration of stay in the hospital was 2.8 day.
A total of 15 stool specimens were collected from the hospitalized patients using sterile cups and transported to the laboratory at NICED, Kolkata for rapid evaluation of cholera using Crystal VC dipstick kits followed by comprehensive analysis of the same set of specimens through conventional microbiological as well as real-time PCR (RT-PCR) assay.
Dipstick assay: The rapid cholera dipstick assay was performed using Crystal VC dipstick kit (Span Diagnostics Ltd., India) following the instructions by the manufacturer. Briefly, 200 μl of diarrhoeal stool was transferred from specimen container to sterile clean glass test tubes properly labelled with respective patient identification code. The specimens were diluted with supplied buffer and the VC dipstick strip was placed into it. After 10 min of incubation, strips were taken out from the tube, observed appearance of coloured line in the specified region, which served as an index to interpret the presence or absence of cholera vibrios in the test specimen.
Microbiological assay: Stool specimens were transported to NICED, Kolkata for microbiological assessment on the presence of V. cholerae, diarrhoeagenic Escherichia coli and Campylobacter sp. A portion of the stool specimens were directly inoculated onto blood agar plate, kept under microaerophillic conditions and transported to the laboratory for the detection of Campylobacter sp. Cary-Blair transport medium was employed for transporting stool specimens and processed within 96 h of collection in the laboratory for the detection of V. cholerae and diarrhoeagenic E. coli22. Selective enrichment of V. cholerae organisms was made by allowing growth in alkaline peptone water (APW) broth for 6 h. This was followed by plating onto highly selective thiosulphate citrate bile salt sucrose (TCBS, Eiken, Japan) pate and plates were incubated overnight at 37°C. The yellow coloured colonies of suspected V. cholerae were further confirmed by routine biochemical tests23. Detected V. cholerae were identified to belong to O1 serogroup through slide agglutination test using antisera kit (BD Difco, USA). Stool specimens were inoculated onto MacConkey agar (BD Difco) plates and incubated overnight for identification of E. coli. Lactose fermenting colonies were further tested in pathogroup specific PCR assays for identification of diarrhoeagenic E. coli. Stool specimens inoculated onto blood agar plates were observed for the presence of typical translucent colonies for presumptive identification of Campylobacter sp. which was subsequently confirmed by PCR assay.
Real-time PCR (RT-PCR) assay: Stool specimens were stabilized using RNALater (Ambion, USA) and transported to NICED, Kolkata, for RT-PCR based assessment on the presence of V. cholerae O1 and O139. Specimens were processed for extraction of total DNA using QIAamp DNA stool mini kit (Qiagen, Germany) within 96 h of collection. The total DNA thus extracted from the stabilized stool specimens were used as a template for RT-PCR assay. V. cholerae O1 and O139 specific RT-PCR assay were performed separately using O1 wbe, and O139 wbf specific primers24. Briefly, the RT-PCR assay was performed in a 7900HT Fast Real-Time PCR instrument (Applied Biosystems, USA) with Power SYBR Green kit (Applied Biosystems). PCR reactions were carried out in 20 μl reaction volume containing 10 μl of 2X Power SYBR Green master mix, 1 μl of template DNA and each of the PCR primers appropriately diluted to 0.6 pmole/ μl. PCR cycles consisted of 35 cycles of denaturation at 94°C for 20 sec, annealing at 55°C for 20 sec and extension at 72°C for 50 sec. After PCR amplification, melting (Tm) curve analysis was performed. The amplified products were adjusted to 60 °C followed by gradual increase to 95°C at a rate of 0.2 °C/sec. During the analyzing time, fluorescence signals were continuously monitored to ascertain amplification specificity. The Tm values of the amplicons were determined by plotting the negative derivative of fluorescence over temperature versus temperature (dF/dT versus T)25. PCR products were differentiated and verified during analysis of melting curve as the melting curves of the products are based on the GC content of the products26.
Quantification of the V. cholerae O1 load in the diarrhoeal specimens was made by comparing the data obtained with reference DNA sets prepared from known number of cells of V. cholerae O1 El Tor Inaba strain N16961. For this, a standard curve was prepared by potting Ct values obtained in the RT-PCR assay against number of V. cholerae cells required to give included amount of template DNA.
Results
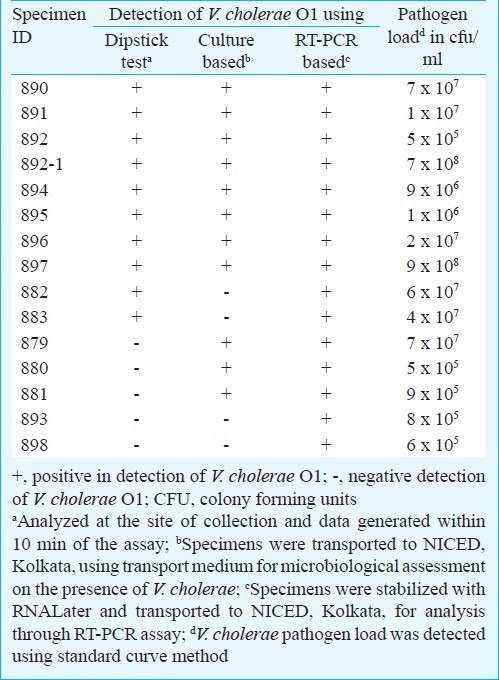
Analysis of the stool specimens using Crystal VC cholera dipstick tests revealed presence of V. cholerae O1 among 10 of 15 (66.6%) specimens and that was within a 10 min of time. None of the specimens were positive for V. cholerae O139. Results of dipstick assay were compared with data obtained through microbiological as well as RT-PCR based assays (Table). Microbiological analysis revealed presence of V. cholerae O1 among 11 of 15 specimens (73.3%). All these V. cholerae strains were of O1 El Tor variant27 and belonged to serotype Ogawa. Presence of V. cholerae O139 was not detected in these specimens. Among culture confirmed 11 V. cholerae positive specimens, only eight were positive by dipstick assay. Further, two specimens that were positive for V. cholerae by the dipstick assay failed to show presence of V. cholerae pathogen in microbiological assay even after applying enrichment techniques. To overcome the apparent discrepancy between data generated by the dipstick and culture based assays, these 15 specimens were further screened by RT-PCR based assay. RT-PCR revealed presence of V. cholerae O1 among all these specimens and the load of V. cholerae O1 varied from specimen to specimen (Table). The result showed that, in general, the specimens, which were positive by dipstick assay or culture based assay contained about 106cfu/ml of bacterial cells or more. The speimens were also analyzed for the presence of diarrhoeagenic E. coli and Campylobacter sp. Of the 15 specimens, three possessed enteroaggregative E. coli as confirmed by multiplex diarrhoeagenic E. coli PCR22. Presence of Campylobacter sp. in two of 15 specimens was also detected.
Table.
Detection of V. cholerae O1 in diarrhoeal stool specimens collected from patients admitted to Gandhi Medical College hospital from outbreak affected area of Bholakpur, Secunderabad, India
Discussion
Outbreaks of cholera usually occur in areas that are affected by the lack of supply of safe drinking water. Early detection of cholera helps in controlling the spread and duration of the outbreak and to restrict the morbidity and mortality rate. Efforts have been made to develop rapid cholera detection kit which can be used either in the laboratory or in the field20,28–30. Several rapid test kits like Medicos™ SMART™, IP dipstick have also been evaluated for their ease of use in detecting cholera29. The availability and successful implementation of such rapid cholera detection kit will have enough significance from the public health point of view. The Crystal VC dipstick assay used in the present study, detects presence of V. cholerae O1 and/or O139 antigens in the specimens through immunochromatographic methods. This dipstick utilizes nitrocellulose membrane coated with V. cholerae O1 and O139 ‘O’ antigen specific monoclonal antibodies that are coupled to colloidal gold. Dipping of the membrane into appropriately diluted V. cholerae positive specimens results into development of discrete bands other than the control band.
The outbreak at Bholakpur, Secunderabad lasted about one week. Hospitalization rate was 25.6 per cent, attack rate was 4.5 per cent and case fatality rate was 0.54 per cent. The patients received oral rehydration solution (ORS) or intravenous fluid (Ringer Lactate Solutions), and were also given antimicrobial agents like doxycycline, co-trimoxazole, etc.; in case of vomiting, anti-emetic and antacids were given. At the onset, Bholakpur outbreak was not considered as a cholera outbreak. Upon arrival of the investigation team and the use of rapid dipstick showed presence of V. cholerae O1 in 67 per cent stool specimens collected from hospitalized patients. Identification of cholera cases through rapid dipstick assay resulted into immediate implementation of cholera outbreak control measures and thus it helped in averting many more diarrhoeal events among the population residing in the affected area. Subsequently, microbiological analysis of the stool specimens resulted into detection of V. cholerae O1 in 73 per cent specimens. Microbiological assessment also showed certain number of cholera victims had concomitant infections with Campylobacter sp. and diarrheagenic E. coli. The stool specimens were also screened for the detection of the V. cholerae by RT-PCR assay. All specimens were identified to possess V. cholerae O1. Variations of pathogen load across the specimens were evident in RT-PCR assay. Comparative analysis of three different methods also pointed out the fact that RT-PCR detection was more sensitive as compared to other methods and therefore, such sensitive assay should be utilized the least, in the referral laboratories. Comparative analysis showed that the simple one step rapid Crystal VC dipstick test detected V. cholerae O1 in specimens with specificity more than 50 per cent when microbiological assay has been considered as referral standard. In our study certain specimens were positive for V. cholerae by microbiological and RT-PCR assays but were turned out to be negative in dipstick assay, pointing towards some limitation of the dipstick assay in detecting cholera. Similar limitations have been indicated in other studies carried out in Bangladesh, Madagaskar and Kolkata20,31. Thus, this kit may not be considered suitable as routine diagnostic kit for detecting V. cholerae O1 infection among hospitalized diarrhoeal patients. Importantly, the test kits can be stored at ambient temperature in humidity proof plastic bags making it easy to transport at remote regions. Therefore, employing such rapid test would be helpful in detecting cholera in the outbreak scenario for prompt mobilization of appropriate health resources.
Acknowledgment
The work was supported in part by Program of Founding Research Centre for Emerging and Reemerging Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan. The first two authors (AS and SSG) are recipient of Research Assistant Fellowship from Okayama University project.
Footnotes
Conflict of interest: Authors declared that there is no conflict of interest.
References
- 1.Suar D, Mandal MK, Khuntia R. Supercyclone in Orissa: an assessment of psychological status of survivors. J Trauma Stress. 2002;15:313–9. doi: 10.1023/A:1016203912477. [DOI] [PubMed] [Google Scholar]
- 2.Taneja N, Kaur J, Sharma K, Singh M, Kalra JK, Sharma NM, et al. A recent outbreak of cholera due to Vibrio cholerae O1 Ogawa in and around Chandigarh, north India. Indian J Med Res. 2003;117:243–6. [PubMed] [Google Scholar]
- 3.Mishra M, Mohammed F, Akulwar SL, Katkar VJ, Tankiwale NS, Powar RM. Re-emergence of ElTor Vibrio in outbreak of cholera in and around Nagpur. Indian J Med Res. 2004;120:478–80. [PubMed] [Google Scholar]
- 4.Usman A, Sarkinfada F, Mufunda J, Nyarango P, Mansur K, Daiyabu TM. Recurrent cholera epidemics in Kano -northern Nigeria. Cent Afr J Med. 2005;51:34–8. [PubMed] [Google Scholar]
- 5.World Health Organization. Cholera: global surveillance summary, 2008. Wkly Epidemol Rec. 2009;84:307–24. [Google Scholar]
- 6.Shikanga OT, Mutonga D, Abade M, Amwayi S, Ope M, Limo H, et al. High mortality in a cholera outbreak in western Kenya after post-election violence in 2008. Am J Trop Med Hyg. 2009;81:1085–90. doi: 10.4269/ajtmh.2009.09-0400. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki S, Suzuki H, Fujino Y, Kimura Y, Cheelo M. Impact of drainage networks on cholera outbreaks in Lusaka, Zambia. Am J Public Health. 2009;99:1982–7. doi: 10.2105/AJPH.2008.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taneja N, Mishra A, Sangar G, Singh G, Sharma M. Outbreaks caused by new variants of Vibrio cholerae O1 El Tor, India. Emerg Infect Dis. 2009;15:352–4. doi: 10.3201/eid1502.080943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharya SK, Bhattacharya MK, Nair GB, Dutta D, Deb A, Ramamurthy T, et al. Clinical profile of acute diarrhoea cases infected with the new epidemic strain of Vibrio cholerae O139: designation of the disease as cholera. J Infect Dis. 1993;27:11–5. doi: 10.1016/0163-4453(93)93488-p. [DOI] [PubMed] [Google Scholar]
- 10.Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deen JL, Von Seidlein L, Sur D, Agtini M, Lucas ME, Lopez AL, et al. The high burden of cholera in children: comparison of incidence from endemic areas in Asia and Africa. PLoS Negl Trop Dis. 2008;2:e173. doi: 10.1371/journal.pntd.0000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. Cholera vaccines: WHO position paper. Wkly Epidemol Rec. 2010;85:117–28. [Google Scholar]
- 13.Siddique AK, Salam A, Islam MS, Akram K, Majumdar RN, Zaman K, et al. Why treatment centres failed to prevent cholera deaths among Rwandan refugees in Goma, Zaire. Lancet. 1995;345:359–61. doi: 10.1016/s0140-6736(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 14.Webster PC. Lack of clean water exacerbates cholera outbreak in Haiti. CMAJ. 2011;183:E83–4. doi: 10.1503/cmaj.109-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunnlaugsson G, Angulo FJ, Einarsdóttir J, Passa A, Tauxe RV. Epidemic cholera in Guinea-Bissau: the challenge of preventing deaths in rural West Africa. Int J Infect Dis. 2000;4:8–13. doi: 10.1016/s1201-9712(00)90059-6. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow DL, Malenga G, Begkoyian G, Nyangulu D, Toole M, Waldman RJ, et al. Epidemic cholera among refugees in Malawi, Africa: treatment and transmission. Epidemiol Infect. 1997;118:207–14. doi: 10.1017/s0950268896007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos FL, Lins-Lainson ZC, Da Silva EL, Proietti Júnior AA, Mareco ML, Lamarão ML. Cholera in north Brazil: on the occurrence of strains of Vibrio cholerae O1 which fail to ferment sucrose during routine plating on thiosulphate-citrate-bile salt-sucrose agar (TCBS). A new problem in diagnosis and control? Rev Latinoam Microbiol. 1997;39:141–4. [PubMed] [Google Scholar]
- 18.Mason PR. Zimbabwe experiences the worst epidemic of cholera in Africa. J Infect Dev Countries. 2009;3:148–51. doi: 10.3855/jidc.62. [DOI] [PubMed] [Google Scholar]
- 19.Harris JR, Cavallaro EC, de Nóbrega AA, Dos S, Barrado JC, Bopp C, et al. Field evaluation of crystal VC Rapid Dipstick test for cholera during a cholera outbreak in Guinea-Bissau. Trop Med Int Health. 2009;14:1117–21. doi: 10.1111/j.1365-3156.2009.02335.x. [DOI] [PubMed] [Google Scholar]
- 20.Nato F, Boutonnier A, Rajerison M, Grosjean P, Dartevelle S, Guénolé A, et al. One-step immunochromatographic dipstick tests for rapid detection of Vibrio cholerae O1 and O139 in stool samples. Clin Diagn Lab Immunol. 2003;10:476–8. doi: 10.1128/CDLI.10.3.476-478.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Global task force on cholera control. First steps for managing an outbreak of acute diarrhoea. WHO/CDS/CSR/NCS/2003.7 Rev.2. Geneva, Switzerland: WHO; 2003. [accessed on April 12, 2012]. World Health Organization. Available from: http://www.who.int/cholera/en/ [Google Scholar]
- 22.Nair GB, Ramamurthy T, Bhattacharya MK, Krishnan T, Ganguly S, Saha DR, et al. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog. 2010;5:2–4. doi: 10.1186/1757-4749-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee S, Ghosh K, Raychoudhuri A, Pan A, Bhattacharya MK, Mukhopadhyay AK, et al. Phenotypic and genotypic traits and epidemiological implication of Vibrio cholerae O1 and O139 strains in India during 2003. J Med Microbiol. 2007;56:824–32. doi: 10.1099/jmm.0.46982-0. [DOI] [PubMed] [Google Scholar]
- 24.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, et al. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20:201–7. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 25.Fukushima H, Tsunomori Y, Seki R. Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J Clin Microbiol. 2003;41:5134–46. doi: 10.1128/JCM.41.11.5134-5146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–60. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 27.Raychoudhuri A, Mukhopadhyay AK, Ramamurthy T, Nandy RK, Takeda Y, Nair GB. Biotyping of Vibrio cholerae O1: time to redefine the scheme. Indian J Med Res. 2008;128:695–8. [PubMed] [Google Scholar]
- 28.Bhuiyan NA, Qadri F, Faruque ASG, Malek MA, Salam MA, Nato F, et al. Use of Dipsticks for rapid diagnosis of cholera caused by Vibrio cholerae O1 and O139 from rectal swabs. J Clin Microbiol. 2003;41:3939–41. doi: 10.1128/JCM.41.8.3939-3941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalluri P, Naheed A, Rahman S, Ansaruzzaman M, Faruque AS, Bird M, et al. Evaluation of three rapid diagnostic tests for cholera: does the skill level of the technician matter? Trop Med Int Health. 2006;11:49–55. doi: 10.1111/j.1365-3156.2005.01539.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang XY, Ansaruzzaman M, Vaz R, Mondlane C, Lucas ME, von Seidlein L, et al. Field evaluation of a rapid immunochromatographic dipstick test for the diagnosis of cholera in a high-risk population. BMC Infect Dis. 2006;6:17. doi: 10.1186/1471-2334-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee P, Ghosh S, Ramamurthy T, Bhattacharya MK, Nandy RK, Takeda Y, et al. Evaluation of a rapid immunochromatographic dipstick kit for diagnosis of cholera emphasizes its outbreak utility. Jpn J Infect Dis. 2010;63:234–8. [PubMed] [Google Scholar]


