Abstract
Background:
The adult female sand flies (Diptera: Psychodidae) of the subgenus Larroussius are important vectors of Leishmania infantum (Kinetoplastida: Tripanosomatidae) in Meshkinshahr district, Northwest of Iran. Four Phlebotomus (Larroussius) species are present in this area, i.e. Phlebotomus (Larroussius) kandelakii, P. (La.) major, P. (La.) perfiliewi and P. (La.) tobbi. The objective of the present study was to identify and distinguish the females of P. perfiliewi, P. major and P. tobbi, in this district.
Methods:
Adult sand flies were collected with sticky papers, CDC light traps, and aspirator in 2006. Individual sand flies of this four species from thirty different locations were characterized morphologically and by comparative DNA sequences analyses of a fragment of mitochondrial gene Cytochrome b (Cyt b) and nuclear gene Elongation Factor 1-alpha (EF-1α). PCR amplification was carried out for all three species P. major, P. perfiliewi and P. tobbi in the subgenus Larroussius.
Results:
Phylogenetic analyses of P. major populations in this study displayed two different populations and genetic diversity. Spermathecal segment number, pharyngeal armature and other morphological characters of these three species were examined and found to present consistent interspecific differences.
Conclusion:
According to our findings, the phylogeny of Cyt b and EF-1α haplotypes confirms the relationships between P. major, P. tobbi and P. perfiliewi as already defined by their morphological similarities.
Keywords: Phlebotomus, Larroussius, Cytochrome b, Elongation Factor-1α, Morphology, Iran
Introduction
The blood-feeding female of phlebotomine sand flies (Diptera: Psychodidae, Phlebotominae) include natural vectors of protozoa of the genus Leishmania (Kinetoplastida: Trypanosomatidae), which are the parasitic causative agents of mammalian leishmaniasis (Killick-kendrick 1990). There are approximately 700 species of phlebotominae sand flies divided among 6 genera, of which only two, i.e. Phlebotomus in the old world (OW) and Lutzomyia in the new world (NW) are medically importance (Lane 1987, Lewis 1982, Lane 1993, Sharma and Singh 2008).
Only 10% of these Phlebotominae sand flies act as disease vector. Further, only 30 species of these are important from public health point (Sharma and Singh 2008). A total of about 21 Leishmania spp. have been identified to be pathogenic to human (Singh 2006). However, recent studies based on molecular data have provided evidence that the genus Phlebotomus is not monophyletic (Depaquit et al. 1998). Furthermore, Rispail and Leger (1998a) have recently revised the definition of morphological characters states. In the Mediterranean Region, species of the subgenus Larroussius are the main vectors of Leishmania infantum, which cause Visceral Leishmaniasis (VL) in humans and dogs (Killick-kenrick 1990, Guernaoui et al. 2005). The subgenus is readily identified by the development long extension of the spermathecal neck in females (Rispail 1990).
Description of the subgenus Larroussius created by Nitzulescu (1931) with P. major Annandale, 1910 as the type species (Perfiliev 1968, Lewis 1982).
Among 27 species of Phlebotominae sand flies in the OW (Artemiev and Neronov 1984, Rispail and Leger 1998a, Rispail and Leger 1998b, Secombe et al. 1993), at least 12 species of the subgenus Larroussius are proven or probable vectors of leishmaniasis (Killick-kendrick 1990). The subgenus Larroussius is closely related to the subgenera Transphlebotomus and Adlerius, and the proliferation of species within Larroussius and Adlerius is probably recent (Rispail and Leger 1998b, Rispail and Leger 1991).
Although, identification of male specimens of Larroussius are not very difficult, but determination of some females of this subgenus has been considered impossible based on morphological characters.
Leger et al. (1983) showed that three sympatric species of Larroussius in Greece could readily be separated by the morphology of base of the spermathecal ducts.
Killick-Kendrick et al. (1991) separated 13 species of Larroussius by the morphology of the base of the spermathecal ducts. Recent studies showed that molecular tools could help resolve phylogenetic relationships between species of subgenus Larroussius (Esseghir et al. 1997, Esseghir et al. 2000, Muccio et al. 2000, Pesson et al. 2004, Parvizi and Assmar 2007).
There are four species of subgenus Larroussius including P. kandelakii, P. perfiliewi, P. major and P. tobbi in Meshkinshahr district, north western Iran. The first species is a proven vector and the second is the probable vector of viscerotropic Leishmania spp. in this area (Rassi and Javadian, 1998, Rassi and Javadian 1999, Rassi et al. 2001a, Rassi et al. 2001b, Rassi et al. 2005).
The objective of the present study was to identify and distinguish the females of P. perfiliewi, P. major and P. tobbi. They are similar in morphological characters and indistinguishable. We analyzed the sequences of cytochrome b (mtDNA) and elongation factor-1α (nDNA) genes of these three species and compared with morphological characteristics. Complete phylogenetic information is available for these genes and they are useful for the study of molecular systematic and speciation.
Material and methods
Study area
Meshkinshahr District (48° 17′ N, 38° 15′ E) is located at 1890 masl in Ardebil Province, Iran. The district occupies the northern foothills of the Sabalan Mountains, which rise to an altitude 4881 masl. Temperature varies from −27 °C in winter to 41 °C in summer. The human population was 156141 in 2006 and the principal economic activity is sheep farming.
Sand fly sampling
Sand fly sampling was carried out from Jun–October 2006 (during the period of peak activity), in 30 villages distributed throughout the Meshkinshahr District. Sand flies were collected once every 15 d from indoor habitats (bedroom, stable, toilet, bath room, hen nest, hay loft and store room) and outdoor habitats (yard, rodent burrow, stone and wall crevices, fox burrows and dog kennel, rubble and riverbanks) using sticky papers, CDC light trap and Aspirator. Sand fly specimens were stored in 96% ethanol.
Dissection, Mounting and Morphological identification
After recording the sampling data and locations, sand fly specimens were washed in 1% detergent then twice in sterile distilled water. Each specimen was then dissected in fresh drop of sterile normal saline by cutting off the head and abdominal terminalia with sterilized forceps and single used mounted needles. The remainder of the body was stored in the sterile eppendorf microtubes.
Specimens were mounted in Puri’s or Berlese’s medium and identified using the identification keys of Theodor and Mesghali (1964), Perfiliev (1968), and Lewis (1982). Morphological characters, which used in this study, included pharyngeal armature, spermathecal segments number, length of spermathecal neck, palpal and ascoids formula. First and second are more important characters.
DNA extraction
DNA was extracted from the dissected thorax and attached anterior abdomen of individual sand flies using the method of Ish-Horowiz (Ready et al. 1991). In the 1.5 ml microtubes, the thorax plus anterior abdomen of each sand fly was frozen and defrosted twice to break up tissue using a sampler tips or pestel, with grinding mix. Then SDS mix was used to denature proteins associated with the DNA, then ice cold 8M KOAc was added to remove effectively the SDS proteins from solution. Cell debris and proteins were separated from the DNA by centrifugation then the DNA in the supernatant was precipitated over night at −20 °C in 96% ethanol. Following ethanol precipitation, the DNA was dissolved in 15μl 1X TE (10mM Tris-HCl, 1mM EDTA pH= 8.0) and stored at −20 °C.
PCR amplification of Cyt b and EF-1α
For Cyt b one pair primers were used. CB3-FC (forward) (5′-CA(C/T) ATTCAA-CC (A/T)GAATGATA-3′) with CB-R06 (reverse) (5′-TATCTAATGGTTTCAAAACA ATTGC-3′) to amplify an overlapping 3′ fragment of 499 bp without primers (CB3 fragment). Also for EF-1α one pair primers were used, EF-F05 (forward) (5′-CCTGG ACAT-CGTGATTTCAT-3′) with EF-F08 (reverse) (5′-CCACCAATCTTGTAGACATCCTG-3′) to amplify of 454 bp without primers.
The PCR reaction conditions were identical for both Cyt b and EF-1α. 2μl 10X PCR buffer, 1.2 μl MgCl2, 0.15μl primers (F and R), 2μl dNTPs, 1.5μl DNA with the reaction volume completed to 20 μl by distilled water, followed by initial denaturation of Cyt b at 94 °C for 3 min. PCR consisted of 35 cycles of denaturation at 94 °C for 30 sec, annealing1 at 40 °C for 30 sec, annealing 2 at 44 °C for 30 sec, extension at 72 °C for 90 sec and then final extension at 72 °C for 10 min. For EF-1α, initial denaturation at 94 °C for 3 min. PCR consisted of 35 cycles of denaturation at 94 °C for 30 sec, annealing at 48°C for 30 sec, extension at 72 °C for 30 sec and then final extension at 72 °C for 10 min.
Direct sequencing of PCR products
One hundred nanograms of each purified DNA sample was cycle-sequenced using an ABI Parsim® Big Dye™ Terminator cycle sequencing Ready Reaction Kit (version 2.0) and ABI 373/377 sequencing systems (ABI, PE Applied Biosystems), with 3.2 Pmol of the same primers that were used for PCR.
Aligning and phylogenetic analysis of DNA sequences
DNA sequences from both strands were aligned and edited using Sequencher Demo 4.7 and BioEdit softwares. Multiple or pairwise sequence alignments of DNA were used with CLUSTAL W PPC: Clustalw version 1.7. Phylogenetic analyses were done using Parsimony PAUP. Relationships were inferred based on genetic distances using the Neighbor Joining (NJ) option with default settings.
Results
Species composition of subgenus Larroussius
Out of the 1743 sand fly specimens collected, 660 specimens (37.9%) belonged to the subgenus Larroussius, including: Phlebotomus (La.) kandelakii (31%), P. (La.) major (1.5%), P. (La.) tobbi (1.5%), P. (La.) perfiliewi (1.7%) and P. (La.) major group (2.1%).
Aligning and phylogenetic analysis of Larroussius DNA sequences
PCR amplification of Cyt b and EF-1α was successfully achieved for all 3 species of the subgenus Larroussius.
Seven sequences for Cyt b and four for EF-1α were compared for P. major. Comparison of pairwise genetic similarity or score of sequences showed 87%–100% similarity for Cyt b and 96%–100% similarity for EF-1α. Also 2.7% genetic diversity for Cyt b and 2.8% for EF-1α was observed.
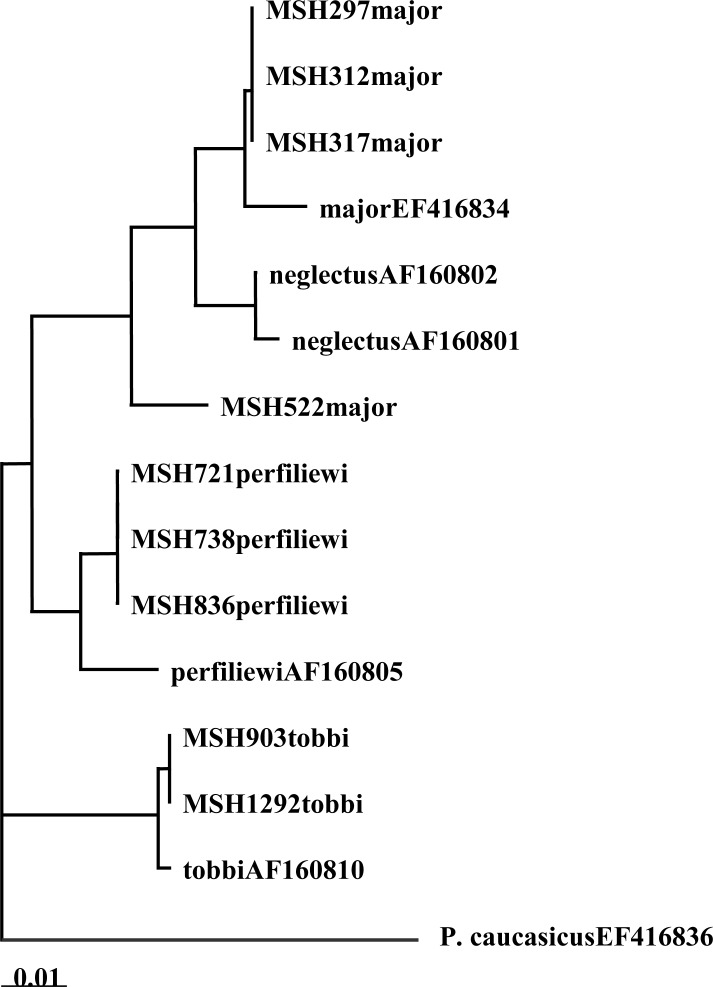
There were two haplotypes for Cyt b from seven sequences (Table 1) and for EF-1α, there were two haplotypes within four sequences (Table 2). The Neighbor-joining (NJ) phylogram for Cyt b showed two lineages, and each of these had subgroups with short branches. One of the lineages had one haplotype from sand flies from the same habitat (Stone crevices) but different locations (Ur kandi, Mueel, Agh daragh). The second lineage had one haplotype from sand flies from different habitats (Rodent burrow and Fox burrows) and locations (Ghurt tappeh, Alni, Niaz suee) (Fig. 1). In addition, phylogenetic tree for EF-1α showed two lineages and only one haplotype had subgroups. One of the lineages had one haplotype from sand flies from the same location (Ur kandi) but different habitats (Stone crevices, bedroom, and hen nest). The second lineage had one haplotype from one specimen in rodent burrow (Fig. 2).
Table 1.
Sequences comparison of Cyt b-mtDNA gene in the subgenus Larroussius
| Species | Total Sequences | Pairwise genetic similarity (%) | Total aplotype | Unique haplotypes | Genetic diversity (%) |
|---|---|---|---|---|---|
| P. major | 7 | 87 – 100 | 2 | 0 | 2.7 |
| P. tobbi | 7 | 100 | 1 | 0 | 0.3 |
| P. perfiliewi | 6 | 100 | 1 | 0 | 0 |
Table 2.
Sequences comparison of EF-1α-nDNA gene in the subgenus Larroussius
| Species | Total Sequences | Pairwise genetic similarity (%) | Total aplotype | Unique haplotypes | Genetic diversity (%) |
|---|---|---|---|---|---|
| P. major | 4 | 96–100 | 2 | 0 | 2.8 |
| P. tobbi | 3 | 100 | 1 | 0 | 0 |
| P. perfiliewi | 3 | 100 | 1 | 0 | 0 |
Fig. 1.
Neighbor-joining phylogenetic tree for DNA haplotypes of Cyt b (mtDNA) of the subgenus Larroussius sand flies species
Fig. 2.
Neighbor-joining phylogenetic tree for DNA haplotypes of EF-1α (nDNA) of the subgenus Larroussius sand flies species
Seven sequences for Cyt b and three for EF-1α were compared for P. tobbi. Comparison of pairwise genetic similarity of sequences indicated the 100% similarity for both Cyt b and EF-1α sequences. Genetic diversity and unique haplotype for Cyt b and EF-1α were not observed. For Cyt b and EF-1α one haplotype was obtained (Table 1 and 2). The Neighbor-joining (NJ) phylogram for both Cyt b and EF-1α showed one lineage for all specimens in the same location and habitat (Fig. 1 and 2).
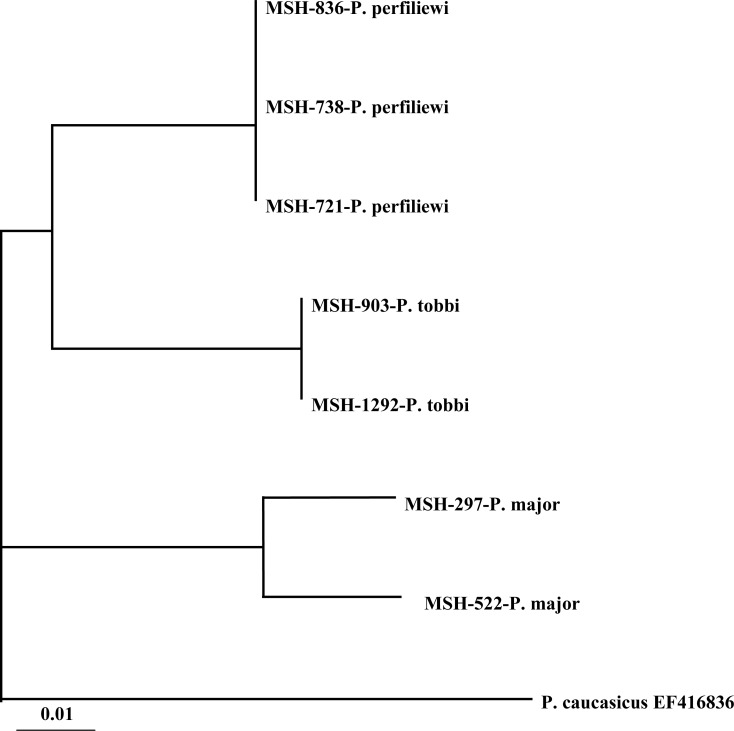
Six sequences for Cyt b and three for EF-1α were compared for P. perfiliewi. Comparison of pair wise genetic similarity of sequences indicated 100% similarity for both Cyt b and EF-1α sequences. In this species genetic diversity for Cyt b and EF-1α were not observed (Table 1 and 2). Phylogenetic tree for both Cyt b and EF-1α showed one lineage for all specimens in the different location and habitats (Fig. 1 and 2). The Neighbor-joining (NJ) phylogram in combination of Cyt b and EF-1α for seven specimens showed two lineages for P. perfiliewi and P. tobbi and one lineage for P. major (Fig. 3).
Fig. 3.
Neighbor-joining phylogenetic tree for DNA haplotypes of combination of Cyt b and EF-1α of the subgenus Larroussius sand flies species.
Identification of the female Larroussius species using morphological characters
In the present study, the morphological characteristics of the three species female of Larroussius1647 were described as follows:
Phlebotomus major and Phlebotomus neglectus
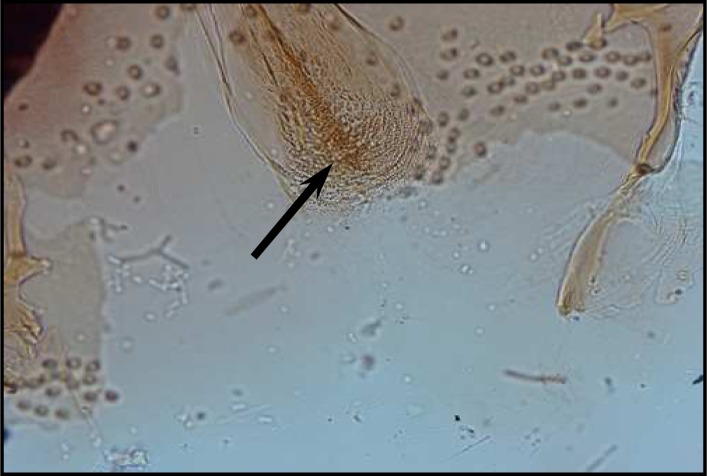
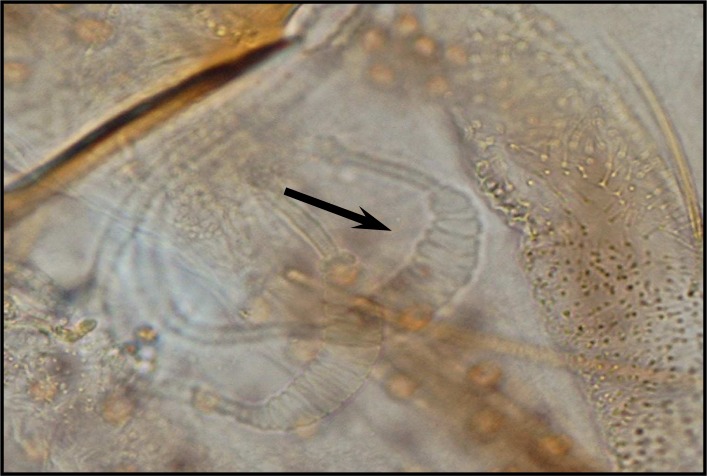
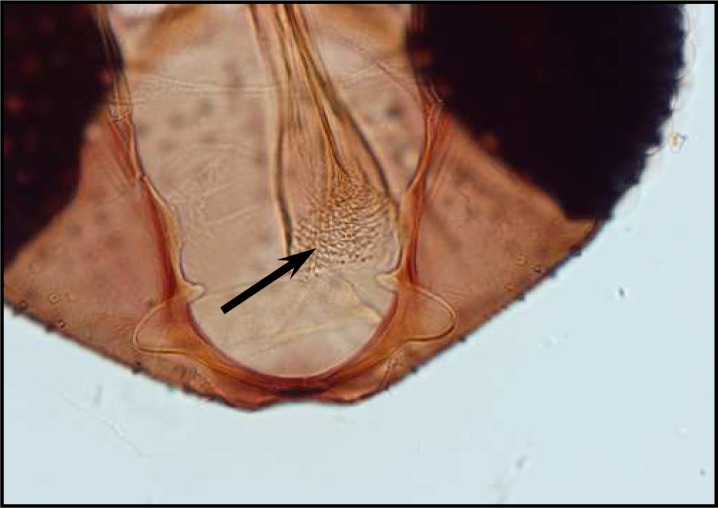
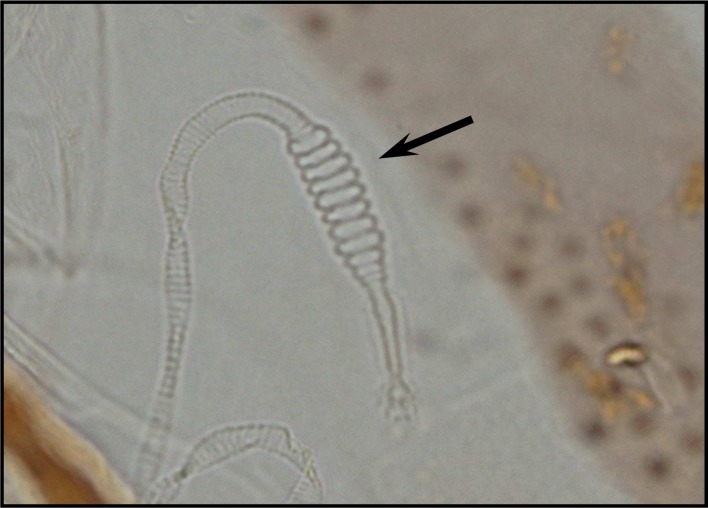
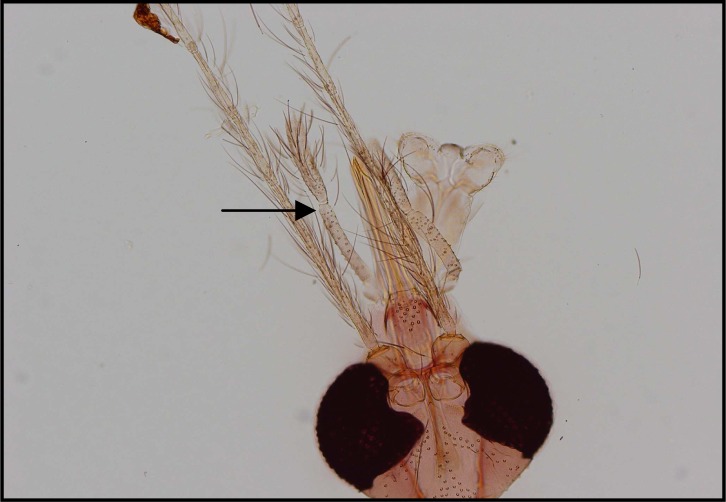
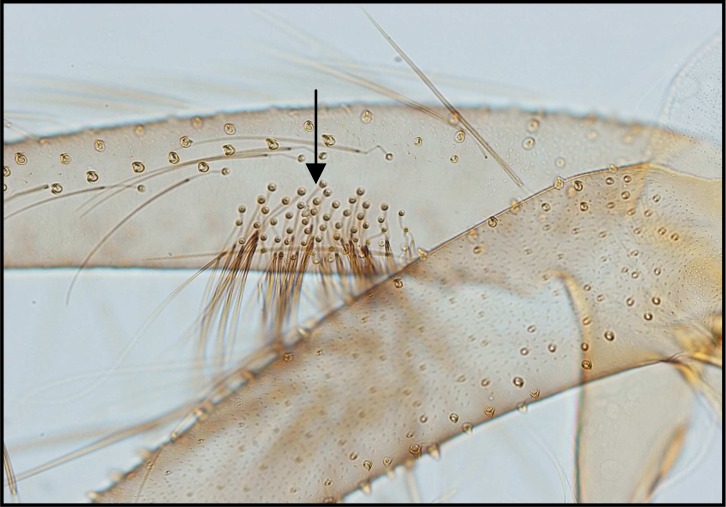
Palpal formula: 1, 4, 2, 3, 5 and the formula of ascoids: 2/3–8, 1/9–5. Pharyngeal armature has occupied ⅓ of pharynx space and punctiform, arranged in several rows and anterior elements have serrated margin (Fig .4). Spermatheca with 14–16 segments and length of spermatheca neck ⅔ of spermatheca capsule (Fig. 5).
Fig. 4.
Pharyngeal armature of P. major
Fig. 5.
Spermatheca of P. major
Phlebotomus tobbi
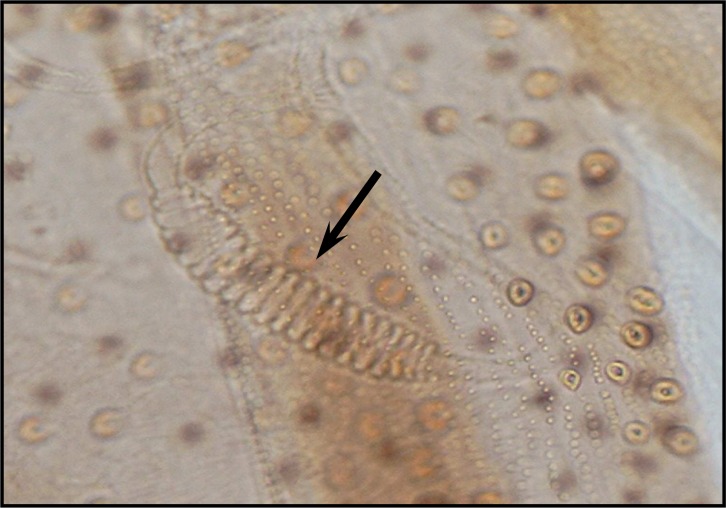
Palpal formula: 1, 4, 2, 3, 5 or 1, (2, 4), 3, 5 and the formula of ascoids: 2/3–8, 1/9–15. Pharyngeal armature has occupied over ½ of pharynx space and punctiform, arranged in several concave irregular rows of large dots and anterior part of pharynx have a fine dots (Fig. 6). Spermatheca with 11–13 segments. Length of spermatheca neck as long as spermatheca capsule (Fig. 7).
Fig. 6.
Pharyngeal armature of P. tobbi
Fig. 7.
Spermatheca of P. tobbi
Phlebotomus perfiliewi
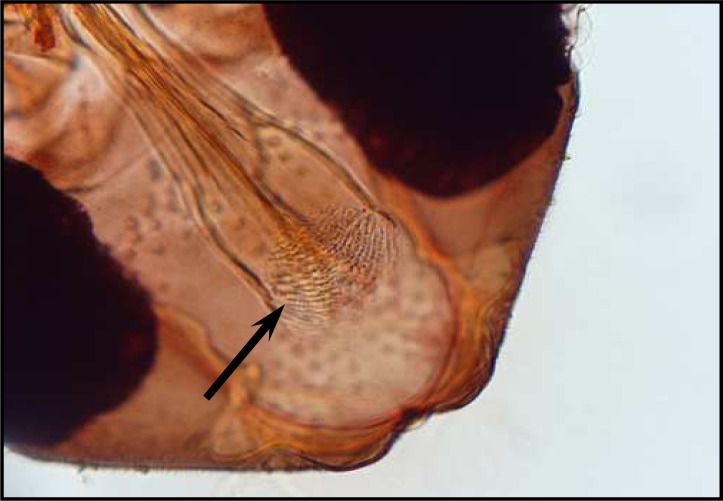
Palpal formula: 1, 4, 2, 3, 5 and the formula of ascoids: 2/3–9, 1/10–15. Pharyngeal armature has occupied over ½ of pharynx space and punctiform, arranged in several concave regular rows of large dots (Fig. 8). Spermatheca with 17–20 segments. Length of spermatheca neck ½ of spermatheca capsule (Fig. 9).
Fig. 8.
Pharyngeal armature of P.perfiliewi
Fig. 9.
Spermatheca of P.perfiliewi
Identification of the male P. major and P. neglectus using morphological characters
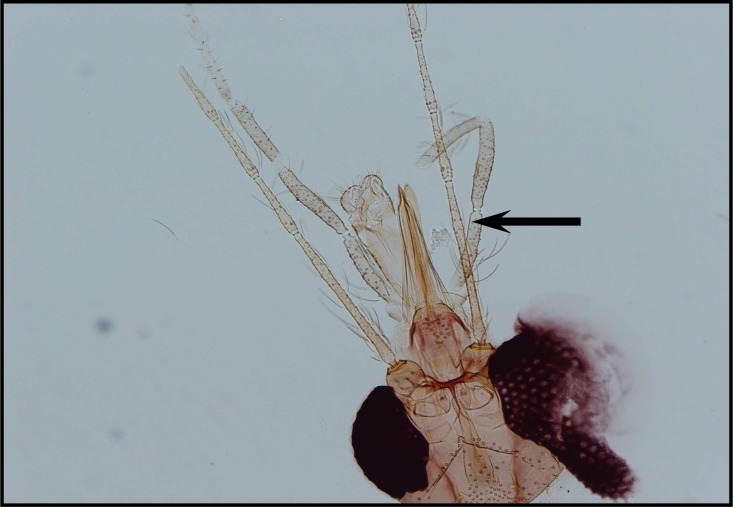
Among 26 male specimens in this species 17 specimens of P. major (65.4%) with 20–30 ventrally directed and long hairs stand densely on coxite and Palpal formula 1, 4, (2, 3), 5 (Fig. 10 and 11). In 9 specimens of P.neglectus (34.6%) with less than 20 ventrally directed hairs, widely spaced and sparser and Paplal formula 1, 4, 2, 3, 5 (Fig. 12 and 13).
Fig. 10.
Palpal formula of P.major
Fig. 11.
Coxite hairs of P.major
Fig. 12.
Palpal formula of P.neglectus
Fig. 13.
Coxite hairs of P.neglectus
Discussion
According to the results of previous studies, the vectors of viscerotropic Leishmania spp. in OW mainly belong to the subgenera Larroussius, Adlerius, Euphlebotomus, Synphlebotomus and Paraphlebotomus. In the subgenus Larroussius, P.perniciosus in Algeria, France, Italy, Malta, Spain, P.ariasi in France, Spain, Italy, P. perfiliewi in Italy, East Mediterranean, North of Africa, Greece, Azerbaydzhan, Tunisia, P. tobbi in Cyprus, East Mediterranean, Sicil, P. kandelakii in Afghanistan, Russia, and P. neglectus in Greece, Albania, Portugal (Killick-kendrick 1990). In North West of Iran P. kandelakii and P. perfiliewi transcaucasicus are proven and probable vector of viscerotropic Leishmania spp. (Rassi et al. 2005).
According to the finding of the present investigations, the phylogeny of Cyt b and EF-1α haplotypes confirms the morphological relationships among the three species P. perfiliewi, P. major and P. tobbi in the subgenus Larroussius. The males of these three species have a several diagnostic morphological characters, whereas the females of these species show very similarities in morphology of spermathecal segment number, pharyngeal armature and other characteristics.
Although morphological characteristics are the most practical methods for species distinguishing, new molecular techniques are very useful to resolve problems of identification in the cases with morphologically similarities.
Access on the genetic diversity and molecular systematic of the Larroussius sand flies species not only useful to find the taxonomic status of them, but also indicates the ecological and geographical differences.
In our analysis, all of the Cyt b and EF-1α sequences, the monophyly of the subgenus Larroussius was confirmed, in concordance with the morphologically and molecularly based phylogenies of Rispail and Leger (1991, 1998b) and Esseghir et al. (1997). All of the branches of the parsimony and distances trees had strong support and showed identical relationships and indicated the validity of many of characters in inferring evolutionary relationships. The trees were also topologically similar to the parsimony tree of Esseghir et al. (1997).
On morphological characteristics, the present study confirmed the observations of Perfiliev (1968), Lewis (1982), Leger et al. (1983), and Killick-Kendrick et al. (1991).
Phylogenetic analyses of P. major populations showed 2 lineages in different locations such as rodents burrow and stone crevices (Table 3).
Table 3.
Collected sand flies of the subgenus Larrossius in different location and habitats in Meshkinshahr District, Iran
| Species | Code No. | Sex | Location of capture | Altitude of capture (masl) | Habitat | Date of capture | Method of catching | Type of Geue | Geubank accession number |
|---|---|---|---|---|---|---|---|---|---|
| P.major | MSH-1201 | Male | Agh daragh | 1390 | Stone crevices | 05.08.06 | SP | Cyt b | GQ169331 |
| P.major | MSH-297 | Male | Ur kandi | 1551 | Stone crevices | 09.07.06 | SP | Cyt b | GQ169332 |
| P.major | MSH-619 | Male | Mueel | 2780 | Stone crevices | 10.07.06 | SP | Cyt b | GQ169333 |
| P.major | MSH-861 | Male | Niaz suee | 1080 | Fox burrows | 22.07.06 | SP | Cyt b | GQ169334 |
| P.major | MSH-007 | Female | Alni | 1267 | Fox burrows | 16.06.06 | SP | Cyt b | GQ169335 |
| P.major | MSH-052 | Male | Ghurt tappeh | 1547 | Rodent burrow | 17.06.06 | SP | Cyt b | GQ169336 |
| P.major | MSH-1397 | Male | Mueel | 2780 | Stone crevices | 07.08.06 | SP | Cyt b | GQ169337 |
| P. neglectus (GenBank) | Cyt b | AF161191 | |||||||
| P. neglectus (GenBank) | Cyt b | AF161193 | |||||||
| P.perfiliem | MSH-721 | Male | Dushan lu | 1209 | Rodent burrow | 22.07.06 | SP | Cyt b | GQ169338 |
| P.perfiliewi | MSH-738 | Male | Dushan lu | 1209 | Rodent burrow | 22.07.06 | SP | Cyt b | GQ169339 |
| P.perfiliewi | MSH-756 | Female | Dushan lu | 1209 | Yard | 22.07.06 | CDC | Cyt b | GQ169340 |
| P.perfiliewi | MSH-795 | Female | Niaz suee | 1080 | Yard | 22.07.06 | CDC | Cyt b | GQ169341 |
| P.perfiliewi | MSH-812 | Female | Niaz suee | 1080 | Yard | 22.07.06 | CDC | Cyt b | GQ169342 |
| P.perfiliewi | MSH-836 | Male | Niaz suee | 1080 | Stone crevices | 22.07.06 | SP | Cyt b | GQ169343 |
| P.perfiliewi (GenBauk) | Cyt b | AF161201 | |||||||
| P.tobbi | MSH-480 | Male | Ghurt tappeh | 1547 | Rodent burrow | 09.07.06 | SP | Cyt b | GQ169344 |
| P.tobbi | MSH-551 | Male | Ghurt tappeh | 1547 | Stone crevices | 09.07.06 | SP | Cyt b | GQ169345 |
| P.tobbi | MSH-903 | Male | Ghurt tappeh | 1547 | Stone crevices | 23.07.06 | SP | Cyt b | GQ169346 |
| PJobbi | MSH-1292 | Male | Ghurt tappeh | 1547 | Stone crevices | 06.08.06 | SP | Cyt b | GQ169347 |
| P.tobbi | MSH-1481 | Male | Ghurt tappeh | 1547 | Stone crevices | 20.08.06 | SP | Cyt b | GQ169348 |
| P.tobbi | MSH-1657 | Male | Ghurt tappeh | 1547 | Stone crevices | 03.09.06 | SP | Cyt b | GQ169349 |
| P.tobbi | MSH-899 | Female | Ghurt tappeh | 1547 | Stone crevices | 23.07.06 | SP | Cyt b | GQ169350 |
| P,tobbi (GenBank) | Cyt b | AF161212 | |||||||
| P.tobbi (GenBank) | Cyt b | AF161210 | |||||||
| P.caucasicus (GenBank/Out group) | Cyt b | FJ217390 | |||||||
| P.sergenti (GenBank/Out group) | Cyt b | EU980367 | |||||||
| P.major | MSH-297 | Male | Ur kandi | 1551 | Stone crevices | 09.07.06 | SP | EF-1α | GQ169351 |
| P.major | MSH-312 | Male | Ur kandi | 1551 | Bed room | 09.07.06 | ASP | EF-1α | GQ169352 |
| P.major | MSH-317 | Male | Ur kandi | 1551 | Hen nest | 09.07.06 | SP | EF-1α | GQ169353 |
| P. major | MSH-522 | Male | Ghurt tappeh | 1547 | Rodent burrow | 09 07 06 | SP | EF-1α | GQ169354 |
| P. major (GenBauk) | EF-1α | EF416834 | |||||||
| P.neglectus (GenBank) | EF-1α | AF160802 | |||||||
| P.neglectus (GenBank) | EF-1α | AF160801 | |||||||
| P.perfiliewi | MSH-721 | Male | Dushan lu | 1209 | Rodent burrow | 22.07.06 | SP | EF-1α | GQ169355 |
| P.perfiliewi | MSH-738 | Male | Dushan lu | 1209 | Rodent burrow | 22.07.06 | SP | EF-1α | GQ169356 |
| P.perfiliewi | MSH-836 | Male | Niaz suee | 1080 | Stone crevices | 22.07.06 | SP | EF-1α | GQ169357 |
| P.perfiliewi (GenBauk) | EF-1α | AF160805 | |||||||
| P.tobbi | MSH-903 | Male | Ghurt tappeh | 1547 | Stone crevices | 23.07.06 | SP | EF-1α | GQ169358 |
| P.tobbi | MSH-1292 | Male | Ghurt tappeh | 1547 | Stone crevices | 06.08.06 | SP | EF-1α | GQ169359 |
| P.tobbi | MSH-1481 | Male | Ghurt tappeh | 1547 | Stone crevices | 20.08.06 | SP | EF-1α | GQ169360 |
| P.tobbi (GenBank) | EF-1α | AF160810 | |||||||
| P.caucasicus (GenBank/Out group) | EF-1α | EF416836 | |||||||
SP = Sticky paper ; CDC = CDC Light trap ; Asp = Aspirator
Cyt b = Cytochrome b ; EF-lα = Elongation Factor 1-α
According to our molecular and morphological finding, it seems that there are both P. major and P. neglectus in Meshkinshahr district. This is the first report of P. neglectus in this area as well as in Iran (Table 3)
Further studies needs to resolve the taxonomic status of this species. P. major has many geographical variants the females of which have conventionally been distinguished by differences in pharyngeal armature. Adler (1933) stressed the tendency of P. major to form geographical races. He showed (1946) that they differ mainly in the form of the pharyngeal armature of the females. He also mentioned ecological differences between the races and stresses that some races may have a different importance in the epidemiology of VL.
Our study on phylogenetic analyses of P. tobbi populations showed one lineage with single haplotype for either cytB or EF-1α gene. P. tobbi was first found in Iran and Israel, and was described as a variety of P. perniciosus. Parrot (1934) reported that it is a single species with the name of P. tobbi. This was accepted also by Theodor (1948).
Phlebotomus tobbi is mainly found in burrows and is rather common in Transcaucasia and it seems to be identical with the sand flies from Iran and southwest Asia. Therefore, P. tobbi is considered a separate species.
Phlebotomus perfiliewi is the probable vector of VL in the north west of Iran. In the present study, this species had one lineage with a single haplotype. Phylogenetic tree retrieved from cytB or combination of cytB and EF-1α showed close relationship of P. perfiliewi with P. tobbi.
Perfiliev (1966) recognized three subspecies of P. perfiliewi (perfiliewi, galilaeus and transcaucasicus) which were distinguished by minor morphological differences in the aedeagus of the male. Lewis (1982) listed the same three subspecies but commented that the distinction of the male galilaeus from transcaucasicus uncertain. Artemiev and Neronov (1984) raised galilaeus and transcaucasicus to level species. The spermatheca of galilaeus and perfiliewi are indistinguishable, suggesting that these allopatric sand flies are taxonomically very close and are perhaps two subspecies (Lewis, 1982).
To find an exact distinction and identification of the female species of Larroussius, needs more investigations in different parts of the world as well as Iran.
Acknowledgments
The author would like to appreciate very much for kind collaboration of Health of Center staffs of Meshkin-shahr district. This study was M.Sc thesis and financially was supported by the School of Public Health. Tehran University of Medical Science: Project No.: 240. 6129.
References
- Artemiev MM, Neronov VM. Distribution and ecology of sandflies of the old world (genus Phlebotomus) Moscow: Instituteof Evolution, Morphology and Ecology of Animals; 1984. p. 207. [in Russian] [Google Scholar]
- Depaquit J, Perrotey S, Lecintre G, Tillier S, Ferte H, Kaltenbach M, Leger N. Systematique moleculaire des Phlebotominae: etude pilote. Paraphylie du genre Phlebotomus. 1998;321:849–855. doi: 10.1016/s0764-4469(99)80025-0. [DOI] [PubMed] [Google Scholar]
- Esseghir S, Ready PD, Killick-kendrick R, Ben-Ismail R. Mitochondrial haplotypes and geographical vicariance of Phlebotomus vectors of Leishmania major. Insect Mol Biol. 1997;6:211–225. doi: 10.1046/j.1365-2583.1997.00175.x. [DOI] [PubMed] [Google Scholar]
- Esseghir S, Ready PD, Killick-kendrick R, Ben-Ismail R. Speciation of Phlebotomus Sandflies of the subgenus Larroussius coincided with the late Miocence-Pliocence aridification of the Mediterranean subregion. Biol J Linn Soc. 2000;70:189–219. [Google Scholar]
- Guernaoui S, Pesson B, Boumezzough A, Pichon G. Distribution of Phlebotomine Sandflies, of the subgenus Larroussius, in Morocco. Med Vet Entomol. 2005;19:111–115. doi: 10.1111/j.0269-283X.2004.00548.x. [DOI] [PubMed] [Google Scholar]
- Killick-kendrick R. Phlebotomine vectors of leishmaniasis: a review. Med Vet Entomol. 1990;4:1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Killick-kendrick R, Tang Y, Killick-kendrick M, Sang DK, Sirdar MK, Ke L, et al. The identification of female sandflies, of the subgenus Larroussius by the morphology of the spermathecal ducts. Parasitologia. 1991;33(Suppl. 1):335–347. [PubMed] [Google Scholar]
- Killick-kendrick R. The biology and control of Phlebotominae Sandflies. Med Vet Entomol. 1999;17:279–289. [Google Scholar]
- Lane RP. Phlebotominae Sandflies, Family: Psychodidae, Subfamily Phlebotominae, Important genus, Phlebotomus (old word) and Lutzomyia (new world) In: Manson-Bahr PEC, Bell DR, editors. Manson’s Tropical Disease. 19th ed. Bailiere Tindall/WB Sanders; London: 1987. pp. 1395–1404. [Google Scholar]
- Lane RP. Sandflies (Phlebotominae) In: Lane RP, Crosskey RW, editors. Medical insects and Arachnids. Chapman and Hall; 1993. pp. 78–119. [Google Scholar]
- Leger N, Pesson B, Madulo-Leblond G, Abonnence E. Sur la differenciation des femelles du sous-genre Larroussius Nitzulescu, 1931. (Diptera-Phlebotomidae) de la region Mediterraneene Annales de Parasitologie Humaine et Comparee. 1983;58:611–623. [PubMed] [Google Scholar]
- Lewis DJ. A taxonomic review of the genus Phlebotomus. Bull Brit Mus Nat Hist Entomol. 1982;45:121–209. [Google Scholar]
- Muccio TD, Marinucci M, Frusteri L, Maroli M, Pesson B, Gramiccia M. Phylogenetic analysis of Phlebotomus species belonging to the subgenus Larroussius (Diptera, Psychodidae) by ITS2-rDNA sequences. Insect Mol Biol. 2000;30:387–393. doi: 10.1016/s0965-1748(00)00012-6. [DOI] [PubMed] [Google Scholar]
- Parvizi P, Assmar M. Nuclear Elongation Factor-1α Gene, A molecular marker for Iranian Sandfly identification. Iranian J Publ Health. 2007;36(2):25–37. [Google Scholar]
- Pefiliev PP. In: Fauna of U.S.S.R. Diptera: Phlebotomidae (Sandflies) Scientific translation, editor. Jerusalem: 1968. pp. 1–363. [Google Scholar]
- Pesson B, Ready JS, Benabdennbi I, Martin-Sanchez J, Esseghir S, Cadi-Soussi M, Morillas-Marquez F, Ready PD. Sandflies of the Phlebotomus perniciosus complex: mitochondrial introgression and a new sibling species of P. longicuspis in the Moroccan Rif. Med Vet Entomol. 2004;18:25–37. doi: 10.1111/j.0269-283x.2004.0471.x. [DOI] [PubMed] [Google Scholar]
- Rassi Y, Javadian E. Ecology of Phlebotomus kandelakii, the probable vector of visceral leishmaniasis (VL) in Northwest of Iran, IX International congress of Parasitology (ICOPA); 1998. Abstract book, O-0567. [Google Scholar]
- Rassi Y, Javadian E. Study on Phlebotomus perfiliewi, the probable vector of visceral leishmaniasis (VL) in Northwest of Iran (1995–1998), 3rd International symposium on Phlebotomine Sandflies; 23–27 August; Montpellier, France. 1999. p. 1. [Google Scholar]
- Rassi Y, Javadian E, Jalali A. Ecology of Phlebotomus kandelakii the main vector of visceral leishmaniasis in Northwest of Iran, 3rd International congress of Vector Ecology; September 2000.2001a. [Google Scholar]
- Rassi Y, Javadian E, Jalali A. Phlebotomus kandelakii, the main vector of visceral leishmaniasis in Northwest of Iran, World Leish 2; May 20–24; Greece. 2001b. Abstract book, P.242. [Google Scholar]
- Rassi Y, Javadian E, Nadim A, Zahraii AR, Vatandoost H, Moatazedian H, Azizi K, Mohebali M. Phlebotomus (Larroussius) kandelakii the principal and proven vector of visceral leishmaniasis in Northwest of Iran. Pakistan J Biol Sci. 2005;8(12):1802–1806. [Google Scholar]
- Ready PD, Lainson R, Shaw JJ, Souza AA. DNA probes for distinguishing Psychodopygus wellcomi from Psychodopygus complexus (Diptera: Psychodidae) Mem Inst Oswaldo Cruz. 1991;86(1):41–49. doi: 10.1590/s0074-02761991000100008. [DOI] [PubMed] [Google Scholar]
- Rispail P. Approche phenetique et cladistique du genre Phlebotomus Rondani and Berte, 1840 (Diptera: Psychodidae), Apport des caracteres morphologiques imaginaux. These de Doctorat, Universite de Sciences et Techniques du Languedoc; Montpellier, France: 1990. [Google Scholar]
- Rispail P, Leger N. Application of numerical taxonomic methods on Phlebotominae. In: Maroli M, editor. Proceeding of the first international symposium on Phlebotominae Sandflies. Parasitologia. Suppl.1. Vol. 33. 1991. pp. 485–492. [PubMed] [Google Scholar]
- Rispail P, Leger N. Numerical taxonomy of old world Phlebotominae (Diptera: Psychodidae). 1. Consideration of morphological characters in the genus Phlebotomus Rondani and Berte 1840. Mem Inst Oswaldo Cruz. 1998a;93(6):773–785. doi: 10.1590/s0074-02761998000600015. [DOI] [PubMed] [Google Scholar]
- Rispail P, Leger N. Numerical taxonomy of old world Phlebotominae (Diptera: Psychodidae). 2. Restatement of classification upon subgeneric morphological characters. Mem Inst Oswaldo Cruz. 1998b;93(6):787–793. doi: 10.1590/s0074-02761998000600016. [DOI] [PubMed] [Google Scholar]
- Secombe AK, Ready PD, Huddleston LM. A catalogye of the old world Phlebotominae Sandflies (Diptera, Psychodidae, Phlebotominae) Occasional papers on systemat entomol. 1993;8:1–57. [Google Scholar]
- Sharma U, Singh S. Insect vector of Leishmania: distribution, physiology and their control. J Vector Borne Dis. 2008;45:255–278. [PubMed] [Google Scholar]
- Singh S. New developments in diagnosis of leishmaniasis. Indian J Med Res. 2006;123:311–330. [PubMed] [Google Scholar]
- Theodor O. Classification of the old world species of the subfamily Phlebotominae (Diptera: Psychodidae) Bull Ent Res. 1948;39:85–115. doi: 10.1017/s0007485300024305. [DOI] [PubMed] [Google Scholar]
- Theodor O, Mesghali A. On the Phlebotominae of Iran. J Med Entomol. 1964;1(3):285–300. doi: 10.1093/jmedent/1.3.285. [DOI] [PubMed] [Google Scholar]