Abstract
There is compelling evidence about the manifest effects of inbreeding depression on individual fitness and populations' risk of extinction. The majority of studies addressing inbreeding depression on wild populations are generally based on indirect measures of inbreeding using neutral markers. However, the study of functional loci, such as genes of the major histocompatibility complex (MHC), is highly recommended. MHC genes constitute an essential component of the immune system of individuals, which is directly related to individual fitness and survival. In this study, we analyse heterozygosity fitness correlations of neutral and adaptive genetic variation (22 microsatellite loci and two loci of the MHC class II, respectively) with the age of recruitment and breeding success of a decimated and geographically isolated population of a long-lived territorial vulture. Our results indicate a negative correlation between neutral genetic diversity and age of recruitment, suggesting that inbreeding may be delaying reproduction. We also found a positive correlation between functional (MHC) genetic diversity and breeding success, together with a specific positive effect of the most frequent pair of cosegregating MHC alleles in the population. Globally, our findings demonstrate that genetic depauperation in small populations has a negative impact on the individual fitness, thus increasing the populations' extinction risk.
Keywords: MHC, heterozygosity–fitness correlation, inbreeding, breeding success, long-lived species, Egyptian vulture
1. Introduction
Reduced and isolated populations are highly exposed to genetic drift and to the loss of genetic variation, which may make them more vulnerable to inbreeding depression [1]. Inbreeding depression is the decline in mean fitness owing to the increased expression of recessive deleterious alleles and the loss of heterozygous advantage at functionally important genes [2]. Compelling evidence has demonstrated the negative effects of inbreeding on the performance of individuals through reduced survival, breeding success and resistance to environmental stress [3], all of which contribute, in turn, to enhance the risk of extinction of natural populations. Effects of inbreeding in the wild should ideally be assessed by estimating individual inbreeding coefficients from detailed pedigrees [4]. However, this information is inherently difficult to obtain, especially for long-lived species. Consequently, a number of studies have used molecular markers to obtain indirect estimates of inbreeding, many of them finding significant heterozygosity–fitness correlations (HFCs) [5–9]. Two main hypotheses are proposed to explain HFCs: (i) the local effect hypothesis suggests that neutral loci can be linked to loci influencing fitness and hence heterozygotes exhibit an advantage via dominance (i.e. heterozygotes experience lower expression of recessive deleterious mutations) and/or overdominance (i.e. heterozygotes are superior per se [10]); and (ii) the general effect hypothesis suggests that multi-locus heterozygosity (MLH) reflects genome-wide variation or inbreeding [5,11]. Nevertheless, for marker loci to indicate general inbreeding, the studied population may have gone through any mechanism representing some form of variation in inbreeding (e.g. consanguineous mating, drift and/or bottleneck) and then generating identity disequilibrium (ID) [12]. Correlations between neutral genetic diversity and fitness still remain, however, very weak and inconsistent, and are expected to depend on population demography and life history [2–13]. Furthermore, recent findings suggest that losses of genetic variability in bottlenecked populations can be more dramatic in adaptive genes than at neutral markers [14–15] and, hence, the analysis of adaptive loci may be more appropriate when estimating genome-wide functional diversity in bottlenecked populations [14]. Genes of the major histocompatibility complex (MHC) are good candidates for these kinds of analyses. MHC genes play a crucial role in pathogen confrontation and clearance, and, consequently, MHC diversity has been associated with individual fitness [16]. MHC molecules present foreign antigens to specialized T cells, which triggers the production of antibodies or the destruction of pathogen-infected cells. The antigen-binding domains of MHC molecules are clearly under selection as a result of the coevolutionary arms race between pathogens and their hosts; therefore, MHC markers are excellent candidates to investigate adaptive variation in vertebrates and its relationship with fitness [17].
Many long-lived vertebrate species are highly threatened owing to their conservative life-history strategies and their vulnerability to human-related pressures [18]. However, obtaining the individualized information necessary for the proper management of these species is difficult and involves long-term monitoring. Although there is an increasing number of studies addressing inbreeding depression in threatened populations of long-lived species [5,19], few incorporate the analysis of functional genes [11,20]. In this study, we investigate the occurrence of inbreeding depression in a highly reduced and potentially inbred insular population of a globally endangered bird of prey, the Egyptian vulture (Neophron percnopterus). We use genetic and ecological data from the entire population to explore relationships between neutral (using 22 microsatellites) and adaptive (MHC class II β chain, exon 2) genetic diversity with two demographic parameters believed to greatly affect individual fitness: age of recruitment into the breeding population and breeding success [21,22]. We specifically test whether there is a negative correlation between genetic variability and age of recruitment, and whether there is a positive correlation with respect to breeding success. Additionally, and in order to assess specific effects, we related MHC composition with fitness parameters to specifically test whether frequent haplotypes are selectively advantageous.
2. Material and methods
(a). Study species and population
The Egyptian vulture (Neophron percnopterus) is a long-lived (species generation time: 13 years), medium-sized (mean weight: 2 kg) scavenger with a broad distribution range in dry areas of Europe, Asia and Africa, where populations are migratory. When breeding, it maintains exclusive territories; nests are placed on cliffs and usually two eggs are laid. Populations are declining worldwide as a consequence of non-natural mortality [23]. At present, the number of mature individuals is only 30 000 to 40 000, and thus it is listed as ‘Endangered’ [23]. Insular sedentary populations survive in the Atlantic Ocean and the Mediterranean and Arabic Seas, but have also suffered drastic declines [24]. A relic population remains in the Canary Islands, where it has been described as a differentiated subspecies (Neophron percnopterus majorensis). Although it was very abundant in the past [25,26], it has also suffered a precipitous decline during the second half of the twentieth century [26]. At present, most of the Canarian population survives in Fuerteventura (in the southeastern part of the archipelago). An intensive monitoring programme over the last 12 years has revealed the presence, on average, of 30 breeding territories/year (s.d. = 6.4) with a mean productivity (number of fledglings/number of breeding pairs) of 0.53 (s.d. = 0.05) and a total population of approximately 200 birds in 2009 (J. A. Donázar, R. Agudo & L. Gangoso 2009, unpublished data).
(b). Population monitoring and sampling
From 1998 to 2009, 175 fledglings were captured at nests, and 82 immature and adult birds were trapped by cannon netting. All birds were marked by using both metal and plastic rings with an individual alphanumeric code, and released after measuring (wing-chord length, in millimetres), weighing (in grams) and bleeding (5 ml) them. Blood samples were preserved in absolute ethanol and kept at 4°C until their processing in the laboratory. Adult and immature birds were aged on the basis of plumage features. We estimated that about 85 per cent of the population was individually marked by 2009. During the same period, we intensively monitored the breeding population (38 territories in 2009) through the reproductive season (February to June) to determine the presence and identity of breeders as well as their breeding success [22]. Additionally, naturally moulted feathers were also collected at breeding territories in order to obtain DNA from non-banded adult breeding birds.
(c). Genetic characterization of individuals
DNA was isolated from blood samples using a standard phenol–chloroform extraction [27]. Genomic DNA from the blood clot contained within the feather shaft was extracted according to Horváth et al. [28]. Sex profiles (following the study of Fridolfsson & Ellegren [29]), individual genotypes at 22 microsatellites and two MHC class II loci (β chain, exon 2) were available from our previous studies [30–32]. Overall, the successful genotyping and molecular sexing of 135 feathers (fewer than 10% of feathers did not amplify) revealed 26 unbanded breeding individuals. We used the program genalex v. 6.2 [33] to detect repeated genotypes and to assign genetically profiled feathers to individual adults. Feathers from territories occupied by banded birds matched in 99 per cent of the cases with already resolved genotypes, which suggests that the collection of moulted feathers from adults other than the territorial birds is unlikely.
Bayesian analyses [34,35] did not indicate any substructure within the Canarian population, hence neglecting a potential effect of population structure on the potential HFCs [36] (data not shown). Neutral genetic diversity was measured by calculating a corrected MLH [37] estimate: homozygosity by loci (HL) [38] and the internal relatedness (IR) [39] obtained from the multi-locus genotypes of 22 microsatellite loci (n = 242) by using the software cernicalin [38]. We used HL rather than MLH because HL is believed to improve heterozygosity estimates by weighing the contribution of each locus to the overall homozygosity value [38]. IR is based on allele sharing, and the frequency of each allele counts towards the final inbreeding value, with shared rare alleles being more heavily weighted than shared common alleles [39].
(d). Paternity assessment and relatedness
Parentage was evaluated through both field observations and the genotypes of the 22 microsatellite loci [32] using the likelihood-based approach implemented in cervus v. 3.0 [40]. This method calculates statistical confidence based on the difference in the logarithm of the likelihood ratio (LOD) scores of candidate parents, and confidence is determined using criteria that are generated through simulation. For the simulation step, we used 10 000 cycles (‘offspring’), 5 pairs of candidate parents (i.e. 5 mothers and 5 fathers), 75 per cent of candidate parents sampled, 1 per cent of missing genotype data and 1 per cent of sampling errors. Confidence levels were set to 80 per cent (relaxed) and 95 per cent (strict), but only parents assigned with 95 per cent confidence were considered. This molecular method allowed us to corroborate field information, resolve the kinship of individuals with unknown parents (i.e. those that were captured as immature or adults), identify cases of extra pair paternity and resolve the identity of the parents in the case of breeding trios (n = 4).
Relatedness among individuals was assessed by calculating maximum-likelihood estimates of pairwise relatedness coefficients from multi-locus genotypes with the method implemented for microsatellite data in the software ml-relate [41].
(e). Local versus general effect
To discriminate whether the observed HFCs were driven by local or general effect, we measured the correlation in heterozygosity across loci (ID) by calculating the parameter g2 (and if it significantly differs from zero (no correlation) based on 1000 iterations) using the software rmes [42]. This parameter is a measure of the excess of double heterozygotes at two loci relative to the expectation under a random association (i.e. covariance in heterozygosity), standardized by average heterozygosity [12]. Additionally, we ran a multiple regression following Szulkin et al. [12], where each locus (n= 22) was included as an individual predictor and coded as 0 or 1 as homozygous or heterozygous, respectively. Missing genotypes were replaced with the mean heterozygosity for that locus [12]. If this model explains more variation than a basic model, where MLH is included as a single predictor, then this lends support to the local effects hypothesis. To compare models, we calculated the F ratio following the formula outlined in Szulkin et al. [12]. Finally, we calculated a heterozygosity–heterozygosity correlation [43] using the R package ‘Rhh’ [44] in R v. 2.12.2 [45] and the ‘h_cor’ function, which repeatedly and randomly divides the loci in half and calculates the correlation between them. If microsatellites carry information about genome-wide levels of heterozygosity, then comparing two random subsets of such markers should yield a positive, significant correlation [43,44]. We ran 250 randomizations.
(f). Genetic effects on fitness
We used generalized linear models (GLMs) to explore, at the individual level, the relationships between both measures of neutral genetic diversity (IR and HL; normal error distribution and identity link function), and between neutral (IR and HL) and adaptive genetic diversity (number of MHC different alleles in the individual; normal error distribution and identity link function). Then, the relationship between genetic diversity and individual fitness components was assessed through generalized linear mixed models (GLMMs), including ‘individual’ and ‘year’ as random terms to control for non-independence in data. Year was included as a random variable because preliminary analysis testing for temporal changes on HFCs (including year as an explanatory factor) yielded non-significant differences among years. Models for the age of recruitment (log-transformed, normal error distribution and identity link function) and breeding success (number of successful reproduction attempts/number of reproductive attempts; binomial error distribution and logit link function) were built by including as independent variables the individual's sex and measures of neutral (HL and IR) and adaptive genetic diversity and composition (i.e. number of different MHC alleles, MHC genotype and presence/absence of three particular linkage groups (LGs) of alleles: LG1 = Nepe1 + Nepe5; LG2 = Nepe18 + Nepe19 and LG3 = Nepe1 + Nepe7). The other three LGs observed in the population (LG4 = Nepe1 + Nepe6; LG5 = Nepe1 + Nepe20 and LG6 = Nepe8 + Nepe9) were not included in the models because they are present in less than 2 per cent of the total population [31]. We considered LGs rather than individual alleles based on our previous observations that indicated the cosegregation of MHC alleles from two gene duplicates in this insular population [31].
To model the breeding success, we also considered non-genetic terms known to affect breeding output in other long-lived species, such as age and body condition (BC, calculated, for completely grown individuals, as the residuals of a log–log least-squares linear regression of body mass against wing chord, controlling by sex [46]). Owing to the reduction in sample sizes (we have information on age and BC for less than 40% of reproductive individuals; table 1), genetic effects on the breeding success were tested both including and excluding these two parameters.
Table 1.
Results of model comparison to assess the effects of genetic diversity on age of recruitment and breeding success. Smaller AICc values suggest a better fit of the model to data while also penalizing for complexity (k, number of parameters). Models whose AIC values differ from that of the top model (ΔAICc) by more than two are considered to lack explanatory power relative to the top model. IR, internal relatedness; HL, individual homozygosity; NoMHCAlle, individual number of different MHC alleles; NoTotalMHCAlle, number of different MHC alleles in the breeding pair; LG2, presence/absence of linkage group 2 in the individuals and in and female (f) of the breeding pairs.
| model | sample size (inds./paris) | sample size (observations) | AICc | ΔAICc | k | weights | relative likelihood | |
|---|---|---|---|---|---|---|---|---|
| age of recruitment | IR | 40 | 40 | −68.77 | 0.00 | 2 | 0.35 | 1 |
| HL | 40 | 40 | −68.14 | 0.63 | 2 | 0.25 | 0.73 | |
| individual reproductive success | age | 41 | 181 | 173.07 | 0.00 | 2 | 1 | 1 |
| LG2 | 96 | 502 | 625.54 | 0.00 | 5 | 0.26 | 1 | |
| NoMHCAlle | 96 | 502 | 626.61 | 1.07 | 4 | 0.15 | 0.59 | |
| reproductive success of the breeding pair | LG2f | 39 | 181 | 213.10 | 0.00 | 2 | 0.22 | 1 |
| NoTotalAlleMHC, LG2f | 39 | 181 | 214.14 | 1.04 | 6 | 0.13 | 0.59 |
Finally, we assessed whether the combination of the genetic constitution of birds affected the breeding success at the level of breeding pairs. In these models, we included as independent variables HL and IR of each mate, relatedness coefficients between the male and female of each breeding pairs and the MHC characteristics of the pair (i.e. genotype of each mate, total number of different MHC alleles in the breeding pair, and presence/absence of LG1, LG2 and LG3 in the male and female, respectively). ‘Breeding pair’ and ‘year’ were included as random terms. The relationship between genetic diversity and productivity was not assessed, given that almost 95 per cent the breeding attempts resulted in a single fledgling (see §3).The relative explanatory power of models was compared, penalizing for complexity, using differences in their Akaike information criterion corrected for small sample sizes (AICc; lower scores indicated greater statistical support [47,48]). Models with ΔAICc higher than two (i.e. models differing from that of the lowest score by more than two) were considered to be unsupported statistically [48]. The relative likelihood of models was assessed through AICc weights [47]. Models were run in sas v. 9.2 [49].
3. Results
(a). Neutral and adaptive genetic diversity
The average values for individual microsatellite HL and IR were 0.40 (s.e. = 0.007) and 0.01 (s.e. = 0.012), respectively. Measures of HL and IR were positively correlated (p < 0.0001; r = 0.89). Pairwise relatedness coefficients between mates ranged from 0 to 0.63, with an average value of 0.07 (s.e. = 0.017). This value was similar to the average value within the whole population (0.08 (s.e. = 0.008)).
The individual survey of MHC variability reported 2, 3 or 4 different alleles (exon 2) per individual, which is in agreement with the simultaneous amplification of two gene duplicates in the Egyptian vulture [31]. Of the 236 birds, 103 individuals presented two MHC class II alleles, 19 birds presented three different alleles and 114 had four alleles. Overall, 10 different alleles comprising a total of nine different genotypes were distinguished, with three genotypes (G1–G3) accounting for 89 per cent of the genotypic variation in this population (see electronic supplementary material, table S1). Our previous results [31] hinted at the coevolution of gene duplicates maintaining different pairs of divergent alleles in linkage disequilibrium (LD). There, we identified six different LGs: LG1 (Nepe1 + Nepe3), LG2 (Nepe18 + Nepe19), LG3 (Nepe1 + Nepe7), LG4 (Nepe1 + Nepe6), LG5 (Nepe1 + Nepe20) and LG6 (Nepe8 + Nepe9; see electronic supplementary material, table S1).
Neutral and adaptive genetic diversity were positively linked, as shown by the significant relationship between both HL and IR and the number of MHC alleles (F = 10.91, p = 0.0011 and F = 10.62, p = 0.0013 respectively; figure 1).
Figure 1.
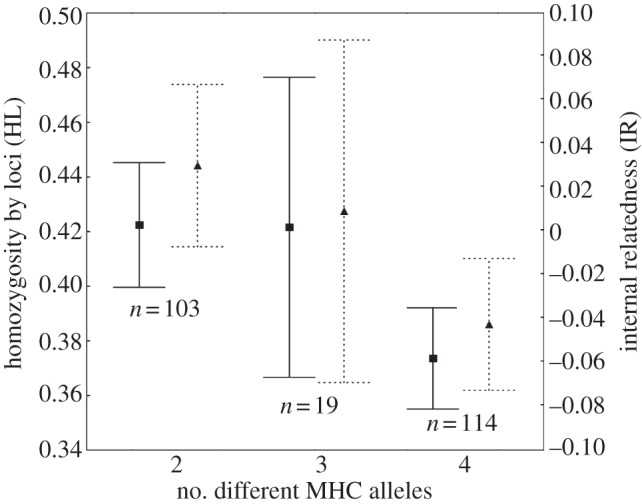
Individual homozygosity by loci (HL; left axis, squares) and internal relatedness (IR; right axis, triangles) in individuals with two, three and four different MHC alleles. Bars show mean ±95% CI.
(b). Local versus general effect
Results from the analysis of ID indicated a significant correlation in heterozygosity across loci (g2 = 0.007, s.d. = 0.005; p = 0.03). The F-test ratio comparing the multiple regression model containing all loci as predictors with the single regression model containing only MLH (i.e. HL) as the sole predictor revealed no significant difference (F21, 24 = 1.505, p = 0.17, n = 47). Finally, following the method of Balloux et al. [43], we did not find a significant correlation between randomly assigned subsets of loci: mean r for HL = 0.04 (−0.07, 0.15; 95% quantiles) and mean r for IR = 0.05 (−0.06, 0.15; 95% quantiles).
(c). Heterozygosity–fitness correlations
Age of recruitment into the breeding population ranged from 4 to 10 years (average = 5.8, s.e. = 0.11). The two highest-ranked models show that IR and HL were significant factors shaping the variability in this fitness component (estimates = 0.17, s.e. = 0.07 and 0.25, s.e. = 0.12, respectively; table 1), earlier recruiters presenting lower levels of inbreeding than later recruiters (figure 2). As HL and IR were highly related (i.e. individuals with high HL also showed high IR), models could be considered both statistically and biologically as alternatives. The other variables did not produce statistically supported models (ΔAICc > 2).
Figure 2.
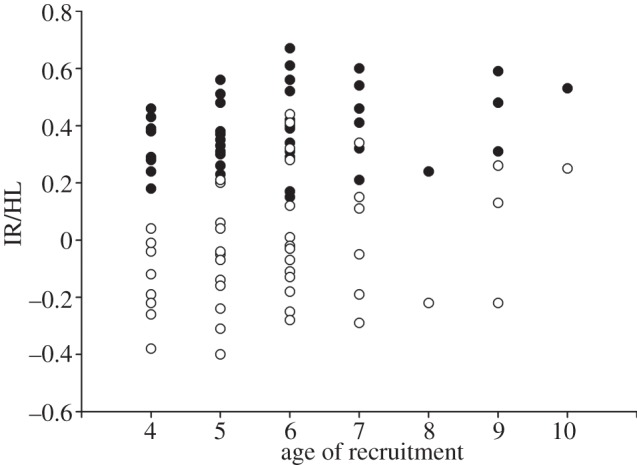
Correlation of neutral genetic diversity (black dots, HL; white dots, IR) and age of recruitment in Canarian Egyptian vultures.
We followed 502 reproductive attempts during the study period; 37.7 per cent of them were successful, from which 94.6 per cent resulted in a single fledgling. Model selection criteria supported age as the main driver of individual breeding success (estimate = 0.33, s.e. = 0.11, n = 41 birds, 181 breeding observations; table 1). Successful breeding attempts corresponded to individuals 1 year older on average (mean age = 8.7, s.e. = 0.43) than those individuals failing to breed (mean age = 7.4, s.e. = 0.19). Although two alternative models were initially considered by following the AIC criteria, we discarded them because they included variables (specifically BC and IR) whose estimates showed very high standard errors (s.e. ≥ 2 × estimate, i.e. their effects are non-significantly different from zero). Therefore, no alternative models were obtained when analysing breeding success of individuals on known age. Models constructed without considering the effect of age allowed us to fit models with larger sample sizes (n = 96 individuals, 502 breeding observations), showing an effect of adaptive genetic diversity. Individual breeding success was positively linked to the presence of LG 2 (estimate for absence = −0.76, s.e. = 0.45). Furthermore, the number of MHC alleles was also positively related to individual breeding success (estimate = 0.30, s.e. = 0.22), successful reproduction attempts corresponding to individuals with higher number of MHC alleles (mean = 3.12 alleles, s.e. = 0.11 and mean = 2.95, s.e. = 0.10 different alleles for successful and unsuccessful breeding individuals respectively; figure 3 and table 1). Note, nevertheless, that the relatively high standard errors obtained for the estimates may suggest a weak effect of these two variables, and even though the raw data seem to be supportive, these results should be interpreted with caution.
Figure 3.
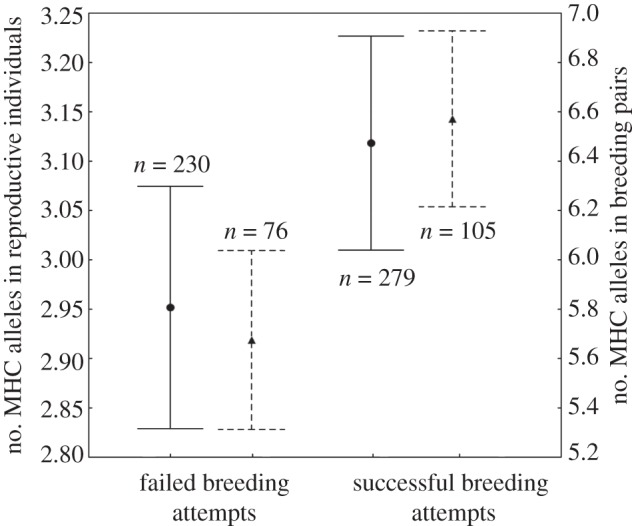
The number of MHC alleles (mean and 95% CI) in relation to successful and failed breeding attempts (pooled data from the entire study period). Data for individual birds are shown by circles with continuous lines; breeding pairs are shown by triangles with dashed lines.
Two alternative models were obtained for the breeding success of pairs, including female LG2 (estimate: −1.48, s.e. = 0.77) alone and the total number of MHC alleles in the pair (i.e. the sum of different MHC alleles of both mates; estimate = 0.51, s.e. = 0.33) combined with female LG2 (estimate for absence = −1.21, s.e. = 0.80). Thus, breeding success seemed to be higher for pairs with a higher number of different MHC alleles (mean = 6.57 alleles, s.e. = 0.22 and mean = 5.66, s.e. = 0.21 for successful and unsuccessful pairs, respectively), and when females hold LG2. Our observations are in accordance with these results. We observed homozygous individuals for LG2 (n = 20) having more reproductive attempts (n = 110) and succeeding more (n = 61) than homozygous individuals holding LG1 (n = 18; total number of reproductions = 88 and successful ones = 40). Furthermore, those pairs formed by homozygous mates sharing LG2 (n = 2 breeding pairs) succeed in three out of a total of five reproduction attempts, whereas pairs of homozygous mates sharing LG1 (n = 3 breeding pairs) always failed and did not produce any offspring out of the observed nine reproduction attempts.
4. Discussion
To our knowledge, this study represents the first attempt to link neutral and adaptive genetic diversity to individual fitness components in a long-lived, threatened avian species. Our findings support a positive effect of neutral and adaptive genetic variability on two components of fitness, so that heterozygous birds at neutral loci recruit earlier, while those with a higher number of different MHC alleles present higher breeding success. Additionally, we identified fitness benefits derived from the possessing of the most frequent pair of divergent MHC alleles in LD in the population.
Our analyses on neutral loci support the general effect hypothesis (i.e. a relationship between individual heterozygosity and inbreeding). First, we detect a significant correlation in heterozygosity across loci by calculating the parameter g2, which is the central measure of ID in HFC theory [12]. Even though the method proposed by Balloux et al. [43] indicated non-significant heterozygosity–heterozygosity correlation between random sets of markers, we must recall that this procedure (to divide a set of loci into two halves and to compute the correlation of MHL between them) yields a complicated distribution of heterozygosity–heterozygosity correlation coefficients not independent from one another and providing no synthetic measure that can be related to HFC theory [12]. Even low significant values of g2 are more powerful measures of ID than those obtained by the Balloux method [12] and we may conclude that our observed value (0.007), although low, is significant to detect a weak ID. Second, the comparison between a model containing all loci as individual predictors and the simple model containing only an MHL measure did not show significant differences, thus rejecting the local effect hypothesis. Third, we observed a significant relationship between neutral and adaptive genetic diversity. Finally, the characteristics of the studied population may also suggest that HFCs may be generated as a result of the effects of homozygosity at loci genome-wide [10]: the population is partially inbred and consanguineous mating may occur, as indicated by the high relate coefficient observed in some breeding pairs (relate greater than 0.2 in 8 out of 42 breeding pairs, or 19%), and the population is subjected to drift and has been affected by bottlenecks [25,31,32].
(a). Genetic diversity and age of recruitment
Under the general effect hypothesis, we expect heterozygous individuals to be the highest-quality birds in a population. In this sense, our results indicate that heterozygous individuals recruit earlier (at 4–5 years old) than more homozygous birds (recruiting at 6–10 years old). This may suggest that recruiting earlier is the best reproductive strategy in this species, as previously proposed for other long-lived species [50]. Consequently, the studied population may be subjected to inbreeding depression if increased inbreeding determines a delay in the age of recruitment. Mate choice could also be reinforcing this pattern of early recruitment. It has been widely discussed that the relationship between genome-wide heterozygosity or degree of inbreeding and sexual characters suggest a directional preference for heterozygous mates [51]. Theory indicates that heterozygous individuals are more efficient at acquiring mates, thus breeding younger than those more inbred.
(b). Genetic diversity and breeding success
Analyses of reproductive individuals of known age show that age is the main factor determining breeding success. The effects of age on reproductive performance of long-lived species have been described in many avian species [52], even raptors [21], suggesting that this is a general pattern. The main innovation of our results is the additive effect of genetic diversity: individuals with a higher number of MHC alleles show higher breeding success. It is worth noting that the models including age were performed with a reduced sample size of the youngest individuals of the population, which were monitored from birth or immature stages. When expanding our sample to include individuals of unknown exact age, MHC genetic diversity (number of MHC alleles) and configuration, particularly the presence of LG2 (alleles Nepe18 + Nepe19), became the main determinants of individual breeding success: birds holding four alleles and those holding LG2 are significantly more successful than others. Although models obtained should be cautiously interpreted (see §3), raw data are supportive (figure 3). Analyses considering the breeding success of pairs yielded similar results, with breeding success mainly explained by MHC diversity of the pair and by the presence of LG2 in females.
Although some studies point out intermediate rather than maximal levels of MHC diversity to be more advantageous [53,54], the observed relationship with MHC diversity in this study is in line with recent findings suggesting that higher MHC heterozygosity enhances fitness, either through survival or reproductive performance. For example, Banks et al. [55] described a positive correlation between heterozygosity at MHC-linked markers and survival in a long-lived mammal species (the mountain brushtail possum Trichosurus cunninghami). In males of fat-tailed dwarf lemur (Cheirogaleus medius) and grey mouse lemur (Microcebus murinus), MHC individual allelic diversity was associated with increased reproductive success [56]. Worley et al. [20] observed a higher survival rate of MHC heterozygous individuals in red junglefowl (Gallus gallus) and also identified a particular MHC genotype strongly affecting survival probabilities. Also, Thoss et al. [57] described an increment in fecundity by MHC heterozygosity in house mice. The most parsimonious explanation for these findings is that MHC heterozygosity improves mating success or fecundity by enhancing health and disease resistance [20,57].
Whereas we cannot totally rule out a potential general effect from inbreeding, given the correlation between neutral and MHC variability [13], our findings that indicate a positive effect of LG2 and a lack of effect of neutral variability and relatedness between breeders (within breeding pairs) strongly support a particular MHC effect on breeding success. In a previous study [31], we hypothesized that the coevolution of gene duplicates, favouring high frequencies of different pairs of divergent alleles in LD, would be counteracting to some extent the loss of genetic diversity in this insular population. Furthermore, we suggested the existence of local adaptation favouring certain LGs, based on the observed frequency of these LGs among different populations [31]. Our present results seem to support these hypotheses, suggesting positive selection acting on individuals holding LG2, the most common pair of cosegregating MHC alleles in the Canary Islands, but lacking from other insular populations [31]. This apparent advantage of LG2 is strengthened by results regarding breeding success of pairs that indicate that the presence of LG2 in females plays an important role in the reproductive success of breeding pairs. Cost of reproduction is higher for females during egg production. Experimental studies point towards the existence of a trade-off between immune response and reproduction [58], indicating that reproductive effort can have relevant consequences in immune responses [59]. In this sense, genetically advantageous individuals (here, individuals holding LG2) may better afford the cost of reproduction [60].
Overall, our results are very relevant as they may support that positive frequency-dependent selection, probably driven by local pathogens, is presently acting in this bottlenecked population. This may determine the fixation of frequent LGs and the loss of rare alleles by negative selection and drift [61]. This process may determine a higher-than-expected rate of loss of adaptive genetic variability (a scenario that may be common in bottlenecked populations [14]), potentially having important consequences for the long-term persistence of this highly threatened insular population. Selection has seemed to favour common alleles, but emerging pathogen challenges can overcome the most extended MHC-based immune response in the populations and then select for rare genotypes [62]. However, the low MHC variability combined with the loss of rare alleles and the rapid invasion of new pathogens owing to human action [24] may seriously compromise the adaptive capability of this genetically impoverished population, strongly affecting its future viability. In fact, Canarian Egyptian vultures present low immune response capabilities and high susceptibility to infection [24].
(c). The whole picture: falling into the extinction vortex
Our results indicate that the genetic deterioration in this small population has a negative impact on individual fitness [3,19] through three main avenues. First, reduced genetic diversity delays age of recruitment, reducing population growth rates and increasing extinction risk [22,63]. Second, impoverished levels of genetic diversity at important functional genes affect individual fitness, decreasing population productivity. Third, reduced genetic variability lowers individual ability to respond to new environmental challenges such as the arrival of new pathogens [31]. The Canarian Egyptian vultures show the lowest breeding success known for the species throughout the world [26]. Similar trends have also been described in other long-lived raptor populations [64], but the causes of such low productivity on islands are widely unknown and have been included in the so-called ‘insular syndrome’ [65]. Present results and the observed lower levels of neutral and adaptive genetic diversity in the insular population compared with its continental counterpart [31,32] may support inbreeding as one of the main causes determining the comparatively lower insular productivity. Therefore, the combined effect of genetic depauperation and the deterministic factors related to human activity (i.e. adult unnatural mortality owing to persecution [26,66] and the arrival of new pathogens [24,31]) can have catastrophic consequences in the long-term maintenance of this endangered population by inevitably increasing its risk of extinction.
Acknowledgements
Manuel de la Riva, Laura Gangoso, Angel de Pazo, Ana Trujillano, Juan José García, Carmen Díez, Marcos Mallo, Juan Manuel Grande, Ainara Cortés-Avizanda, Olga Ceballos and José Ramón Benítez helped with the field work and provided blood samples. This research was funded by the projects REN 2000-1556 GLO, CGL2004-00270 and CGL2009-12753-C02-02. The Cabildo Insular of Fuerteventura, the Canarian Government and the Spanish Ministry of Environment provided logistic and economic support. M.A. was funded by a postdoctoral fellowship from the Spanish Ministry of Science and Innovation (www.micinn.es).
References
- 1.Frankham R. 2003. Genetics and conservation biology. C. R. Biol. 326, S22–S29 10.1016/s1631-0691(03)00023-4 (doi:10.1016/s1631-0691(03)00023-4) [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth D., Charlesworth B. 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18, 237–268 10.1146/annurev.es.18.110187.001321 (doi:10.1146/annurev.es.18.110187.001321) [DOI] [Google Scholar]
- 3.Keller L. F., Waller D. M. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 10.1016/S0169-5347(02)02489-8 (doi:10.1016/S0169-5347(02)02489-8) [DOI] [Google Scholar]
- 4.Saccheri I., Kuussaari M., Kankare M., Vikman P., Fortelius W., Hanski I. 1998. Inbreeding and extinction in a butterfly metapopulation. Nature 392, 491–494 10.1038/33136 (doi:10.1038/33136) [DOI] [Google Scholar]
- 5.Luikart G., Pilgrim K., Visty J., Ezenwa V. O., Schwartz M. K. 2008. Candidate gene microsatellite variation is associated with parasitism in wild bighorn sheep. Biol. Lett. 4, 228–231 10.1098/rsbl.2007.0633 (doi:10.1098/rsbl.2007.0633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Silva A., Luikart G., Yoccoz N. G., Cohas A., Allaine D. 2006. Genetic diversity–fitness correlation revealed by microsatellite analyses in European alpine marmots (Marmota marmota). Conserv. Genet. 7, 371–382 10.1007/s10592-005-9048-y (doi:10.1007/s10592-005-9048-y) [DOI] [Google Scholar]
- 7.Jamieson I. G., Tracy L. N., Fletcher D., Armstrong D. P. 2007. Moderate inbreeding depression in a reintroduced population of North Island robins. Anim. Conserv. 10, 95–102 10.1111/j.1469-1795.2006.00078.x (doi:10.1111/j.1469-1795.2006.00078.x) [DOI] [Google Scholar]
- 8.Chapman J. R., Nakagawa S., Coltman D. W., Slate J., Sheldon B. C. 2009. A quantitative review of heterozygosity–fitness correlations in animal populations. Mol. Ecol. 18, 2746–2765 10.1111/j.1365-294X.2009.04247.x (doi:10.1111/j.1365-294X.2009.04247.x) [DOI] [PubMed] [Google Scholar]
- 9.Harrison X. A., Bearhop S., Inger R., Colhoun K., Gudmundsson G. A., Hodgson D., McElwaine G., Tregenza T. O. M. 2011. Heterozygosity–fitness correlations in a migratory bird: an analysis of inbreeding and single-locus effects. Mol. Ecol. 20, 4786–4795 10.1111/j.1365-294X.2011.05283.x (doi:10.1111/j.1365-294X.2011.05283.x) [DOI] [PubMed] [Google Scholar]
- 10.Hansson B., Westerberg L. 2002. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 11, 2467–2474 10.1046/j.1365-294X.2002.01644.x (doi:10.1046/j.1365-294X.2002.01644.x) [DOI] [PubMed] [Google Scholar]
- 11.Da Silva A., et al. 2009. Heterozygosity–fitness correlations revealed by neutral and candidate gene markers in roe deer from a long-term study. Evolution 63, 403–417 10.1111/j.1558-5646.2008.00542.x (doi:10.1111/j.1558-5646.2008.00542.x). [DOI] [PubMed] [Google Scholar]
- 12.Szulkin M., Bierne N., David P. 2010. Heterozygosity–fitness correlations: a time for reappraisal. Evolution 64, 1202–1217 10.1111/j.1558-5646.2010.00966.x (doi:10.1111/j.1558-5646.2010.00966.x). [DOI] [PubMed] [Google Scholar]
- 13.Radwan J., Biedrzycka A., Babik W. 2009. Does reduced MHC diversity decrease viability of vertebrate populations? Biol. Conserv. 143, 537–544 10.1016/j.biocon.2009.07.026 (doi:10.1016/j.biocon.2009.07.026). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton J. T., Nakagawa S., Robertson B. C., Jamieson I. G. 2011. Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Mol. Ecol. 20, 4408–4420 10.1111/j.1365-294X.2011.05292.x (doi:10.1111/j.1365-294X.2011.05292.x) [DOI] [PubMed] [Google Scholar]
- 15.Ejsmond M., Radwan J. 2011. MHC diversity in bottlenecked populations: a simulation model. Conserv. Genet. 12, 129–137 10.1007/s10592-009-9998-6 (doi:10.1007/s10592-009-9998-6) [DOI] [Google Scholar]
- 16.Spurgin L. G., Richardson D. S. 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B 277, 979–988 10.1098/rspb.2009.2084 (doi:10.1098/rspb.2009.2084). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piertney S. B., Oliver M. K. 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 10.1038/sj.hdy.6800724 (doi:10.1038/sj.hdy.6800724). [DOI] [PubMed] [Google Scholar]
- 18.Groombridge B., Jenkins M. D. 2002. World atlas of biodiversity: Earth's living resources in the 21st century. Berkeley, CA: University of California Press [Google Scholar]
- 19.Blomqvist D., Pauliny A., Larsson M., Flodin L. A. 2010. Trapped in the extinction vortex? Strong genetic effects in a declining vertebrate population. BMC Evol. Biol. 10, 33. 10.1186/1471-2148-10-33 (doi:10.1186/1471-2148-10-33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worley K., Collet J., Spurgin L. G., Cornwallis C., Pizzari T., Richardson D. S. 2010. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 19, 3064–3075 10.1111/j.1365-294X.2010.04724.x (doi:10.1111/j.1365-294X.2010.04724.x) [DOI] [PubMed] [Google Scholar]
- 21.Blas J., Sergio F., Hiraldo F. 2009. Age-related improvement in reproductive performance in a long-lived raptor: a cross-sectional and longitudinal study. Ecography 32, 647–657 10.1111/j.1600-0587.2008.05700.x (doi:10.1111/j.1600-0587.2008.05700.x). [DOI] [Google Scholar]
- 22.Grande J. M., Serrano D., Tavecchia G., Carrete M., Ceballos O., Diaz-Delgado R., Tella J. L., Donazar J. A. 2009. Survival in a long-lived territorial migrant: effects of life-history traits and ecological conditions in wintering and breeding areas. Oikos 118, 580–590 10.1111/j.1600-0706.2009.17218.x (doi:10.1111/j.1600-0706.2009.17218.x). [DOI] [Google Scholar]
- 23.BirdLife International 2008. Action plan for the Egyptian Vulture Neophron percnopterus in the European Union. Madrid, Spain: BirdLife International [Google Scholar]
- 24.Gangoso L. 2006. Insularidad y Conservación: el caso del Alimoche en Canarias. Seville, Spain: Universidad de Sevilla [Google Scholar]
- 25.Agudo R., Rico C., Vila C., Hiraldo F., Donazar J. 2010. The role of humans in the diversification of a threatened island raptor. BMC Evol. Biol. 10, 384. 10.1186/1471-2148-10-384 (doi:10.1186/1471-2148-10-384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donazar J. A., Palacios C. J., Gangoso L., Ceballos O., Gonzalez M. J., Hiraldo F. 2002. Conservation status and limiting factors in the endangered population of Egyptian vulture (Neophron percnopterus) in the Canary Islands. Biol. Conserv. 107, 89–97 10.1016/S0006-3207(02)00049-6 (doi:10.1016/S0006-3207(02)00049-6) [DOI] [Google Scholar]
- 27.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd edn. New York, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 28.Horváth M. B., Martínez-Cruz B., Negro J. J., Kalmár L., Godoy J. A. 2005. An overlooked DNA source for non-invasive genetic analysis in birds. J. Avian Biol. 36, 84–88 10.1111/j.0908-8857.2005.03370.x (doi:10.1111/j.0908-8857.2005.03370.x) [DOI] [Google Scholar]
- 29.Fridolfsson A., Ellegren H. 1999. A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol. 30, 116–121 10.2307/3677252 (doi:10.2307/3677252) [DOI] [Google Scholar]
- 30.Agudo R., Roques S., Galarza J. A., Rico C., Hiraldo F., Donazar J. A. 2008. Isolation and characterization of 18 microsatellite loci in the Egyptian vulture (Neophron percnopterus). Conserv. Genet. 9, 1345–1348 10.1007/s10592-007-9486-9 (doi:10.1007/s10592-007-9486-9). [DOI] [Google Scholar]
- 31.Agudo R., Alcaide M., Rico C., Lemus J. A., Blanco G., Hiraldo F., Donázar J. A. 2011. Major histocompatibility complex variation in insular populations of the Egyptian vulture: inferences about the roles of genetic drift and selection. Mol. Ecol. 20, 2329–2340 10.1111/j.1365-294X.2011.05107.x (doi:10.1111/j.1365-294X.2011.05107.x) [DOI] [PubMed] [Google Scholar]
- 32.Agudo R., Rico C., Hiraldo F., Donázar J. A. 2011. Evidence of connectivity between continental and differentiated insular populations in a highly mobile species. Divers. Distrib. 17, 1–12 10.1111/j.1472-4642.2010.00724.x (doi:10.1111/j.1472-4642.2010.00724.x) [DOI] [Google Scholar]
- 33.Peakall R., Smouse P. E. 2006. GENALEX 6: genetic analysis in Excel. Population genetics software for teaching and research. Mol. Ecol. Notes 6, 288–295 10.1111/j.1471-8286.2005.01155.x (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falush D., Stephens M., Pritchard J. K. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slate J., Pemberton J. 2006. Does reduced heterozygosity depress sperm quality in wild rabbits (Oryctolagus cuniculus)? Curr. Biol. 16, R790–R791 10.1016/j.cub.2006.08.047 (doi:10.1016/j.cub.2006.08.047) [DOI] [PubMed] [Google Scholar]
- 37.Castric V., Bernatchez L., Belkhir K., Bonhomme F. 2002. Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchill (Pisces, Salmonidae): a test of alternative hypotheses. Heredity 89, 27–35 10.1038/sj.hdy.6800089 (doi:10.1038/sj.hdy.6800089) [DOI] [PubMed] [Google Scholar]
- 38.Aparicio J. M., Ortego J., Cordero P. J. 2006. What should we weigh to estimate heterozygosity, alleles or loci? Mol. Ecol. 15, 4659–4665 10.1111/j.1365-294X.2006.03111.x (doi:10.1111/j.1365-294X.2006.03111.x). [DOI] [PubMed] [Google Scholar]
- 39.Amos W., Worthington Wilmer J., Fullard K., Burg T. M., Croxall J. P., Bloch D., Coulson T. 2001. The influence of parental relatedness on reproductive success. Proc. R. Soc. Lond. B 268, 2021–2027 10.1098/rspb.2001.1751 (doi:10.1098/rspb.2001.1751). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalinowski S. T., Taper M. L., Marshall T. C. 2007. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106 10.1111/j.1365-294X.2007.03089.x (doi:10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 41.Kalinowski S. T., Wagner A. P., Taper M. L. 2006. ML-RELATE: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 6, 576–579 10.1111/j.1471-8286.2006.01256.x (doi:10.1111/j.1471-8286.2006.01256.x). [DOI] [Google Scholar]
- 42.David P., Pujol B., Viard F., Castella V., Goudet J. 2007. Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 16, 2474–2487 10.1111/j.1365-294X.2007.03330.x (doi:10.1111/j.1365-294X.2007.03330.x). [DOI] [PubMed] [Google Scholar]
- 43.Balloux F., Amos W., Coulson T. 2004. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 13, 3021–3031 10.1111/j.1365-294X.2004.02318.x (doi:10.1111/j.1365-294X.2004.02318.x). [DOI] [PubMed] [Google Scholar]
- 44.Alho J. S., Välimäki K., Meril Ä. J. 2010. Rhh: an R extension for estimating multilocus heterozygosity and heterozygosity–heterozygosity correlation. Mol. Ecol. Resour. 10, 720–722 10.1111/j.1755-0998.2010.02830.x (doi:10.1111/j.1755-0998.2010.02830.x) [DOI] [PubMed] [Google Scholar]
- 45.R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 46.Schulte-Hostedde A. I., Zinner B., Millar J. S., Hickling G. J. 2005. Restitution of mass-size residuals: validating body condition indices. Ecology 86, 155–163 10.1890/04-0232 (doi:10.1890/04-0232) [DOI] [Google Scholar]
- 47.Burnham K. P., Anderson D. R. 2002. Model selection and multimodel inference. New York, NY: Springer [Google Scholar]
- 48.Richards S. A. 2005. Testing ecological theory using the information-theoretic approach: examples and cautionary results. Ecology 86, 2805–2814 10.1890/05-0074 (doi:10.1890/05-0074) [DOI] [Google Scholar]
- 49.SAS Institute 2004. SAS/STAT 9.1. user's guide. Cary, NC: SAS Institute Inc [Google Scholar]
- 50.Becker P. H., Bradley J. S. 2007. The role of intrinsic factors for the recruitment process in long-lived birds. J. Ornithol. 148, S377–S384 10.1007/s10336-007-0157-x (doi:10.1007/s10336-007-0157-x). [DOI] [Google Scholar]
- 51.Fromhage L., Kokko H., Reid J. M. 2009. Evolution of mate choice for genome-wide heterozigosity. Evolution 63, 684–694 10.1111/j.1558-5646.2008.00575.x (doi:10.1111/j.1558-5646.2008.00575.x). [DOI] [PubMed] [Google Scholar]
- 52.Lewis S., Elston D. A., Daunt F., Cheney B., Thompson P. M. 2009. Effects of extrinsic and intrinsic factors on breeding success in a long lived seabird. Oikos 118, 521–528 10.1111/j.1600-0706.2009.17308.x (doi:10.1111/j.1600-0706.2009.17308.x). [DOI] [Google Scholar]
- 53.Kalbe M., Eizaguirre C., Dankert I., Reusch T. B. H., Sommerfeld R. D., Wegner K. M., Milinski M. 2009. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc. R. Soc. B 276, 925–934 10.1098/rspb.2008.1466 (doi:10.1098/rspb.2008.1466). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wegner K. M., Kalbe M., Milinski M., Reusch T. 2008. Mortality selection during the 2003 European heat wave in three-spined sticklebacks: effects of parasites and MHC genotype. BMC Evol. Biol. 8, 124. 10.1186/1471-2148-8-124 (doi:10.1186/1471-2148-8-124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banks S. C., Dubach J., Viggers K. L., Lindenmayer D. B. 2010. Adult survival and microsatellite diversity in possums: effects of major histocompatibility complex-linked microsatellite diversity but not multilocus inbreeding estimators. Oecologia 162, 359–370 10.1007/s00442-009-1464-0 (doi:10.1007/s00442-009-1464-0). [DOI] [PubMed] [Google Scholar]
- 56.Schwensow N., Fietz J., Dausmann K., Sommer S. 2008. MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair-living primate. Evol. Ecol. 22, 617–636 10.1007/s10682-007-9186-4 (doi:10.1007/s10682-007-9186-4) [DOI] [Google Scholar]
- 57.Thoss M., Ilmonen P., Musolf K., Penn D. J. 2011. Major histocompatibility complex heterozygosity enhances reproductive success. Mol. Ecol. 20, 1546–1557 10.1111/j.1365-294X.2011.05009.x (doi:10.1111/j.1365-294X.2011.05009.x). [DOI] [PubMed] [Google Scholar]
- 58.Bonneaud C., Mazuc J., Chastel O., Westerdahl H., Sorci G., Poulin R. 2004. Terminal investment induced by immune challenge and fitness traits associated with major histocompatibility complex in the House sparrow. Evolution 58, 2823–2830 [DOI] [PubMed] [Google Scholar]
- 59.Knowles S. C. L., Nakagawa S., Sheldon B. C. 2009. Elevated reproductive effort increases blood parasitaemia and decreases immune function in birds: a meta-regression approach. Funct. Ecol. 23, 405–415 10.1111/j.1365-2435.2008.01507.x (doi:10.1111/j.1365-2435.2008.01507.x) [DOI] [Google Scholar]
- 60.Gustafsson L., Nordling D., Andersson M. S., Sheldon B. C., Qvarnstrom A. 1994. Infectious diseases, reproductive effort and the cost of reproduction in birds. Phil. Trans. R. Soc. Lond. B 346, 323–331 10.1098/rstb.1994.0149 (doi:10.1098/rstb.1994.0149). [DOI] [PubMed] [Google Scholar]
- 61.Ejsmond M., Radwan J. 2009. MHC diversity in bottlenecked populations: a simulation model. Conserv. Genet. 12, 129–137 10.1007/s10592-009-9998-6 (doi:10.1007/s10592-009-9998-6). [DOI] [Google Scholar]
- 62.Bernatchez L., Landry C. 2003. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377 10.1046/j.1420-9101.2003.00531.x (doi:10.1046/j.1420-9101.2003.00531.x) [DOI] [PubMed] [Google Scholar]
- 63.Eberhardt L. L. 2002. A paradigm for population analysis of long-lived vertebrates. Ecology 83, 2841–2854 10.1890/0012-9658(2002)083[2841:APFPAO]2.0.CO;2 (doi:10.1890/0012-9658(2002)083[2841:APFPAO]2.0.CO;2) [DOI] [Google Scholar]
- 64.Thibault J.-C., Patrimonio O., Torre J. 1992. Does the diurnal raptor community of Corsica (Western Mediterranean) show insular characteristics? J. Biogeogr. 19, 363–373 10.2307/2845564 (doi:10.2307/2845564) [DOI] [Google Scholar]
- 65.Blondel J. 2000. Evolution and ecology of birds on islands: trends and prospects. Vie et Milieu Life Environ. 50, 205–220 [Google Scholar]
- 66.Gangoso L., Alvarez-Lloret P., Rodriguez-Navarro A. A. B., Mateo R., Hiraldo F., Donazar J. A. 2009. Long-term effects of lead poisoning on bone mineralization in vultures exposed to ammunition sources. Environ. Pollut. 157, 569–574 10.1016/j.envpol.2008.09.015 (doi:10.1016/j.envpol.2008.09.015) [DOI] [PubMed] [Google Scholar]


