Abstract
The most important proteins involved in olfaction include odorant binding protein (OBP), chemosensory protein (CSP), olfactory receptor (OR), and gustatory receptor (GR). Despite that the exhaustive genomic analysis has revealed a large number of olfactory genes in a number of model insects, it is still poorly understood for most nonmodel species. This is mostly due to the reason that the small antenna is challenging for collection. We can generally isolate one or few genes at a time by means of the traditional method. Here, we present the large-scale identifying members of the main olfactory genes from the head of Tomicus yunnanensis using Illumina sequencing. In a single run, we obtained over 51.8 million raw reads. These reads were assembled into 57,142 unigenes. Nearly 29,384 of them were functionally annotated in the NCBI nonredundant database. By depth analysis of the data, 11 OBPs, 8 CSPs, 18 ORs, and 8 GRs were retrieved. Sequences encoding full length proteins were further characterised for one OBP and two CSPs. The obtained olfactory genes provide a major resource in further unraveling the molecular mechanisms of T. yunnanensis chemoperception. This study indicates that the next generation sequencing is an attractive approach for efficient identification of olfactory genes from insects, for which the genome sequence is unavailable.
1. Introduction
Olfaction plays a role in almost every aspect of insect life, which represents one of the key interfaces between insects and the environment. It is used by insects to recognize a huge variety of airborne molecules for providing them with information about food, predators, and potential mates [1]. Olfaction is a complex process requiring the interaction of numerous proteins to generate a neuronal signal. The first step in the recognition of chemical signals is the odorants that enter the insects' antennae and other sensory organs via pores and travel across the hydrophobic space to the chemosensory receptors, where the response of the insect to the signal is initiated [2]. Chemoreceptor of insects is mainly formed by the olfactory receptors (ORs) and gustatory receptors (GRs), which are located in the dendritic membrane of neurons [3, 4]. As common odorants are hydrophobic molecules [5], it is difficult for them passing the aqueous barrier of the sensillar lymph surrounding the dendrites of neuronal cells [6]. During the passing process, odorants are thought to be translocated from the air to the chemoreceptors by a variety of protein mainly existing in the sensillar lymph, including odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) [7–9]. Studying the genes that code for olfactory proteins could provide valuable insight into the molecular mechanisms of olfactory function.
Since the first discovery of insect OBP in the male antennae of the giant moth Antheraea polyphemus [7], a large number of olfactory genes have now been identified in numerous insect species through molecular cloning, cDNA library sequencing, and genome-wide analyses [10–12]. With regard to molecular cloning, olfactory genes are assigned to divergent gene families with relatively low sequence identity (overall 20% protein sequence identity), which results in difficulty in designing appropriate primers for successful cloning [13, 14]. To some extent, cDNA library sequencing is also unable for large scale identifying olfactory genes. This is partly due to the fact that it is difficult to collect enough sensory tissues that olfactory genes restrictedly expressed for constructing the tissue targeted cDNA library because of the tiny size of sensory tissues. In addition, sequencing of limited number of randomly selected cDNA clones often have insufficient coverage of less abundant transcripts [15]. Among the available insect olfactory genes, the majority of them have been obtained by means of bioinformatic approaches based on the characteristic features of the protein families from the completed genome sequences [9, 16].
Over the past several years, the next generation sequencing technology has emerged as a cutting edge approach for high-throughput sequence determination, promptly improved the efficiency and speed of gene discover, and provided fascinating opportunities in the life sciences with dramatically reduced time, labor, and cost in nonmodel organisms [17–19]. We here took advantage of this technology to present the global transcriptome characterization of the head of Tomicus yunnanensis, a serious pest of Pinus yunnanensis occurred in southwestern China [20], with focus on comprehensively revealing the olfactory genes using Illumina sequencing. In a single run, we identified 51,822,228 raw reads, which were assembled into 57,142 unigenes. From the transcriptome database, many genes encoding major olfactory proteins were mined. These data provide an invaluable resource as a first step for grounding future studies investigating chemosensory processes. Furthermore, this study presents the simple method to efficiently uncover the divergent olfactory gene family from tiny insects.
2. Materials and Methods
2.1. Insect
Adult beetles identified based on morphological characters [21] as T. yunnanensis by their trunk attacking phase on P. yunnanensis were collected from Qujing city, Yunnan province, China.
2.2. RNA Extraction and Sequencing
Head of both female and male adults were cut off and transferred to an Eppendorf tube. After grinding them under liquid nitrogen, total RNA was extracted by using TRIzol Reagent (Invitrogen) according to the manufacturer's protocol. Total RNA quantity and quality were assessed using the Agilent 2100 Bioanalyzer (Agilent technologies) with a minimum RNA integrated number value of 8. The samples for transcriptome analysis were prepared using Illumina's kit following manufacturer's recommendations. Briefly, mRNA was selected using oligo(dT) probes and then fragmented using divalent cations. cDNA was synthesized using random primers, modified and enriched for attachment to the Illumina flowcell, and then sequenced on the Illumina GA II platform. Raw data have been deposited in the NCBI Short Read Archive (SRA).
2.3. Bioinformatics
Prior to assembly, adaptor sequences, empty reads, and low quality sequences (reads with unknown sequences “N”) were filtered from the short raw reads. De novo assembly of the short reads was performed using SOAPdenovo [22] at default parameters. The generated unigenes were analyzed by searching the nonredundant (NR) database in NCBI with the BLASTx algorithm using an E-value cut-off of 10−5. Gene Orthology (GO) annotation was performed by Blast2GO [23] through a search of the NR database. Individual unigene with at least one match similar to olfactory genes was identified using custom databases for Blast search. ClustalX (version 1.83) [24] was used to conduct multiple sequence alignments. The presence and location of signal peptide cleavage sites in amino acid sequences was predicted by SignalP 4.0 server [25]. The software package MEGA5 [26] was used for phylogenetic analysis. Bootstrap analysis was performed using Neighbor-Joining and the Poisson correction model with 1000 replicates. Positions containing alignment gaps and missing data were eliminated with pairwise deletion.
2.4. PCR
To validate and extend fragments of several candidate gene sequences to encode full proteins, 5′ gene-specific primers (3′-TTCTCAATCACACACTTCAAGAAAC-5′ and 3′-TTGGTAGAATATTGGTCATCAGGTC-5′) were designed based on the sequences of Unigene41600_YNNT and Unigene26497_YNNT. They were used to clone the 5′ ends in conjunction with adapter primer provided in the SMART RACE cDNA amplification kit (Clontech). Two specific primer pairs (3′-ATGAAAACATTCGTGCTTGTTGCTT-5′ and 3′-TTACAATTTTAAACCTTCTTTTTCG-5′) were designed according to the sequence of Unigene55391_YNNT. They were employed for amplifying the open reading frame of this putative gene. cDNA template was synthesized with a SMART RACE cDNA amplification kit (Clontech) using the total RNA extracted above. PCR was performed using Advantage 2 Polymerase Mix (Clontech) according to recommended conditions and following the manufacturer's instructions.
3. Results and Discussion
3.1. Overview of the Head Transcriptome
After cleaning and quality checks, we generated over 4,600 million bases sequence information. The total number of reads was 51,822,228 with an average length of 90 bp (Table 1). The GC percentage of the reads is 42.53%. The number is comparable with other insect and eukaryote sequencing projects that have GC content between 38.7% and 56.5% [27]. Using SOAPdenovo, these reads were assembled into 42,678 contigs, 140,311 scaffolds, and 57,142 unigenes. The average length of contigs, scaffolds and unigenes were 113 bp, 226 bp, and 355 bp, respectively. The longest unigene had 4,762 bases. The lengths of the 5,136 unigenes were ≥700 bp (Figure 1).
Table 1.
Summary of Illumina sequencing of Tomicus yunnanensis head transcripts.
| Reads | Contigs | Scaffolds | Unigenes | |
|---|---|---|---|---|
| Number of sequences | 51,822,228 | 671,467 | 140,311 | 57,142 |
| Mean length (bp) | 90 | 113 | 226 | 355 |
| Total length (bp) | 4,664,000,520 | 75,947,228 | 31,643,690 | 20,291,442 |
Figure 1.
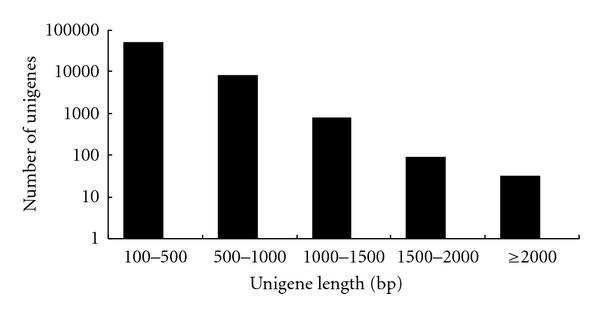
Length distribution of assembled unigenes. Note the logarithmic y-axis.
After assembly, all unigenes were aligned to NCBI NR databases with a cut-off E-value of 10−5 for annotation. 29,384 unigenes had at least one significant alignment to existing gene model in Blastx searches. The remaining 27,758 unigenes with no matches with any known sequences and likely represent novel. This might be due to the relatively short length of distinct gene sequences [17] and lack of genetic information in Tomicus. In addition, this might be resulted partly by the transcripts derived from the cDNA of untranslated regions, chimerical sequences (assemblage errors) and nonconserved areas of proteins where homology is not detected [28]. The species distribution of the best match result for each sequence is shown in (Figure 2). The T. yunnanensis sequences produced 20,896 hits to Tribolium castaneum, 1,396 hits to Apis mellifera, and 1,363 hits to Drosophila. Overall, a strong preference for matches is against T. castaneum genes, composing of 71.11%. It might be due to the relatively near evolutionary relationship of these two species, belonging to the same order Coleoptera, and the available complete genome sequence of T. castaneum.
Figure 2.
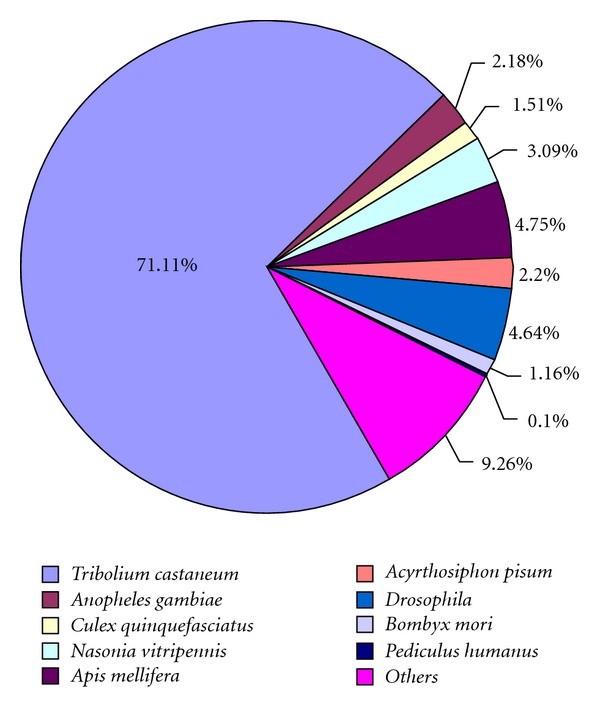
Species distribution of the top Blastx hits for unigenes from the head of Tomicus yunnanensis with a cutoff E-value 105 to searches from the NCBI nonredundant database. The first hit of each sequence was used for analysis.
For functional comparisons, all unigenes were assigned for GO terms based on BLAST matches with sequences whose function is previously known. A total of 54,427 unigenes were able to map to GO terms. These transcripts were assigned for biological process (25,348 sequences), cellular component (17,537 sequences), and molecular function (11,542 sequences) (Figure 3). Cellular process (19.58%) and metabolic process (15.77%) were the main subcategories of biological process, indicating the important metabolic activities in T. yunnanensis head. Under the category of cellular component, cell (31.16%), cell part (31.16%), and organelle (17.23%) were among the most highly represented subcategories. The molecular function category was mainly comprised of proteins involved in binding (46.44%) and catalytic activities (36.54%).
Figure 3.
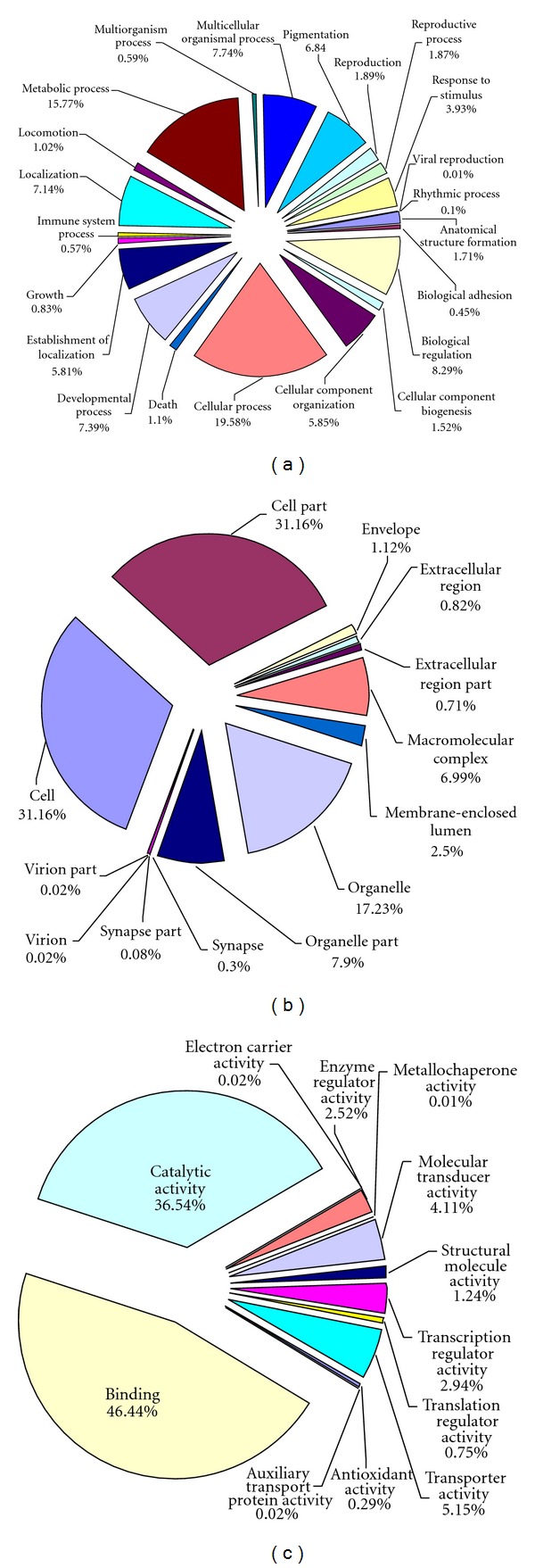
GO categories of the unigenes. (a) Molecular function GO terms, (b) biological process GO terms, and (c) cellular component GO terms. GO annotation was done with the Blast2GO tool. The data presented represent the level 2 analysis, illustrating general functional categories.
3.2. Discovery of Genes Encoding Olfactory Proteins
Of particular interest to detect the sequences encoding olfactory proteins, the head transcriptome of T. yunnanensis was analyzed more in detail. Unigenes with at least one match to olfactory proteins with an E-value of 10−5 or lower were selected. As shown in Table 2, we identified many sequences with homology to OBP, CSP, OR, and GR. Overall, 45 potential olfactory genes out of 57,142 unigenes were identified. The proportion is similar to what was recorded in antennal transcriptome of Manduca sexta and in the combined EST and transcriptome data of Solenopsis invicta [19, 29]. Among these unigenes, 11 encoded for OBP, 8 for CSP, 18 for OR, and 8 for GR. The data in this study presents some information on the molecular basis of olfaction of a Coleoptera species besides T. castaneum. Analysis of fully sequenced genome has identified 49 OBPs, 20 CSPs, 299 ORs, and 220 GRs in T. castaneum [1, 30]. Compared with the gene number of olfactory genes reported from this genome, the current number of T. yunnanensis is at the lower end of the range of T. castaneum. Additional olfactory genes may await discovery due to their absence from the current transcriptomic dataset. The remainders not obtained from this dataset might be due to the following reasons: (1) only a single run carried out in this study, which results in the dataset that do not cover all genes expressed in the head of T. yunnanensis, and (2) the tissues excluded the head that some olfactory genes abundantly located are not included as the target tissue. Increasing the number of olfactory genes can discovery from transcriptome of the samples that are greatly restricted to the main olfactory organs. In addition, as the ability of Illumina sequencing is for short reads (<120 bp) [31], only one unigene is appeared to complete in all the identified putative olfactory transcripts. More candidates encoding full sequence could be determined by 454 pyrosequencing, which can generate longer sequence reads (~450 bases) than those obtained by Illumina sequencing [32, 33]. Using 454 sequencing, the majority of olfactory genes identified from the transcriptomic database of M. sexta antennae and S. invicta encoded the full length proteins [19, 29]. However, the latter technology needs much more cost.
Table 2.
Putatively identified olfactory genes in Tomicus yunnanensis.
| Gene family | Unigene ID | Length (bp) | First hit | E_value | Blast annotation/Organism |
|---|---|---|---|---|---|
| Ordorant binding protein | Unigene13495_YNNT | 279 | EFA12066 | 6.00E-06 | Odorant binding protein 15 [Tribolium castaneum] |
| Unigene14057_YNNT | 415 | EFA07430 | 1.00E-06 | Odorant binding protein (subfamily minus C) C04 [Tribolium castaneum] | |
| Unigene2588_YNNT | 282 | EEZ97740 | 2.00E-07 | Odorant binding protein C12 [Tribolium castaneum] | |
| Unigene41600_YNNT | 256 | EFA05793 | 5.00E-06 | Odorant binding protein 20 [Tribolium castaneum] | |
| Unigene41993_YNNT | 258 | EFA10713 | 5.00E-19 | Odorant binding protein 09 [Tribolium castaneum] | |
| Unigene44812_YNNT | 277 | EFA07430 | 3.00E-06 | Odorant binding protein (subfamily minus C) C04 [Tribolium castaneum] | |
| Unigene4914_YNNT | 231 | EFA10803 | 2.00E-07 | Odorant binding protein 23 [Tribolium castaneum] | |
| Unigene54663_YNNT | 466 | EFA07430 | 2.00E-13 | Odorant binding protein (subfamily minus C) C04 [Tribolium castaneum] | |
| Unigene6724_YNNT | 361 | EFA01425 | 5.00E-10 | Odorant binding protein C20 [Tribolium castaneum] | |
| Unigene7827_YNNT | 311 | AAD31883 | 4.00E-09 | Odorant binding protein RpalOBP2′ [Rhynchophorus palmarum] | |
| Unigene9635_YNNT | 315 | EFA05695 | 7E-28 | Odorant binding protein 11 [Tribolium castaneum] | |
|
| |||||
| Chemosensory protein | Unigene10309_YNNT | 360 | NP_001039280 | 2.00E-15 | Chemosensory protein 12 [Tribolium castaneum] |
| Unigene13359_YNNT | 219 | NP_001039274 | 8.00E-17 | Chemosensory protein 20 [Tribolium castaneum] | |
| Unigene19380_YNNT | 498 | BAF91720 | 1.00E-22 | Chemosensory protein [Papilio xuthus] | |
| Unigene24674_YNNT | 250 | NP_001039290 | 8.00E-11 | Chemosensory protein 8 [Tribolium castaneum] | |
| Unigene24747_YNNT | 1891 | NP_001039288 | 6.00E-26 | Chemosensory protein 6 [Tribolium castaneum] | |
| Unigene2482_YNNT | 235 | NP_001039288 | 2.00E-12 | Chemosensory protein 6 [Tribolium castaneum] | |
| Unigene26497_YNNT | 356 | NP_001039278 | 2.00E-34 | Chemosensory protein 10 [Tribolium castaneum] | |
| Unigene55391_YNNT | 512 | NP_001039278 | 1E-60 | Chemosensory protein 10 [Tribolium castaneum] | |
|
| |||||
| Olfactory receptor | Unigene10465_YNNT | 223 | XP_001507235 | 7.00E-07 | Predicted: similar to olfactory receptor MOR18-3 [Ornithorhynchus anatinus] |
| Unigene1058_YNNT | 236 | XP_001512662 | 1.00E-17 | Predicted: similar to olfactory receptor OR19-14, partial [Ornithorhynchus anatinus] | |
| Unigene16616_YNNT | 426 | XP_001511079 | 4.00E-13 | Predicted: similar to olfactory receptor Olr4 [Ornithorhynchus anatinus] | |
| Unigene17391_YNNT | 522 | XP_002932395 | 2.00E-06 | Predicted: olfactory receptor 10A4-like [Xenopus (Silurana) tropicalis] | |
| Unigene22269_YNNT | 303 | XP_001512662 | 2.00E-20 | Predicted: similar to olfactory receptor OR19-14, partial [Ornithorhynchus anatinus] | |
| Unigene25110_YNNT | 837 | XP_001507235 | 3.00E-43 | Predicted: similar to olfactory receptor MOR18-3 [Ornithorhynchus anatinus] | |
| Unigene25351_YNNT | 285 | XP_001368212 | 1.00E-12 | Predicted: similar to C-family odorant receptor OLFCT1 [Monodelphis domestica] | |
| Unigene27701_YNNT | 391 | EFA09172 | 2.00E-08 | Odorant receptor 12 [Tribolium castaneum] | |
| Unigene30603_YNNT | 205 | XP_001513226 | 8.00E-19 | Predicted: similar to olfactory receptor MOR253-9 [Ornithorhynchus anatinus] | |
| Unigene30802_YNNT | 206 | CAM84014 | 1.00E-27 | Olfactory receptor 16 [Tribolium castaneum] | |
| Unigene3238_YNNT | 419 | XP_001516663 | 2.00E-46 | Predicted: similar to olfactory receptor MOR218-3 [Ornithorhynchus anatinus] | |
| Unigene45804_YNNT | 286 | CAM84014 | 2.00E-40 | Olfactory receptor 16 [Tribolium castaneum] | |
| Unigene47392_YNNT | 300 | XP_001516663 | 3.00E-30 | Predicted: similar to olfactory receptor MOR218-3 [Ornithorhynchus anatinus] | |
| Unigene51999_YNNT | 372 | CAM84014 | 2.00E-30 | Olfactory receptor 16 [Tribolium castaneum] | |
| Unigene53119_YNNT | 403 | BAG12817 | 2.00E-11 | Olfactory receptor-like receptor [Bombyx mori] | |
| Unigene53416_YNNT | 412 | XP_002932395 | 3.00E-08 | Predicted: olfactory receptor 10A4-like [Xenopus (Silurana) tropicalis] | |
| Unigene54915_YNNT | 479 | ACD40044 | 3.00E-66 | Odorant receptor [Phyllotreta striolata] | |
| Unigene9988_YNNT | 617 | XP_002932775 | 0.0000006 | Predicted: olfactory receptor 11L1-like [Xenopus (Silurana) tropicalis] | |
|
| |||||
| Gustatory receptor | Unigene13393_YNNT | 259 | NP_001138958 | 2.00E-18 | Gustatory receptor Gr85 [Tribolium castaneum] |
| Unigene14213_YNNT | 261 | CAL23158 | 6.00E-06 | Gustatory receptor candidate 25 [Tribolium castaneum] | |
| Unigene1791_YNNT | 463 | NP_001138958 | 3.00E-25 | Gustatory receptor Gr85 [Tribolium castaneum] | |
| Unigene24414_YNNT | 906 | NP_001138948 | 2.00E-69 | Gustatory receptor Gr83 [Tribolium castaneum] | |
| Unigene31152_YNNT | 207 | EFA13587 | 2.00E-09 | Gustatory receptor 21 [Tribolium castaneum] | |
| Unigene35383_YNNT | 223 | EFA02924 | 2.00E-18 | Gustatory receptor 2 [Tribolium castaneum] | |
| Unigene4039_YNNT | 493 | NP_001138958 | 2.00E-29 | Gustatory receptor Gr85 [Tribolium castaneum] | |
| Unigene44392_YNNT | 274 | NP_001138958 | 2.00E-20 | Gustatory receptor Gr85 [Tribolium castaneum] | |
3.3. Validation of Several Detected Olfactory Genes
Based on the fragments derived from the transcriptome database, primers were designed to clone the full coding sequences for one OBP (Unigene41600_YNNT) (designated as TyunOBP1) and one CSP (Unigene26497_YNNT) (designated as TyunCSP1) by RACE-PCR, and assure the acute of one CSP sequence (Unigene55391_YNNT) (designated as TyunCSP2) that appeared to be complete after raw reads assembly by RT-PCR. We have successfully performed the amplification.
TyunOBP1 contained a 384 bp open reading frame (ORF) for a polypeptide of 128 amino acids. It was with a molecular mass of 14,15 Da and an isoelectric point (pI) of 5.15. The deduced protein sequence was in accordance with other OBPs, generally composed of subunits of about 14 kDa as small hydrophilic proteins with acidic isoelectric points [34]. Insect OBPs have also been classified into long chain (~160 aa), medium chain (~120 aa), and short chain (~110 aa) classes, which relates to their potential structure and function [35]. According to this hypothesis, TyunOBP1 belongs to the medium chain class. A signal peptide is a common characteristic of OBPs [36]. Similar to that of other insects, the initial 25 amino acids of TyunOBP1 were predicted as a signal peptide. In addition, OBPs are characterized by conserved cysteines that are believed to form three disulfide bridges, and the hydrophobic domains [37]. Multiple amino acid sequence alignment revealed that the six cysteine residues are highly conserved in TyunOBP1 (Figure 4). Moreover, it indicated that TyunOBP1 displayed low similarity to that of other insects, which are typically around 20% or below, and the lengths of the N- and C-termini of OBPs are highly divergent. This highly diverse insect gene family is divided into nine major subclasses: Classic, Minus-C, Plus C, Dimer, PBP/GOBP, ABPI, ABPII, CRLBP, and D7 [16, 38]. Among the currently available genomes, the dipterans consistently have large expansions of these genes [39], and the body louse, Pediculus humanus, appears to possess the smallest set of OBPs [40]. A phylogenetic tree was constructed using the amino acid sequences of TyunOBP1 and T. castaneum OBP (TcasOBP) (Figure 5). According to the phylogenetic tree and the features of OBP subclasses, TyunOBP1 is near to TcasOBP42, belonging to the Classic clad.
Figure 4.
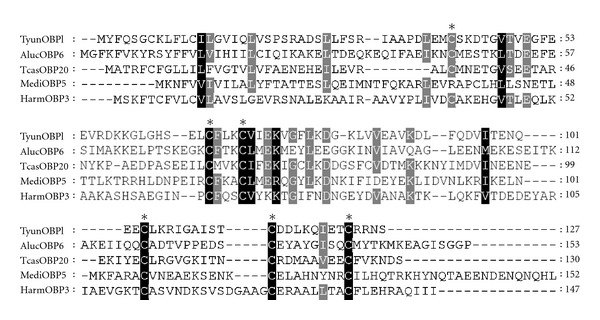
Alignment of the predicted amino acid sequence of TyunOBP1 with OBPs from other insects. The conserved cystein residues are indicated by asterisks. Black and gray indicate residues that are identical or conserved, respectively. The abbreviation and GenBank accession number for the sequences from other insect aligned are AlucOBP6, Apolygus lucorum (AEP95762); TcasOBP20, Tribolium castaneum (EFA05793); MediOBP5, Microplitis mediator (ABM05972), and HarmOBP3, Helicoverpa armigera (AEB54582).
Figure 5.
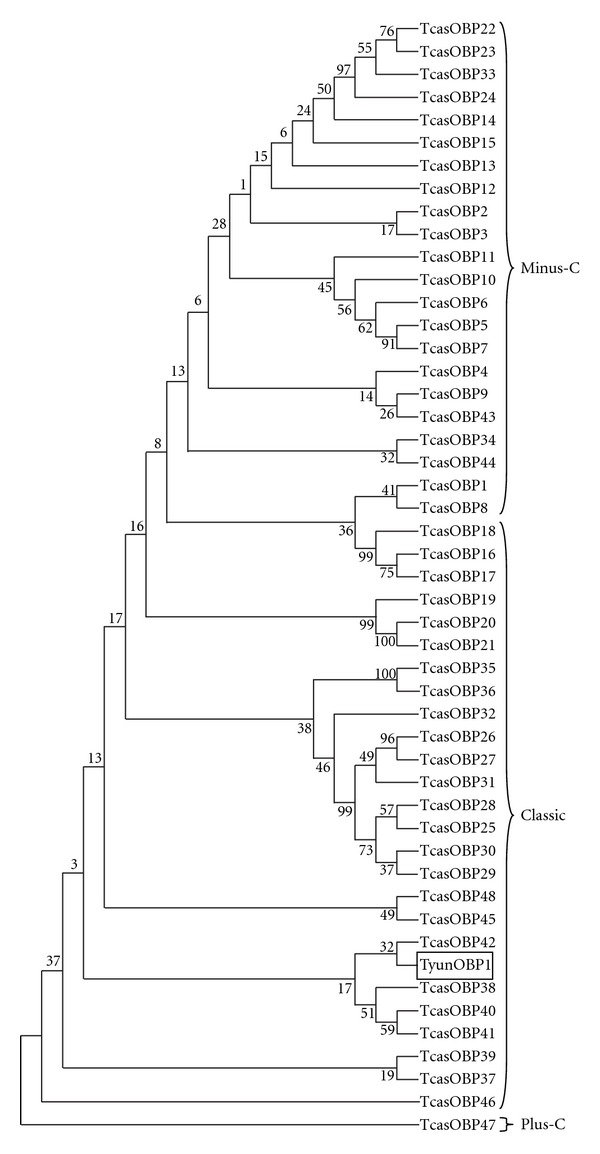
Neighbour-joining phylogenetic analysis of OBPs from Tomicus yunnanensis (Tyun) and Tribolium castaneum (Tcas). Sequences from T. yunnanensis are marked by a box. Numbers at nodes are bootstrap values shown as percentages. The T. castaneum OBP gene family was classified according to Sánchez-Gracia et al. [16].
The ORF for TyunCSP1 and TyunCSP2 were 390 bp. Both of them consisted of 130 amino acids. TyunCSP1 was with a calculated molecular weight of 15.11 kDa and pI of 5.10, whereas TyunCSP2 was with a molecular mass of 15.17 Da and a pI of 8.02. All CSPs appear to have a hydrophobic N-terminus of about 16–20 amino acids that are predicted to encode a signal peptide [41]. The deduced amino acid sequences for both proteins possessed a putative signal sequence of 18 amino acids. A typical feature for CSP is the remarkably conserved 4-cysteine motif that forms disulphide bridges in folded proteins [42]. A comparison between the sequences of TyunCSP1, TyunCSP2, and those of other insects clearly showed that a conserved four-cysteine signature is present at the characteristic positions within the CSP family (Figure 6). CSPs are more conserved than OBPs across evolution, with about a 50% identity even between members of phylogenetically distant species [43]. These two TyunCSPs shared 54% identity, but were with 26–58% identity to CSPs from other species used in the multiple alignments. To characterize the molecular evolution relationships between the CSPs of T. yunnanensis and those of others insects in Coleoptera, we constructed a phylogenetic tree including 2 CSPs of T. yunnanensis identified in this study, and 20 CSPs of T. castaneum (Figure 7). The tree showed that five proposed groups of CSPs were resolved. TyunCSP1, TyunCSP2, and most of the CSPs of T. castaneum were grouped into the largest distinctive phylogenetic clade.
Figure 6.

Alignment of the predicted amino acid sequence of TyunCSP1 and TyunCSP2 with CSPs from other insects. The conserved cystein residues are indicated by asterisks. Black and gray indicate residues that are identical or conserved, respectively. The abbreviation and GenBank accession number for the sequences from other insect aligned are DmelCSP2, Drosophila melanogaster (NP_524966); AgamCSP3, Anopheles gambiae (EAA12338); BmorCSP10, Bombyx mori (NP_001037069), and AmelCSP3, Apis mellifera (NP_001011583).
Figure 7.
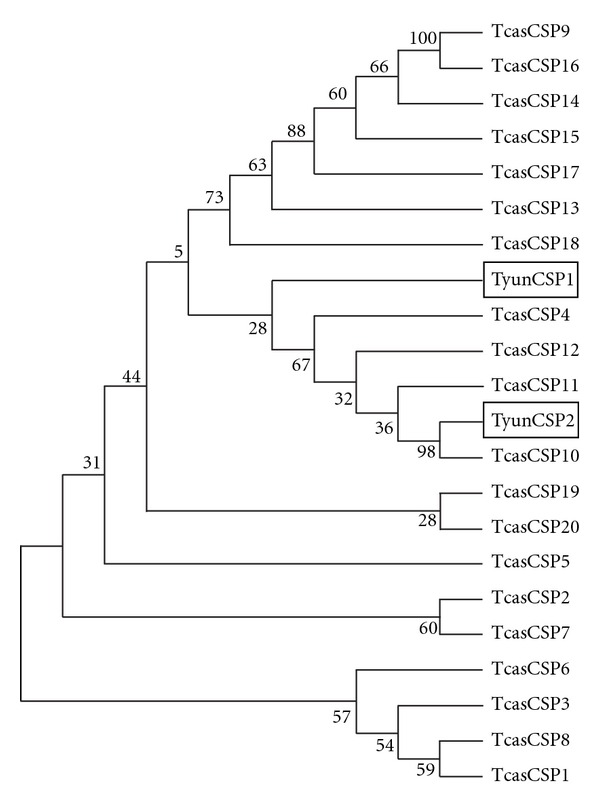
Neighbour-joining phylogenetic analysis of CSPs from Tomicus yunnanensis (Tyun) and Tribolium castaneum (Tcas). Sequences from T. yunnanensis are marked by a box. Numbers at nodes are bootstrap values shown as percentages.
4. Conclusion
Historically, identification of a comprehensive list of candidate olfaction genes for understanding the molecular mechanisms of olfaction has been mostly limited by the difficulty in collecting antenna because it is small and encased in a hard cuticle, and successful cloning as the olfactory gene families are divergent [44]. Next generation short-read DNA sequencing has made it possible to explore genome-level questions in nonmodel organisms, regardless of their phylogenetic proximity to model species [45, 46]. This study produced a large amount of transcriptome data from T. yunnanensis head by Illumina sequencing. An extensive list of candidate genes involved in olfactory signal transduction has been generated from this valuable data platform. This is an important resource for further study of the chemical communication between T. yunnanensis and the environment. This study demonstrated that the Illumina sequencing technology could be applied as a particularly rapid, cost-effective, and fruitful approach to accelerate our understanding of the molecular basis of chemosensory pathways in nonmodel insects that have no previous genome data.
Conflict of Interests
All authors declare that there is no conflict of interests.
Acknowledgments
This work was supported by the Special Fund for Forest Scientific Research in the Public Interest of China (201004067), the Science Foundation for the Excellent Youth Academic and Technological Scholars of Yunnan Province of China (2007YP01-18), and the Science Foundation of Southwest Forestry University (111128).
References
- 1.Engsontia P, Sanderson AP, Cobb M, Walden KKO, Robertson HM, Brown S. The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum . Insect Biochemistry and Molecular Biology. 2008;38(4):387–397. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Zhou JJ, Kan Y, Antoniw J, Pickett JA, Field LM. Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chemical Senses. 2006;31(5):453–465. doi: 10.1093/chemse/bjj050. [DOI] [PubMed] [Google Scholar]
- 3.Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nature Reviews Neuroscience. 2010;11(3):188–200. doi: 10.1038/nrn2789. [DOI] [PubMed] [Google Scholar]
- 4.Vieira FG, Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biology and Evolution. 2011;3:476–490. doi: 10.1093/gbe/evr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horst R, Damberger F, Luginbühl P, et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong ZJ, Zhou WW, Yu HZ, et al. Cloning, expression and functional analysis of a general odorant-binding protein 2 gene of the rice striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Pyralidae) Insect Molecular Biology. 2009;18(3):405–417. doi: 10.1111/j.1365-2583.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- 7.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293(5828):161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 8.Danty E, Michard-Vanhée C, Huet JC, Genecque E, Pernollet JC, Masson C. Biochemical characterization, molecular cloning and localization of a putative odorant-binding protein in the honey bee Apis mellifera L. (Hymenoptera: apidea) FEBS Letters. 1997;414(3):595–598. doi: 10.1016/s0014-5793(97)01048-x. [DOI] [PubMed] [Google Scholar]
- 9.Steinbrecht RA. Odorant-binding proteins: expression and function. Annals of the New York Academy of Sciences. 1998;855:323–332. doi: 10.1111/j.1749-6632.1998.tb10591.x. [DOI] [PubMed] [Google Scholar]
- 10.Rützler M, Zwiebel LJ. Molecular biology of insect olfaction: recent progress and conceptual models. Journal of Comparative Physiology A. 2005;191(9):777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 11.Ha TS, Smith DP. Insect odorant receptors: channeling scent. Cell. 2008;133(5):761–763. doi: 10.1016/j.cell.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Menini A. The Neurobiology of Olfaction. Boca Raton, Fla, USA: CRC Press; 2010. [PubMed] [Google Scholar]
- 13.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. Journal of Human Genetics. 2006;51(6):505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Gracia A, Rozas J. Divergent evolution and molecular adaptation in the Drosophila odorant-binding protein family: inferences from sequence variation at the OS-E and OS-F genes. BMC Evolutionary Biology. 2008;8(1, article 323) doi: 10.1186/1471-2148-8-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu YL, He P, Zhang L, et al. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics. 2009;10, article 632 doi: 10.1186/1471-2164-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103(3):208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- 17.Wang XW, Luan JB, Li JM, Bao YY, Zhang CX, Liu SS. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics. 2010;11(1, article 400) doi: 10.1186/1471-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue J, Bao YY, Li BL, et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens . PLoS ONE. 2010;5(12) doi: 10.1371/journal.pone.0014233.e14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotzek D, Robertson HM, Wurm Y, Shoemaker D. Odorant binding proteins of the red imported fire ant, Solenopsis invicta an example of the problems facing the analysis of widely divergent proteins. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0016289.e16289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Zhang Z, Ye H, Wang H, Clarke SR, Jun L. Response of Tomicus yunnanensis (Coleoptera:Scolytinae) to infested and uninfested Pinus yunnanensis bolts. Journal of Economic Entomology. 2010;103(1):95–100. doi: 10.1603/ec09080. [DOI] [PubMed] [Google Scholar]
- 21.Kirkendall LR, Faccoli M, Ye H. Description of the Yunnan shoot borer, Tomicus yunnanensis Kirkendall & Faccoli sp. n. (Curculionidae, Scolytinae), an unusually aggressive pine shoot beetle from southern China, with a key to the species of Tomicus . Zootaxa. 2008;1819:25–39. [Google Scholar]
- 22.Li R, Zhu H, Ruan J, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Research. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price DP, Nagarajan V, Churbanov A, et al. The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre- and post- blood meal. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0022573.e22573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai X, Mamidala P, Rajarapu SP, Jones SC, Mittapalli O. Transcriptomics of the bed bug (Cimex lectularius) PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0016336.e16336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta . Proceedings of the National Academy of Sciences of the United States of America. 2011;108(18):7449–7454. doi: 10.1073/pnas.1017963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum . Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 31.Gogol-Döring A, Chen W. An overview of the analysis of next generation sequencing data. Methods in Molecular Biology. 2012;802:249–257. doi: 10.1007/978-1-61779-400-1_16. [DOI] [PubMed] [Google Scholar]
- 32.Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochemistry and Molecular Biology. 2008;38(6):677–682. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karatolos N, Pauchet Y, Wilkinson P, et al. Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes. BMC Genomics. 2011;12, article 56 doi: 10.1186/1471-2164-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelosi P. Odorant-binding proteins. Critical Reviews in Biochemistry and Molecular Biology. 1994;29(3):199–228. doi: 10.3109/10409239409086801. [DOI] [PubMed] [Google Scholar]
- 35.Pesenti ME, Spinelli S, Bezirard V, et al. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. Journal of Molecular Biology. 2008;380(1):158–169. doi: 10.1016/j.jmb.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 36.Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster . Genome Research. 2002;12(9):1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou JJ. Odorant-binding proteins in insects. Vitamins and Hormones. 2010;83:241–272. doi: 10.1016/S0083-6729(10)83010-9. [DOI] [PubMed] [Google Scholar]
- 38.Vieira FG, Sánchez-Gracia A, Rozas J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biology. 2007;8(11, article R235) doi: 10.1186/gb-2007-8-11-r235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou JJ, He XL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Molecular Biology. 2008;17(2):147–163. doi: 10.1111/j.1365-2583.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 40.Kirkness EF, Haas BJ. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proceedings of the National Academy of Sciences of the United States of America. 2011;107(27):12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forêt S, Wanner KW, Maleszka R. Chemosensory proteins in the honey bee: insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochemistry and Molecular Biology. 2007;37(1):19–28. doi: 10.1016/j.ibmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Dani FR, Michelucci E, Francese S, et al. Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori . Chemical Senses. 2011;36(4):335–344. doi: 10.1093/chemse/bjq137. [DOI] [PubMed] [Google Scholar]
- 43.Ban L, Scaloni A, Brandazza A, et al. Chemosensory proteins of Locusta migratoria . Insect Molecular Biology. 2003;12(2):125–134. doi: 10.1046/j.1365-2583.2003.00394.x. [DOI] [PubMed] [Google Scholar]
- 44.Anholt RRH, Williams TI. The soluble proteome of the Drosophila antenna. Chemical Senses. 2009;35(1):21–30. doi: 10.1093/chemse/bjp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudson ME. Sequencing breakthroughs for genomic ecology and evolutionary biology. Molecular Ecology Resources. 2008;8(1):3–17. doi: 10.1111/j.1471-8286.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- 46.Crawford JE, Guelbeogo WM, Sanou A, et al. De novo transcriptome sequencing in anopheles funestus using illumina RNA-seq technology. PLoS ONE. 2010;5(12) doi: 10.1371/journal.pone.0014202.e14202 [DOI] [PMC free article] [PubMed] [Google Scholar]


