Abstract
Parasite diversity is a constant challenge to host immune systems and has important clinical implications, but factors underpinning its emergence and maintenance are still poorly understood. Hosts typically harbour multiple parasite genotypes that share both host resources and immune responses. Parasite diversity is thus shaped not only by resource competition between co-infecting parasites but also by host-driven immune-mediated competition. We investigated these effects in an insect–trypanosome system, combining in vivo and in vitro single and double inoculations. In vivo, a non-pathogenic, general immune challenge was used to manipulate host immune condition and resulted in a reduced ability of hosts to defend against a subsequent exposure to the trypanosome parasites, illustrating the costs of immune activation. The associated increase in available host space benefited the weaker parasite strains of each pair as much as the otherwise more competitive strains, resulting in more frequent multiple infections in immune-challenged hosts. In vitro assays showed that in the absence of a host, overall parasite diversity was minimal because the outcome of competition was virtually fixed and resulted in strain extinction. Altogether, this shows that parasite competition is largely host-mediated and suggests a role for host immune condition in the maintenance of parasite diversity.
Keywords: parasite, diversity, co-infection, competition, trypanosome, immune challenge
1. Introduction
Parasite genetic diversity is a constant challenge to host immune systems and hampers important health initiatives combating infectious diseases [1–3]. To understand the factors underpinning the emergence and maintenance of parasite genetic diversity is therefore of great importance. Currently, insights into host–parasite interactions are still largely based on studies involving single parasite strains infecting single hosts. Yet, in the real world, hosts are often infected by numerous parasite genotypes of the same or different species simultaneously, and an approach integrating this variation into both experimental systems and models of infection is required [4].
One approach is to consider that co-infecting parasite species or strains will share not only host resources, but are also probably subject to the same host immune responses. Such interactions with the host's immune system can amplify or reverse inherent differences in competitive ability of co-infecting parasite genotypes. This considerably complicates any attempts to predict the outcome of co-infection. Classical ecological theory provides a useful framework to approach the question [5–9]. In this framework, co-infecting parasites can be seen as either regulated by resource-based (i.e. linked to extraction of host resources; ‘bottom-up’) and/or immune-based (‘top-down’) control mechanisms. In immune-mediated apparent competition, for example, a low-density parasite genotype suffers disproportionately from the presence of a high-density strain in the same host when the latter induces a strong, non-specific immune response [10]. Alternatively, a genotype-specific immune response might primarily affect the high-density parasite genotype that has elicited it, providing an advantage to the low-density strain relative to situations where it infects alone. Various degrees of cross-reactivity of the immune response will obviously modulate this effect in various ways [11]. At the level of the host population, the picture is further complicated by the fact that hosts naturally differ in their immune repertoire and condition.
This work investigates the relative importance of ‘bottom-up’ and ‘top-down’ mechanisms in determining the outcome of co-infection in a natural system. With this aim, we studied experimental infections of the intestinal trypanosome, Crithidia bombi, in its bumble-bee host, Bombus terrestris L. Crithidia bombi shows very high natural genetic diversity: virtually all multi-locus genotypes collected from the field over several years are distinct [12]. Hosts are therefore exposed to a wide range of parasite strains, resulting in the considerable frequency of multiple infections measured in natural populations (more than 40%). Experimental infections with multiple parasite strains have shown, however, that exclusion of one or more of the co-inoculated strains is very common [13]. Interestingly, C. bombi has been shown to reproduce mostly clonally (an estimated 84% of cases), even though it is able to recombine and exchange genetic material with co-infecting strains [14]. Furthermore, the parasite population is strongly reduced every year when it is ‘constricted’ in the population of hibernating bumble-bee queens, the only hosts to survive winter. The question thus arises of how strain diversity can be maintained in C. bombi in the face of limited recombination, frequent elimination of strains by individual hosts and drastic seasonal population bottlenecks.
We hypothesized that in co-infected hosts, parasite competition and consequently parasite diversity are controlled by: (i) ‘top-down’, immune-based mechanisms determined by host identity and host immune condition; and (ii) ‘bottom-up’, resource-based interactions between co-infecting parasites, determined by resource availability and parasite identity. In this experiment, the strength of immune-based control (the ‘top-down’ component) was modulated by manipulating host immune condition. Practically, this was achieved by challenging the bees with an injection of heat-killed bacteria in the haemolymph. Such a bacterial immune challenge, or priming, whether with heat-killed bacteria or lipopolysaccharide (LPS) extracted from bacterial surfaces, is routinely used in experiments and is known to increase antibacterial activity in the haemolymph of insects, including B. terrestris [15,16]. Bacterial priming provides surprisingly specific and durable protection upon secondary exposure to related parasites [15,17,18], but becomes costly when the host faces a mismatched parasite, leading, for example, to higher infection levels [16]. Bacterial priming with LPS also decreases host survival under starvation [19], but not when hosts are fed ad libitum (the treatment per se is not detrimental/toxic). These costs are thus thought to be the result of trade-offs in resource allocation to defence versus survival (in the latter case) and/or to different arms of the immune system (in the former case). Considering the above, we hypothesized that a bacterial challenge would decrease the host ability to fight a subsequent exposure to C. bombi. How such a change would affect intra-specific parasite competition is, however, harder to predict. Note that this working hypothesis does not require the trade-off to be purely immunological. The expected effects could come about via a general mismatch between resources that are effectively invested into defence (as a response to the presence of bacterial elicitors in the haemolymph) and the relevant parasite infection (by live trypanosomes in the gut).
Parallel to this in vivo experiment, we studied ‘bottom-up’, resource-based control mechanisms in the absence of a host immune system with an in vitro competition experiment. Given the homogeneous nature of the resource (liquid medium), we expected the outcome of in vitro competition to be simpler and less variable than in vivo. However, because the conditions pertaining to the in vivo and in vitro situations are obviously different (both in terms of quantity and quality resources, as well as the presence/absence of host immunity), more specific predictions could not be made, and only the qualitative pattern of strain persistence during experimental infections was used as a signal to assess the importance of the two hypothesized processes.
2. Material and methods
(a). Bees, bacteria and trypanosomes
Eight colonies of B. terrestris were started from uninfected bumble-bee queens collected in spring 2009 from a population in western Switzerland (Aesch, Switzerland). All bees were kept at 26 ± 2°C under constant red light illumination, with pollen and sugar water (ApiInvert, Südzucker, Ochsenfurt, Germany) provided ad libitum.
The bacteria used for immune challenges were the Gram-positive Arthrobacter globiformis (strain no. DSM 20124) and the Gram-negative Escherichia coli (strain no. DSM 498) obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). We used both Gram types, because it is known that the insect immune pathways are differentially activated by these two Gram specificities. Bacteria were cultured separately at 30°C (A. globiformis) or 37°C (E. coli) in medium (10 g bacto-tryptone, 5 g yeast extract, 10 g NaCl in 1000 ml of distilled water, pH 7.0). Immediately before use, bacterial cells were washed three times by centrifugation (3000 r.p.m., 4°C, 10 min), removal of the supernatant and resuspension in Ringer saline solution. We determined cell concentration of both cultures and mixed the two into a single inoculum so as to reach a final concentration of bacterial cells of 108 cells ml−1 (corresponding to 0.5 × 108 cells ml−1 of each bacterium). The bacteria were then heat-killed (90°C, 15 min). Efficiency of the heat killing was confirmed by plating out samples of the suspension on agar and checking for an eventual growth of bacteria (none occurred; data not shown).
The five C. bombi strains (labelled A, B, C, D, E) used for the experimental infections were obtained from faeces of naturally infected queens collected in spring 2008 (Neunforn, Switzerland). Single infective cells were isolated using a fluorescence-activated cell sorter and subsequently maintained clonally in liquid medium at 27°C and 3 per cent CO2 (R. Salathé 2007, unpublished data). The strains had distinct multi-locus genotypes at four polymorphic microsatellite loci (see below) and could thus be readily differentiated by genetic markers in a mixture.
(b). Bacterial challenge and in vivo inoculations
Workers were collected as callows (freshly hatched workers) from the eight experimental colonies during three to four consecutive days and kept in groups for three additional days (i.e. until they were 3 to 6 days old). Individual workers were then chilled on ice and randomly assigned to the challenged (C) or naive groups (N). Immune challenge of the workers was performed by injecting 2 µl of the inoculum containing 108 cells ml−1 heat-killed bacteria between the first and second abdominal tergites. Naive bees were sham-manipulated in the same way but not injected because the wounding associated with injection itself is known to induce an immune response [20]. Thus, we compared bees given a general immune activation (Gram-positive/Gram-negative bacterial challenge and wounding) with naive bees. All bees were kept individually from that point.
On day 9, all bees were exposed to C. bombi. For this, bees were starved for 4–5 h before being presented with 10 µl of sugar water containing the inoculum of live C. bombi cells. We administered the five C. bombi strains either alone (‘single exposure’, with strains A, … , E, at 10 000 cells each), or in any of the 10 possible pairwise combinations (‘double exposure’, with strain combinations AB, … , DE, at 5000 cells per strain to keep the overall inoculum constant). On days 4, 6 and 8 post-infection, faeces were collected and kept in glass micro-capillaries until further assessment of infection status. All bees were frozen on 8 days post-infection. In order to attain sufficient sample sizes while ensuring that all bees were the same age, the above procedure was repeated several times during colony development (from two to six times, depending on colony productivity), so that bees from each colony were infected in successive batches.
(c). Transmission speed, infection status, composition and intensity
To assess infection status, all collected faeces (three samples per individual) were microscopically checked for the presence of the parasite (infected/uninfected). For each infected individual, transmission speed was coded as ‘fast’, ‘medium’ or ‘slow’, depending on whether the faeces contained the first C. bombi cells from day 4, 6 or 8 post-infection, respectively. All infected individuals had their gut dissected out and homogenized in 100 µl Ringer solution. DNA was extracted from these individual gut preparations using a Qiagen DNeasy 96 tissue kit. The gut extracts were then genotyped at the C. bombi microsatellite markers Cri 4, Cri 1B6, Cri 4G9 and Cri 2F10 following Ulrich et al. [13] to identify the strains present in each infected individual. Previous work shows that the method is sensitive enough to detect very low concentrations of different C. bombi strains in mixed infections [21]. Note that while this method provides reliable information on the presence/absence of strains, it does not allow strain-specific quantification in mixed infections. Instead, the total relative infection intensity was measured from gut extracts using a quantitative PCR (qPCR) reaction amplifying a portion of the C. bombi 18sRNA gene [13].
(d). In vitro inoculations
The same five C. bombi strains used for in vivo infections were also assayed in vitro (that is, with living cells maintained in axenic cultures in media). For this purpose, 10 000 cells of the same 10 pairwise strain combinations as above were inoculated in 2 ml of liquid medium housed in 24-well culture plates and incubated at 27°C and 3 per cent CO2 for 8 days. Each strain pair was cultivated in six replicates; furthermore, two replicates each of single strain inocula were used as positive controls. After removal of the culture medium, the cells were washed by resuspension in 1× PBS, centrifugation (10 000 r.p.m., 5 min) and removal of the supernatant. DNA was extracted from the cell pellets by adding 10 μl Viagen direct PCR lysis reagent (Viagen Biotech Inc., Los Angeles, CA), 90 μl H2O and 1 μl proteinase K to each sample, followed by incubation at 55°C for 45 min and 85°C for 40 min. These cell culture extracts were then genotyped as above.
(e). Data analysis and statistics
(i). Determinants of parasite establishment, infection intensity and transmission speed
All statistical analyses were performed in R v. 2.8.1 [22]. Owing to the overall low infection rate (see §3), we performed separate analyses for parasite establishment success (uninfected/infected) using the entire dataset, and infection intensity (as measured with qPCR) using the subset of infected individuals. Determinants of parasite establishment success and infection intensity were investigated using binomial and Poisson generalized linear-mixed models (GLMMs), respectively (lmer function in lme4 library in R). The models used immune-treatment (challenged versus naive), and infection type (one of 15 double or single exposures: A, … , E, and AB, … , DE) as independent fixed variables, and ‘batch’ and host type (one of eight host genetic backgrounds) as random variables. Additionally, whenever overdispersion was detected, it was taken into account by incorporating individual-level variability as a random effect in the model. We evaluated the significance of fixed effects and their interaction by comparing models using log-likelihood ratio tests (LRTs) following deletion of terms (starting with the interaction). Terms for which deletion did not significantly decrease model fit were omitted, until only significant terms remained in the model (α < 0.05). The above models were also tested with a two-level infection variable for the multiplicity of exposure (single versus double exposure) in place of the 15-level infection type variable. However, the minimized models were identical in both cases because they did not retain the infection variable (see §3) and we thus present the results obtained only with the first approach (15-level infection variable) in the text. Effect sizes for the significant terms of the final models are reported in the text; the effect sizes for all terms (significant and non-significant) and for both types of models (15-level and 2-level infection variables) can be found in the electronic supplementary material.
An ordinal logistic model for transmission speed (slow, medium and fast) with infection type and immune-treatment as predictors was fitted (and minimized by removing non-significant terms) using the lrm function from library design in R.
(ii). Determinants of coexistence and diversity
We used a binomial GLMM to analyse determinants of parasite coexistence (i.e. both strains maintained) versus exclusion (i.e. one strain maintained) in the subset of infected individuals that had received a double exposure. Again, immune treatment and infection type (one of 10 double exposures AB to DE) were used as fixed factors and their interaction investigated, whereas ‘batch’ and host type were used as random factors. Models were minimized as above.
In cases where one of the strains was excluded, we used simple binomial tests to link strain performance in single inoculations (infection intensity) to their success in co-inoculations (frequency of maintenance versus exclusion), for naive and challenged hosts separately.
Furthermore, we tested whether the frequency distribution of double infections (e.g. observed prevalence of AB) differed from what would be expected if the strains infected independently (e.g. prevalence of A in single infections × prevalence of B in single infections) with Kolmogorov–Smirnov tests, for naive and challenged hosts separately.
To investigate strain-specific effects of host immune challenge and co-inoculation, prevalence data (infected/uninfected) were analysed separately for each strain using a GLMM with the multiplicity of exposure (single/double exposure), and immune treatment as fixed factors, and colony and ‘batch’ as random factors. The second-order interaction between fixed effects was investigated, and models were minimized as above. Note that the strain-specific dose differed between single exposures and double exposures: because the total inoculation dose was kept constant across all treatments, the dose of any strain in a double exposure is half that of a single exposure. However, from previous unpublished work on this system, it is known that both prevalence and infection intensity increase with dose to reach a plateau at doses considerably lower (approx. 1000 cells) than those used in this study (see the electronic supplementary material, figure S1).
Finally, we computed Simpson's inverse index of diversity, D = (∑pi2)−1 (where pi is the frequency of strain i in the parasite population), on the subset of double exposures in the naive and challenged hosts, as well as in the in vitro parasite population.
3. Results
(a). Parasite establishment, infection intensity and transmission speed
Overall infection success was low, at around 29 per cent (135 infected out of 472 exposed individuals). This is partly attributable to two colonies, which produced a large number of workers (n = 204) and were almost completely resistant to the C. bombi strains used (19 infected individuals).
We found a general, positive effect of a general immune challenge in the host for C. bombi (figure 1). Immune challenge had a positive effect on C. bombi establishment (LRT for models with versus without the variable ‘immune-treatment’: 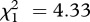 , p = 0.037; see electronic supplementary material, table S1a), with 33 per cent of bees infected when challenged versus 25 per cent in the naive group. A similar analysis performed on the subset of infected individuals revealed that immune-challenged hosts also had higher infection intensities (
, p = 0.037; see electronic supplementary material, table S1a), with 33 per cent of bees infected when challenged versus 25 per cent in the naive group. A similar analysis performed on the subset of infected individuals revealed that immune-challenged hosts also had higher infection intensities (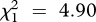 , p = 0.027; see electronic supplementary material, table S2a), with 923 ± 113 cells μl−1 of gut extract (mean ± s.e.) in challenged hosts versus 537 ± 89 cells μl−1 in naive hosts.
, p = 0.027; see electronic supplementary material, table S2a), with 923 ± 113 cells μl−1 of gut extract (mean ± s.e.) in challenged hosts versus 537 ± 89 cells μl−1 in naive hosts.
Figure 1.
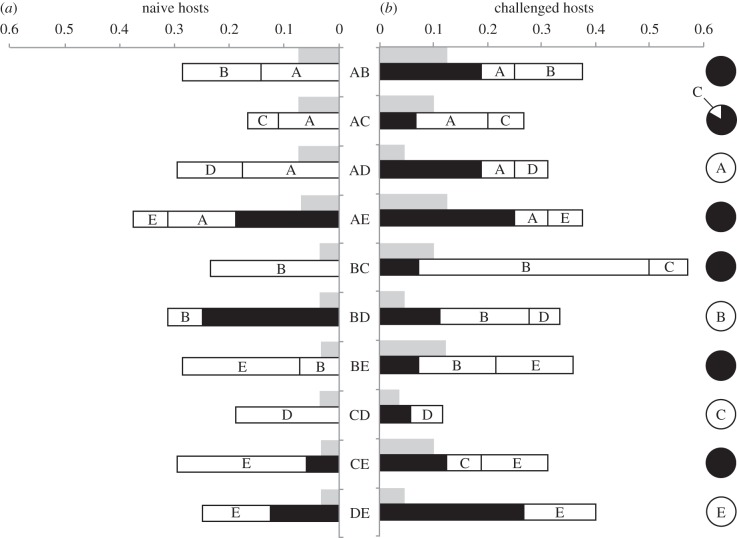
The outcome of infection by pairs of Crithidia bombi strains. For each strain combination (AB–DE), bar length and pie chart area represent the frequency of cases where the two strains coexisted (black), or one of the two strains infected alone (white, the letter indicates the infecting strain), after co-inoculation in (a) naive hosts, (b) challenged hosts and in vitro (pie charts). Grey bars show, for each strain combination, the expected frequency of double infections if both strains infect independently from the other, i.e. the product of their respective prevalences following single exposure: fexp(AB) = f(A) × f(B). One infected host (as determined by faeces check and qPCR) whose infection genotype could not be established is not represented in this figure.
In contrast, neither infection type (one of 15 double or single exposures: A, … , E, and AB, … , DE) nor multiplicity of exposure (single/double) affected C. bombi establishment or infection intensity, as indicated by the fact that these variables were not retained in the respective models, so that the two approaches ‘converged’ to the same minimal model (see electronic supplementary material, tables S1a,b and S2a,b).
Finally, the results from an ordinal logistic model showed that transmission speed was faster in challenged hosts (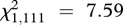 , p = 0.006).
, p = 0.006).
(b). Parasite coexistence and diversity
The outcome of co-inoculation in naive hosts varied across infection types such that it depended on the particular strain combination sharing a host (figure 1a): in two cases (combinations BC, CD), a single strain was consistently maintained, whereas the other was excluded; in four cases (AB, AC, AD, BE), either one or the other strain was maintained but coexistence was never observed; in three cases (BD, CE, DE), a strain was present in all infections, either alone or in coexistence with the other; all three possible outcomes were observed in only one case (AE). Globally, 19 of the 30 possible outcomes were observed (considering three possible outcomes—exclusion of one or the other strain, or coexistence—for each of the 10 different combinations in double exposures).
In challenged hosts (figure 1b), the variability in the outcome of co-inoculation was even greater, with 8 out of 10 strain combinations (all but CD and DE) showing all three possible outcomes. Globally, 28 of the 30 possible outcomes were observed. Thus, the challenge often seemed to allow the otherwise less competitive C. bombi strain to coexist with the other strain, or even to infect alone. To formally test the hypothesis that the challenge disproportionately benefited the less competitive strain, we compared the average increase in the prevalence of the dominant strain—defined for each pair as the strain that more frequently infected alone in naive hosts—with that of the weaker strain in naive versus challenged hosts (in one case where both strains had equal records, we took the dominant strain to be the one with overall higher prevalence and infection intensity in naive hosts). Although the average increase in the prevalence of the weak strain of each pair was higher (mean ± s.e.: 0.097 ± 0.031, n = 10) than that of the dominant strain (mean ± s.e.: 0.054 ± 0.023, n = 10), this difference was not significant (Wilcoxon–Mann–Whitney test: W1 = 35, p = 0.27). Note that in naive hosts, performance in double inoculations appeared not to be associated with success in single infections: in about half of the cases of exclusion (18 out of 33), the strain of the pair with the higher infection intensity in single infections was the one to be excluded. In challenged hosts, however, the strain with the lower infection intensity in single infections was excluded in more than half of the cases (21 out of 31 cases; one-sided binomial test: p = 0.035). Thus, the success of a strain in single infections appeared to be a better predictor of its success in double infections for challenged hosts when compared with naive hosts.
A binomial GLMM on the coexistence/exclusion data showed that the outcome of co-infection differed according to the infection type (LRT for models with versus without the variable ‘infection type’: 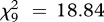 , p = 0.027; see the electronic supplementary material, table S3) and immune treatment (LRT for models with versus without immune treatment:
, p = 0.027; see the electronic supplementary material, table S3) and immune treatment (LRT for models with versus without immune treatment: 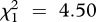 , p = 0.034), with exclusion of one of the strains being less frequent in challenged (57%) compared with naive hosts (76%; figure 1). Kolmogorov–Smirnov tests did not detect any significant difference between the observed and expected distributions of double infection prevalence in challenged hosts (D = 0.4, p = 0.401; figure 1). In naive hosts, the difference between the two distributions was more pronounced, but failed to reach statistical significance (D = 0.6, p = 0.055).
, p = 0.034), with exclusion of one of the strains being less frequent in challenged (57%) compared with naive hosts (76%; figure 1). Kolmogorov–Smirnov tests did not detect any significant difference between the observed and expected distributions of double infection prevalence in challenged hosts (D = 0.4, p = 0.401; figure 1). In naive hosts, the difference between the two distributions was more pronounced, but failed to reach statistical significance (D = 0.6, p = 0.055).
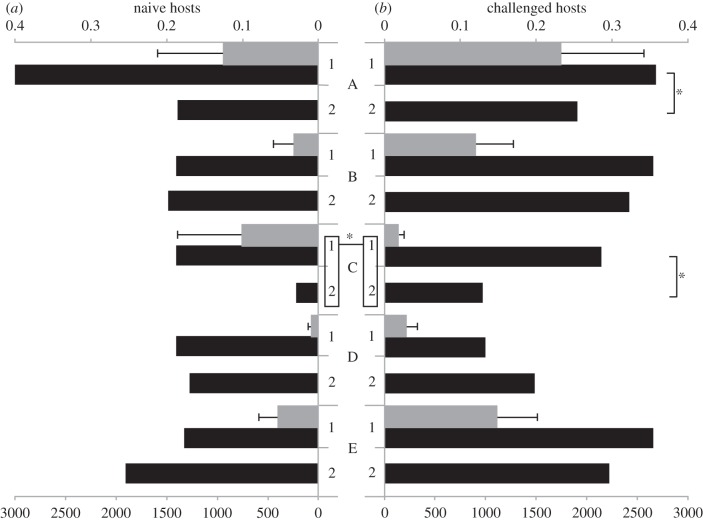
As illustrated in figure 2, strain-specific mixed-effects models detected a negative effect of co-inoculation on the prevalence of strain A (LRT for models with versus without the variable infection multiplicity: 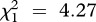 , p = 0.039; see electronic supplementary material, table S4) and strain C (
, p = 0.039; see electronic supplementary material, table S4) and strain C (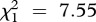 , p = 0.006), as well as a positive effect of the immune challenge on the prevalence of a strain C (LRT for models with versus without the variable immune treatment:
, p = 0.006), as well as a positive effect of the immune challenge on the prevalence of a strain C (LRT for models with versus without the variable immune treatment: 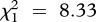 , p = 0.004).
, p = 0.004).
Figure 2.
Strain-specific prevalence following single/double exposures and infection intensity in single infections. Black bars represent the prevalence (top axis) of Crithidia bombi strains A–E in (a) naive hosts and (b) challenged hosts following single (‘1’) and double (‘2’, irrespective of the identity of the co-inoculated strain) in vivo inoculations. Grey bars represent infection intensity (bottom axis, units: cells μl−1 of gut extract, mean ± s.e.) in infected hosts following single inoculation. Data for the strain-specific infection intensity in double infections are not available (*p < 0.05).
The outcome of competition in vitro showed great repeatability: in contrast to the in vivo situation, the outcome of competition between a given pair of strains was virtually constant (see pie charts in figure 1), with only 11 out of 30 possible outcomes observed. Competitive exclusion was relatively rare (42%) and appeared to follow a simple rule: in all but one case, competitive exclusion was observed towards strain D, which consequently disappeared from the in vitro parasite population. Note that this cannot be due to a general failure of this particular strain to grow in liquid medium, because single in vitro inoculations with strain D were successful.
Finally, Simpson's inverse index was lowest in the in vitro parasite population (D = 3.97), intermediate in naive hosts (D = 4.24) and highest in challenged hosts (D = 4.64), supporting the view that a previous, unrelated immune challenge of the host led to the maintenance of C. bombi strain diversity.
4. Discussion
In line with our prediction, a general immune challenge affected the ability of hosts to defend against the trypanosome C. bombi. This resulted in more infected hosts, which carried higher parasite loads than their naive counterparts and started transmitting infective cells earlier. These results mirror those of a study by Sadd & Schmid-Hempel [16], where bumble-bee queens that had received a bacterial challenge produced offspring that were more susceptible to C. bombi (but did not incur a survival cost under starvation). Here, we show that a similar infection cost also materializes in the short term, within the lifetime of an individual. Both cases illustrate, we believe, the negative consequences of an immune ‘mismatch’: because immune activation is costly, responses to immune challenges that are distinct in type (wounding and bacterial versus trypanosomal) and/or location (haemocoel versus gut) cannot necessarily be simultaneously optimized, but must often be traded off against each other [23,24]. These costs could come about directly (via resource allocation trade-offs between different arms of the immune system) or indirectly (if, for example, immune-challenged hosts are generally weaker and more susceptible to new parasites).
Here, the immune challenge was performed under controlled conditions, but immune mismatches are probably not uncommon in nature, where hosts routinely encounter a range of different parasites. Because of such encounters, hosts in natural populations are unlikely to have naive immune systems, and thus the trade-off between defences against different parasites is relevant in the wild, too. The magnitude of its effects is likely to be more dramatic when resources are limited, as must be the case in the field. Our experiment also makes the point that individual differences in the strength and repertoire of immune responses against a particular parasite arise not only from genetic variation in resistance, but also from ‘immune history’. Indeed, we expect that different hosts or categories of hosts (e.g. sex, age, population of origin) encounter different parasites and carry a record of their immune history, either in the form of ‘resident’ live parasites or, if the infection has been cleared, in the form of an altered immune condition. Here, this history is recent, as the exposure to C. bombi occurs shortly after challenging the immune system with bacteria. However, the existence of corresponding trans-generational effects of priming [18,25,26] and the fact that insects such as B. terrestris possess an individual immune memory lasting for weeks [15] suggest that immune history will be relevant even over longer time periods. Recently, Telfer et al. [27] used time-series data from a wild vole population to show very large (positive or negative) effects of some infections on the susceptibility to other parasite species—sometimes even after clearance of the first infection—and invoked immune-mediated mechanisms as one potential explanation. Here, we find an infection cost associated with an unrelated immune challenge involving no live parasite. The situation is thus analogous to a putative vaccine that would affect host susceptibility to unrelated diseases, such that immunization against a parasite would be traded off against protection to other pathogens. This risk has not received much theoretical or empirical attention and could add to the detrimental effects of imperfect vaccines on virulence evolution [28–30].
The increase in parasite load in challenged hosts was associated with an early onset of transmission, suggesting that rapid within-host growth accelerates transmission. Bumble-bees live in dense social groups of frequently interacting individuals, where parasite transmission easily occurs between nest-mates. Accelerated transmission might thus increase within-colony prevalence at the crucial step of parasite transmission to queens, who are the only hosts to transmit the infection from one year to the next (workers and males do not survive winter) and whose fitness is severely reduced by C. bombi infections [31].
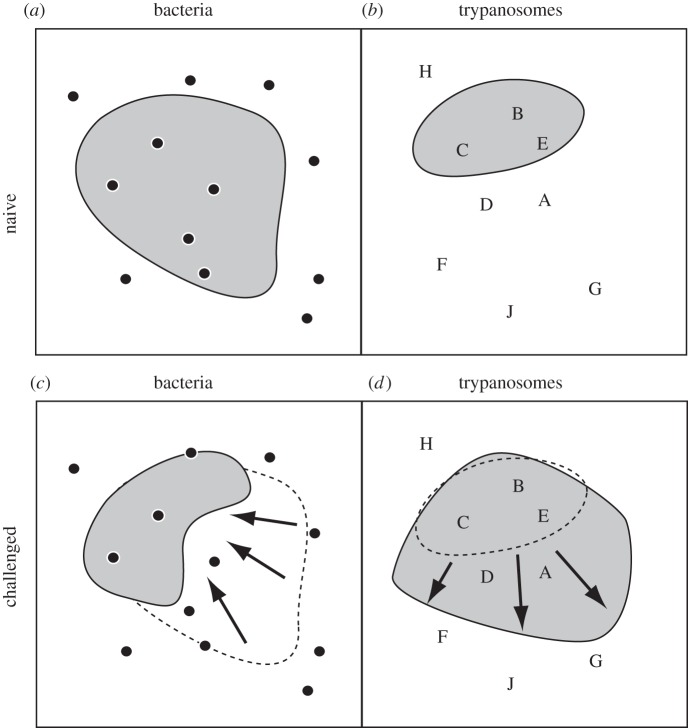
Our findings suggest that the immune challenge acted to increase the host ‘space’ available to the C. bombi population (figure 3). This increase benefited not only the dominant, more competitive C. bombi strain, but also the otherwise weak strain of the co-inoculated pair. The increase in available host space also translated into a more variable outcome of co-infection and more multiple infections. The frequency of multiple infections is of importance for pathogen evolution because it is thought to select for more virulent strains [6,32–35]. In the Bombus–Crithidia system, the multiplicity of infection in workers is associated with the probability of transmission to queens, and therefore linked to host fitness [13].
Figure 3.
Hypothesized scenario for the effect of a bacterial challenge on the immune space available to the trypanosome Crithidia bombi. The squares represent the immune space for (a,c) bacteria and (b,d) trypanosomes. Each dot (for bacteria) or letter (A–J for trypanosome strains) indicates the position a given strain of the parasite would assume in immune space. The grey areas represent the realized immune space where the parasite can infect whereas the remainder (white area) is the space that can be cleared by the host. A bacterial challenge (c,d) changes the immune space when compared with the naive host (a,b; dashed lines in panels c,d) such that the clearable space increases for bacterial infections (i.e. reducing the size of the grey area) but decreases for the trypanosomes (i.e. increasing the grey area) owing to the existence of trade-off within the immune system. The structure of the immune space varies among host individuals.
The consequences of host immune challenge and co-inoculation showed considerable variation across C. bombi strains (figure 2), so that no strain was consistently more successful than the others. For example, strain A performed well alone (with the highest prevalence and infection intensity in both naive and challenged, singly exposed hosts) but suffered in double exposures. Strain C also did poorly in double exposures but this detrimental effect was partially offset in immune-challenged hosts. These strain-specific effects might be at the basis of the lack of a general effect of the multiplicity of exposure on infection characteristics in this study, as well as in a previous experiment using only two strains [36].
The importance of host-mediated effects is reflected by the difference between in vitro and in vivo co-inoculations. In vivo co-inoculation was characterized by frequent exclusion of one of the two strains, whereas in vitro competition occurring in the absence of host-mediated immune effects more often resulted in strain coexistence. This might intuitively lead to the conclusion that host immunity acts to decrease parasite diversity. However, this is not the case. In fact, the variability in outcome and the diversity of the parasite population (as calculated with Simpson's inverse index) were lowest in vitro, intermediate in naive hosts and maximal in challenged hosts. This is because in vitro competition appeared to follow very simple rules, with a virtually fixed outcome per strain combination. One strain was outcompeted to extinction despite the overall infrequent competitive exclusion, resulting in low overall diversity. The host environment thus appears to play a role in maintaining parasite diversity. Furthermore, maximum diversity was found in challenged hosts whose immune condition was altered, which is probably the situation closest to the environment parasites naturally encounter.
Although the numerical differences between the outcomes of in vivo and in vitro co-inoculations cannot be taken as directly representing the difference between immune-mediated competition and resource competition (for instance, because resource competition could also take place in vivo), a qualitative comparison provides insight into the relative importance of ‘bottom-up’ and ‘top-down’ control mechanisms on the diversity of parasite populations. ‘Bottom-up’, resource-based competition alone explained only the simple interactions between parasite strains occurring in the absence of a host. ‘Top-down’, immune-mediated mechanisms associated with the host environment thus appear to be at the source of the complex and diverse parasite population observed in vivo in this study, as well as in the wild. In a recent study on a mouse–trematode system, Beltran et al. [37] showed that the protective effect provided by a repeated light infection with a parasite strain decreased with increasing genetic distance with the later infecting strain, a mechanism by which the vertebrate protective immunity drives parasite genetic diversity. A similar explanation has been proposed in a bovine parasite showing high genetic diversity in the field despite frequent bottlenecks [38].
Acknowledgements
The authors thank R. Schmid-Hempel, H. Koch, N. Rossel, M. Jales Hon and B. Sadd for help with the experiment and/or helpful comments on an earlier draft of the manuscript. Samuel Alizon and an anonymous reviewer provided stimulating comments that helped to improve the manuscript. The qPCR data were generated in the Genetic Diversity Center of ETH Zurich. This work was supported by the Swiss NSF (no. 31003A-116057 to P.S.H.) and the CCES (BioChange, GEDIHAP) projects.
References
- 1.Peeters M., Toure-Kane C., Nkengasong J. N. 2003. Genetic diversity of HIV in Africa: impact on diagnosis, treatment, vaccine development and trials. AIDS 17, 2547–2560 10.1097/00002030-200312050-00002 (doi:10.1097/00002030-200312050-00002) [DOI] [PubMed] [Google Scholar]
- 2.Barry A. E., Leliwa-Sytek A., Tavul L., Imrie H., Migot-Nabias F., Brown S. M., McVean G. A., Day K. P. 2007. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog. 3, e34. 10.1371/journal.ppat.0030034 (doi:10.1371/journal.ppat.0030034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus: 15 years on. J. Gen. Virol. 85, 3173–3188 10.1099/vir.0.80401-0 (doi:10.1099/vir.0.80401-0) [DOI] [PubMed] [Google Scholar]
- 4.Rigaud T., Perrot-Minnot M. J., Brown M. J. 2010. Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc. R. Soc. B 277, 3693–3702 10.1098/rspb.2010.1163 (doi:10.1098/rspb.2010.1163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenton A., Perkins S. E. 2010. Applying predator–prey theory to modelling immune-mediated, within-host interspecific parasite interactions. Parasitology 137, 1027–1038 10.1017/S0031182009991788 (doi:10.1017/S0031182009991788) [DOI] [PubMed] [Google Scholar]
- 6.Nowak M. A., May R. M. 1994. Superinfection and the evolution of parasite virulence. Proc. R. Soc. Lond. B 255, 81–89 10.1098/rspb.1994.0012 (doi:10.1098/rspb.1994.0012) [DOI] [PubMed] [Google Scholar]
- 7.Haydon D. T., Matthews L., Timms R., Colegrave N. 2003. Top-down or bottom-up regulation of intra-host blood-stage malaria: do malaria parasites most resemble the dynamics of prey or predator? Proc. R. Soc. Lond. B 270, 289–298 10.1098/rspb.2002.2203 (doi:10.1098/rspb.2002.2203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham A. L. 2008. Ecological rules governing helminth-microparasite coinfection. Proc. Natl Acad. Sci. USA 105, 566–570 10.1073/pnas.0707221105 (doi:10.1073/pnas.0707221105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown S. P., Le Chat L., Taddei F. 2008. Evolution of virulence: triggering host inflammation allows invading pathogens to exclude competitors. Ecol. Lett. 11, 44–51 10.1111/j.1461-0248.2007.01125.x (doi:10.1111/j.1461-0248.2007.01125.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raberg L., de Roode J. C., Bell A. S., Stamou P., Gray D., Read A. F. 2006. The role of immune-mediated apparent competition in genetically diverse malaria infections. Am. Nat. 168, 41–53 10.1086/505160 (doi:10.1086/505160) [DOI] [PubMed] [Google Scholar]
- 11.Dietz K. 1979. Epidemiologic interference of virus populations. J. Math. Biol. 8, 291–300 10.1007/BF00276314 (doi:10.1007/BF00276314) [DOI] [PubMed] [Google Scholar]
- 12.Salathé R., Schmid-Hempel P. 2011. The genotypic structure of a multi-host bumblebee parasite suggests a role for ecological niche overlap. PLoS ONE 6, e22054. 10.1371/journal.pone.0022054 (doi:10.1371/journal.pone.0022054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulrich Y., Sadd B. M., Schmid-Hempel P. 2011. Strain filtering and transmission of a mixed infection in a social insect. J. Evol. Biol. 24, 354–362 10.1111/j.1420-9101.2010.02172.x (doi:10.1111/j.1420-9101.2010.02172.x) [DOI] [PubMed] [Google Scholar]
- 14.Schmid-Hempel R., Salathé R., Tognazzo M., Schmid-Hempel P. 2011. Genetic exchange and emergence of novel strains in directly transmitted trypanosomatids. Infect. Genet. Evol. 11, 564–571 10.1016/j.meegid.2011.01.002 (doi:10.1016/j.meegid.2011.01.002) [DOI] [PubMed] [Google Scholar]
- 15.Sadd B. M., Schmid-Hempel P. 2006. Insect immunity shows specificity in protection upon secondary pathogen exposure. Curr. Biol. 16, 1206–1210 10.1016/j.cub.2006.04.047 (doi:10.1016/j.cub.2006.04.047) [DOI] [PubMed] [Google Scholar]
- 16.Sadd B., Schmid-Hempel P. 2009. A distinct infection cost associated with trans-generational priming of antibacterial immunity in bumble-bees. Biol. Lett. 5, 798–801 10.1098/rsbl.2009.0458 (doi:10.1098/rsbl.2009.0458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth O., Sadd B., Schmid-Hempel P., Kurtz J. 2009. Strain-specific priming of resistance in the red flour beetle, Tribolium castaneum. Proc. R. Soc. B 276, 145–151 10.1098/rspb.2008.1157 (doi:10.1098/rspb.2008.1157). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tidbury H. J., Pedersen A. B., Boots M. 2011. Within and transgenerational immune priming in an insect to a DNA virus. Proc. R. Soc. B 278, 871–876 10.1098/rspb.2010.1517 (doi:10.1098/rspb.2010.1517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moret Y., Schmid-Hempel P. 2000. Survival for immunity: the price of immune system activation for bumblebee workers. Science 290, 1166–1168 10.1126/science.290.5494.1166 (doi:10.1126/science.290.5494.1166) [DOI] [PubMed] [Google Scholar]
- 20.Korner P., Schmid-Hempel P. 2004. In vivo dynamics of an immune response in the bumble bee Bombus terrestris. J. Invertebr. Pathol. 87, 59–66 10.1016/j.jip.2004.07.004 (doi:10.1016/j.jip.2004.07.004) [DOI] [PubMed] [Google Scholar]
- 21.Schmid-Hempel P., Puhr K., Kruger N., Reber C., Schmid-Hempel R. 1999. Dynamic and genetic consequences of variation in horizontal transmission for a microparasitic infection. Evolution 53, 426–434 10.2307/2640779 (doi:10.2307/2640779) [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 23.Viney M. E., Riley E. M., Buchanan K. L. 2005. Optimal immune responses: immunocompetence revisited. Trends Ecol. Evol. 20, 665–669 10.1016/j.tree.2005.10.003 (doi:10.1016/j.tree.2005.10.003) [DOI] [PubMed] [Google Scholar]
- 24.Siva-Jothy M. T., Tsubaki Y., Hooper R. E., Plaistow S. J. 2001. Investment in immune function under chronic and acute immune challenge in an insect. Physiol. Entomol. 26, 1–5 10.1111/j.1365-3032.2001.00206.x (doi:10.1111/j.1365-3032.2001.00206.x) [DOI] [Google Scholar]
- 25.Sadd B. M., Kleinlogel Y., Schmid-Hempel R., Schmid-Hempel P. 2005. Trans-generational immune priming in a social insect. Biol. Lett. 1, 386–388 10.1098/rsbl.2005.0369 (doi:10.1098/rsbl.2005.0369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadd B. M., Schmid-Hempel P. 2007. Facultative but persistent trans-generational immunity via the mother's eggs in bumblebees. Curr. Biol. 17, R1046–R1047 10.1016/j.cub.2007.11.007 (doi:10.1016/j.cub.2007.11.007) [DOI] [PubMed] [Google Scholar]
- 27.Telfer S., Lambin X., Birtles R., Beldomenico P., Burthe S., Paterson S., Begon M. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330, 243–246 10.1126/science.1190333 (doi:10.1126/science.1190333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackinnon M. J., Gandon S., Read A. F. 2008. Virulence evolution in response to vaccination: the case of malaria. Vaccine 26(Suppl. 3), C42–C52 10.1016/j.vaccine.2008.04.012 (doi:10.1016/j.vaccine.2008.04.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandon S., Mackinnon M., Nee S., Read A. 2001. Imperfect vaccines and the evolution of pathogen virulence. Nature 414, 751–756 10.1038/414751a (doi:10.1038/414751a) [DOI] [PubMed] [Google Scholar]
- 30.Ganusov V. V., Antia R. 2006. Imperfect vaccines and the evolution of pathogens causing acute infections in vertebrates. Evolution 60, 957–969 10.1554/05-504.1 (doi:10.1554/05-504.1) [DOI] [PubMed] [Google Scholar]
- 31.Brown M. J. F., Schmid-Hempel R., Schmid-Hempel P. 2003. Strong context-dependent virulence in a host–parasite system: reconciling genetic evidence with theory. J. Anim. Ecol. 72, 994–1002 10.1046/j.1365-2656.2003.00770.x (doi:10.1046/j.1365-2656.2003.00770.x) [DOI] [Google Scholar]
- 32.van Baalen M., Sabelis M. W. 1995. The dynamics of multiple infection and the evolution of virulence. Am. Nat. 146, 881–910 10.1086/285830 (doi:10.1086/285830) [DOI] [Google Scholar]
- 33.Frank S. A. 1996. Models of parasite virulence. Q. Rev. Biol. 71, 37–78 10.1086/419267 (doi:10.1086/419267) [DOI] [PubMed] [Google Scholar]
- 34.Alizon S., van Baalen M. 2008. Multiple infections, immune dynamics, and the evolution of virulence. Am. Nat. 172, E150–E168 10.1086/590958 (doi:10.1086/590958) [DOI] [PubMed] [Google Scholar]
- 35.Gandon S., van Baalen M., Jansen V. A. 2002. The evolution of parasite virulence, superinfection, and host resistance. Am. Nat. 159, 658–669 10.1086/339993 (doi:10.1086/339993) [DOI] [PubMed] [Google Scholar]
- 36.Imhoof B., Schmid-Hempel P. 1998. Single-clone and mixed-clone infections versus host environment in Crithidia bombi infecting bumblebees. Parasitology 117, 331–336 10.1017/S0031182098003138 (doi:10.1017/S0031182098003138) [DOI] [PubMed] [Google Scholar]
- 37.Beltran S., Gourbal B., Boissier J., Duval D., Kieffer-Jaquinod S., Pierce R. J., Grunau C., Theron A., Mitta G. 2011. Vertebrate host protective immunity drives genetic diversity and antigenic polymorphism in Schistosoma mansoni. J. Evol. Biol. 24, 554–572 10.1111/j.1420-9101.2010.02190.x (doi:10.1111/j.1420-9101.2010.02190.x) [DOI] [PubMed] [Google Scholar]
- 38.McKeever D. 2009. Bovine immunity: a driver for diversity in Theileria parasites? Trends Parasitol. 25, 269–276 10.1016/j.pt.2009.03.005 (doi:10.1016/j.pt.2009.03.005) [DOI] [PubMed] [Google Scholar]