Abstract
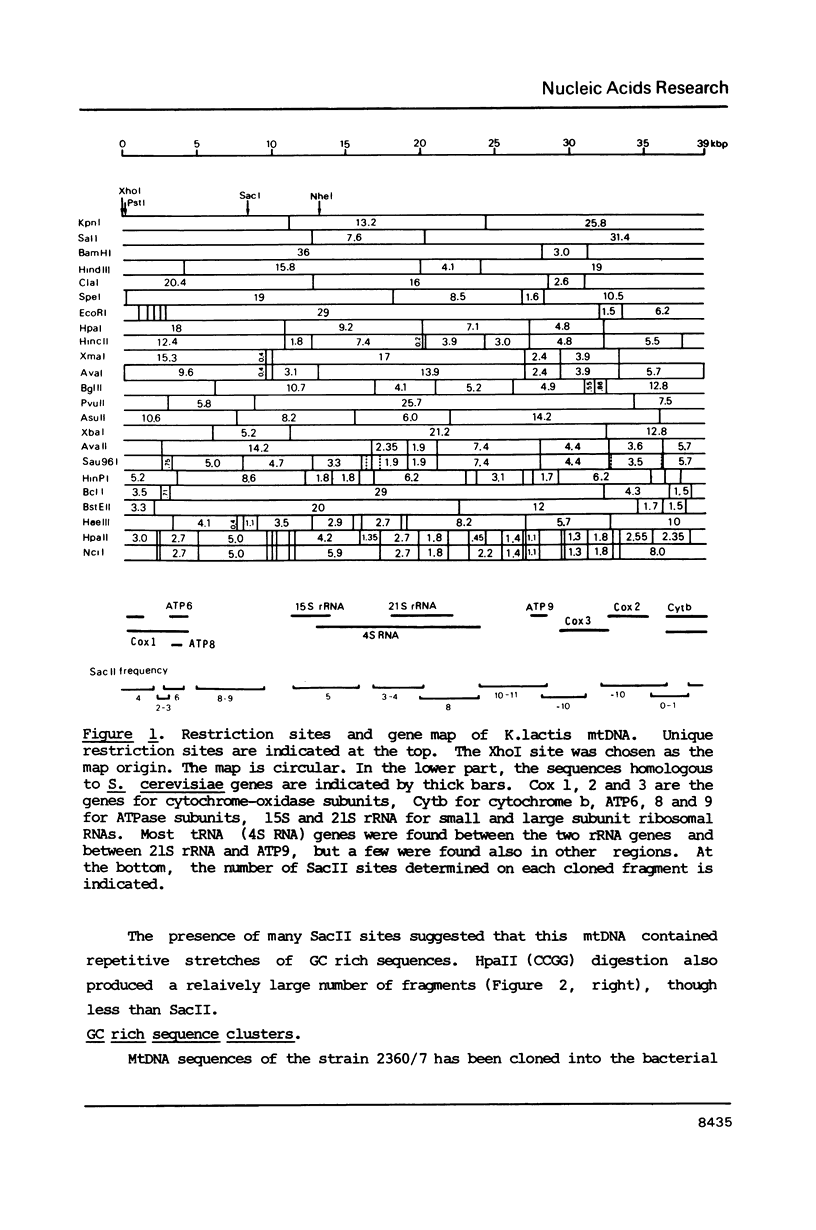
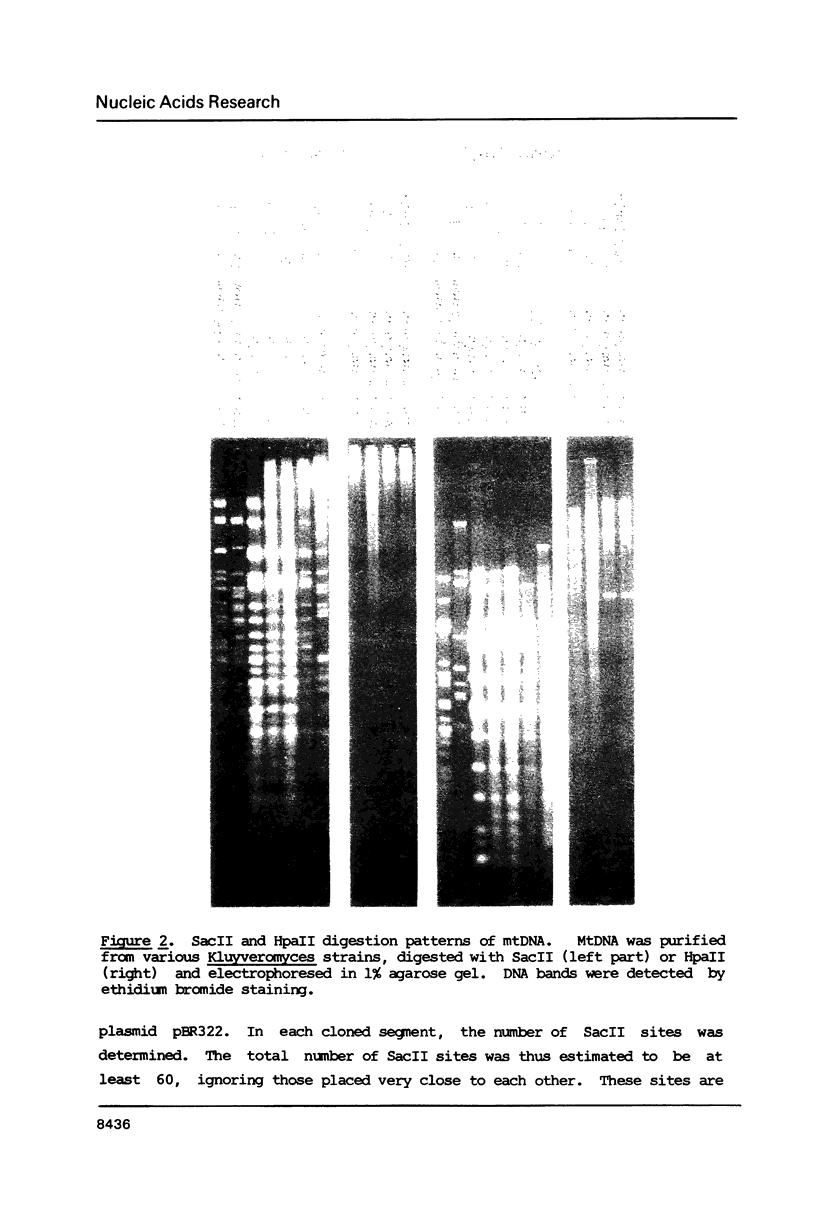
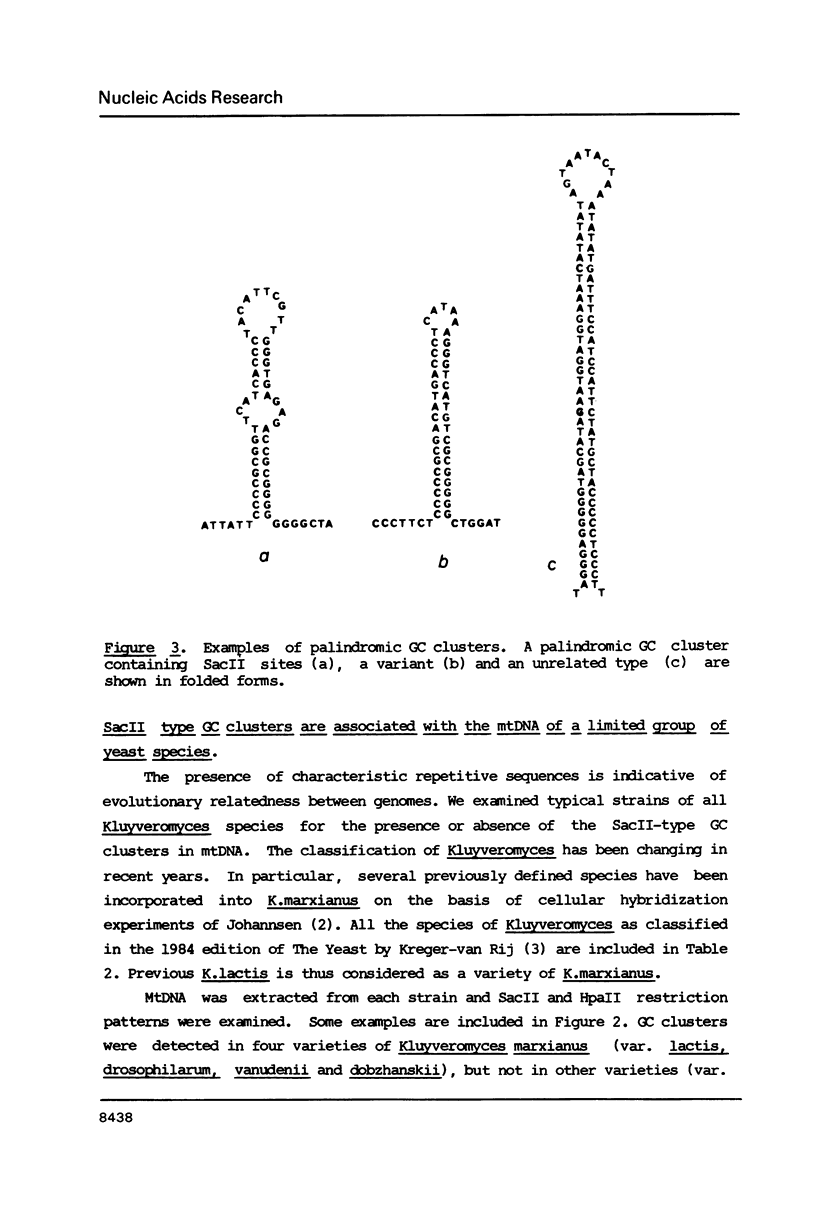
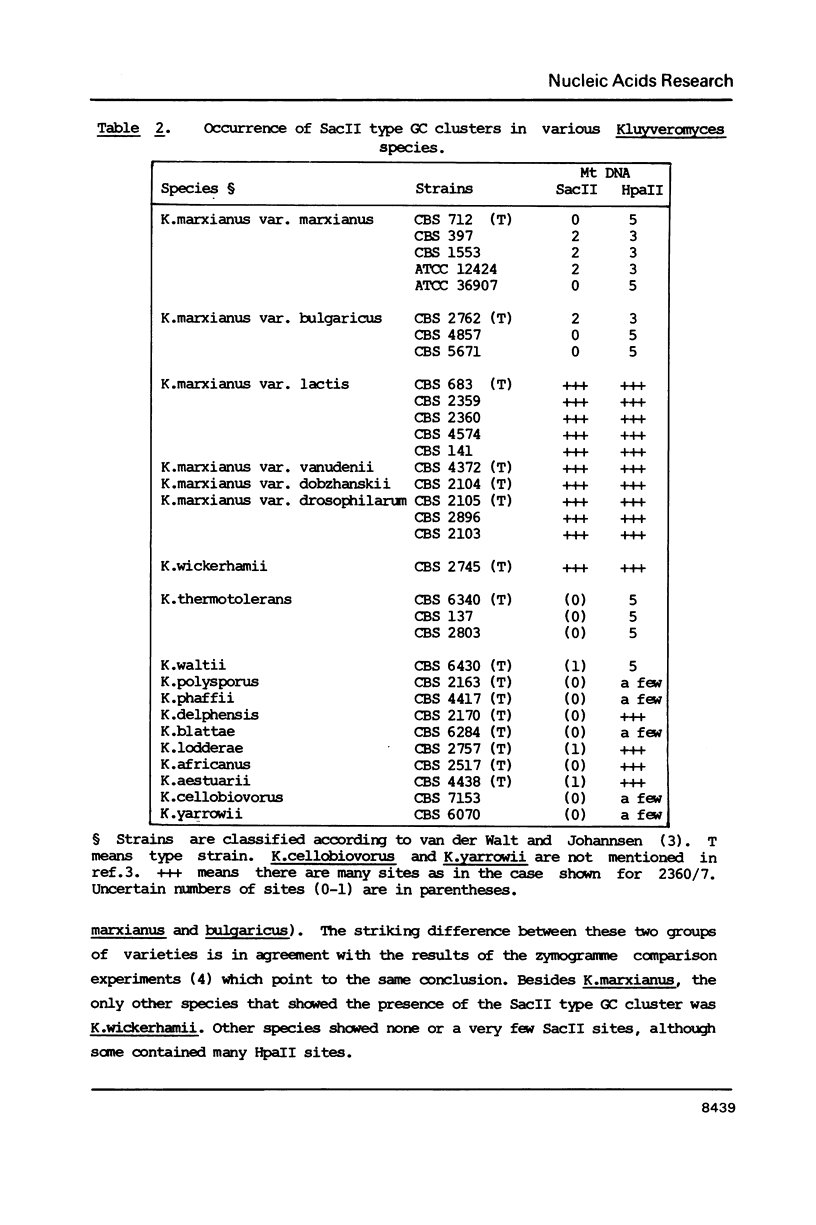
Mitochondrial DNA from the yeast Kluyveromyces marxianus var. lactis (K.lactis) is a circular molecule of 39 kilobase-pairs. A genetic and physical map was constructed. We found that this genome contained a large number of guanine-cytosine (GC)-rich sequence clusters, many of which are characterized by the presence of SacII restriction sites (CCGCGG). The primary sequence of the GC clusters often showed a palindromic structure. These GC clusters were present in several varieties of K.marxianus, but not in others. The presence of these clusters is a major feature that distinguishes K.lactis strains from those of K.marxianus var. marxianus (including K.fragilis).
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Falcone C., Saliola M., Chen X. J., Frontali L., Fukuhara H. Analysis of a 1.6-micron circular plasmid from the yeast Kluyveromyces drosophilarum: structure and molecular dimorphism. Plasmid. 1986 May;15(3):248–252. doi: 10.1016/0147-619x(86)90044-2. [DOI] [PubMed] [Google Scholar]
- Francisci S., Palleschi C., Ragnini A., Frontali L. Analysis of transcripts of the major cluster of tRNA genes in the mitochondrial genome of S. cerevisiae. Nucleic Acids Res. 1987 Aug 25;15(16):6387–6403. doi: 10.1093/nar/15.16.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeben P., Clark-Walker G. D. An approach to yeast classification by mapping mitochondrial DNA from Dekkera/Brettanomyces and Eeniella genera. Curr Genet. 1986;10(5):371–379. doi: 10.1007/BF00418409. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Ainley W. M., Shumard D. S., Butow R. A., Grossman L. I. Location and structure of the var1 gene on yeast mitochondrial DNA: nucleotide sequence of the 40.0 allele. Cell. 1982 Sep;30(2):617–626. doi: 10.1016/0092-8674(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Johannsen E. Hybridization studies within the genus Kluyveromyces van der Walt emend. van der Walt. Antonie Van Leeuwenhoek. 1980;46(2):177–189. doi: 10.1007/BF00444073. [DOI] [PubMed] [Google Scholar]
- Prunell A., Goutorbe F., Strauss F., Bernardi G. Yield of restriction fragments from yeast mitochondrial DNA. J Mol Biol. 1977 Feb 15;110(1):47–52. doi: 10.1016/s0022-2836(77)80097-1. [DOI] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Nature of an inserted sequence in the mitochondrial gene coding for the 15S ribosomal RNA of yeast. Nucleic Acids Res. 1982 Mar 11;10(5):1625–1633. doi: 10.1093/nar/10.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S., Heckman J., RajBhandary U. L. Highly conserved GC-rich palindromic DNA sequences flank tRNA genes in Neurospora crassa mitochondria. Cell. 1981 Nov;26(3 Pt 1):325–332. doi: 10.1016/0092-8674(81)90201-4. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene. 1986;41(1):1–22. doi: 10.1016/0378-1119(86)90262-3. [DOI] [PubMed] [Google Scholar]